Abstract
Late embryogenesis abundant (LEA) proteins were first described as accumulating late in plant seed development. They were also shown to be involved in plant responses to environmental stress and as well as in bacteria, yeast and invertebrates. They are known to play crucial roles in dehydration tolerance. This study describes a genome-wide analysis of LEA proteins and the corresponding genes in Solanum tuberosum. Twenty-nine LEA family members encoding genes in the Solanum genome were identified. Phylogenetic analyses allowed the classification of the potato LEA proteins into nine distinct groups. Some of them were identified as putative orthologs of Arabidopsis and rice LEA genes. In silico analyses confirmed the hydrophilicity of most of the StLEA proteins, whereas some of them can be folded. The in silico expression analyses showed that the identified genes displayed tissue-specific, stress and hormone-responsive expression profiles. Five StLEA classified as dehydrins were selected for expression analyses under salt and drought stresses. The data revealed that they were induced by both stresses. The analyses indicate that several factors such us developmental stages, hormones, and dehydration, can regulate the expression and activities of LEA protein. This report can be helpful for the further functional diversity studies and analyses of LEA proteins in potato. These genes can be overexpressed to improve potato abiotic stress response.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dehydration is a normal process during seed maturation, which is controlled by programmed expression of specific late embryogenesis abundant (LEA) genes during the later embryo development stages (post-abscission) in plant seeds [1–3]. The intensive biosynthesis of LEA proteins at late seed maturation stages and during dehydration, suggest that these proteins are involved in the protection of embryo structures during water deficit conditions [2–5].
LEA expression is regulated by abscisic acid (ABA), a key hormone in the regulation of dehydration [1, 5, 6]. Indeed, the involvement of the ABA signaling pathways was shown for a number of LEA genes [6–8]. The overexpression of genes encoding LEA proteins improves stress tolerance of transgenic plants [5].
LEA proteins are primarily found in plants seeds [1–3] and also in other plant tissues [2].
LEA gene expression can be related to ABA dependant or ABA independent signaling pathways. Their gene promoters can harbor DRE/CRT ABA independent sequence [1–3, 5, 9]. LEA proteins were first detected in cotton (Gossypium hirsutum) seeds [5, 9]. They have subsequently been identified in several plant species to the extent that they can be considered ubiquitous in plants [3, 9, 10].
Moreover, several LEA proteins were in non viridae plantae species such as bacteria [1, 11], mosses [5], fungi [9, 25] and nematodes [9, 12].
LEA proteins are part of evolutionary conserved group of hydrophilic protein called hydrophylin [25]. They are high hydrophilic [1, 3, 9] and heat stabile in solution [1, 13] because of their particular amino acid composition. However, under desiccation or extreme temperature conditions some of them can exhibit particular three-dimensional structures [8].
Some LEA proteins seems to act as chaperones and protect enzyme activities under in vitro conditions [3, 8, 14]. Furthermore, LEA proteins can undergo phosphorylation [8, 15] and can sequester ions [8, 16] or biotin [8]. They are shown to be associated with vacuoles or lipid vesicles [8]. However, a number of Arabidopsis thaliana LEA proteins can be cytosolic, secreted or located in chloroplasts and mitochondria, while others were detected in all cellular compartments [19].
The LEA proteins were first divided into different groups [4, 16] but the classification varies between authors. The deduced amino acid sequences of LEA proteins allowed the identification of six groups based on their similarities [16]. However, Dure [4] labelled the groups according to their molecular weight and identified 8 groups that were named D-7, D-11, D-19, D-29, D-34, D-73, D-95 and D-113.
The identification of other plant LEA genes revealed that their classification is not so clear and some groups were related to other groups [17].
The three major groups are as follows [4, 18, 19]. LEA proteins Group 1 posses a hydrophilic motif repeated of 20 amino acid-long. This motif repetition can exceed four times in some species [19, 20]. These proteins are also characterized by a high proportion of Gly, Glu, and Gln residues. Group 2 protein sequences, corresponding to dehydrins contain three distinct sequence motifs designated Y, S and K [5, 10, 21–24]. These group 2 LEA proteins are the most characterized [2]. These highly hydrophilic proteins are polar amino acids rich while they harbor low hydrophobic residues content. They also lack Trp and frequently Cys residues [4, 9]. Hence, they can also be considered as hydrophilins [2, 25]. The Group 3 proteins display multiple copies of an 11 amino acid-long motif [4].
Most of the LEA proteins belong these three main groups. However, some others are members of minor groups [16]: Group 4 (D-113), Group 5 (D-29) and Group 6 (D-34). The LEA5 (D-73) and LEA14 (D-95) [26] were not classified in LEA numbered groups.
This study focused on the identification of potato LEA (StLEA) genes throughout the potato genome in addition to their evolution analysis. The expression patterns of these genes were investigated using RNA-seq data and semi-quantitative RT-PCR analysis of some selected LEA genes was performed.
Materials and methods
BLAST searches and sequence analysis
BLAST searches and sequence analyses were performed by BLASTp on the potato genome (http://www.phytozome.net/potato.php) using known A. thaliana LEA genes as queries. The selected potato genes were used for a second BLAST round on the potato genome; this additional step allowed the identification of potato paralogs that had been excluded by their dissimilarity to the Arabidopsis orthologs.
Domain and motif searches were performed on the protein sequences using the Pfam software (http://www.sanger.ac.uk/software/pfam). Proteins without any appropriate motif were re-predicted and corrected using Fgenesh software (http://linux1.softberry.com/berry.phtml) using dicotyledon plant models. Proteins with no reliable prediction were removed.
For phylogenetic analyses, deduced full length protein sequences of the identified potato LEA gene models and those from Arabidopsis and rice (www.phytozome.net) were aligned with Clustal W software [27]. The phylogenetic tree was conducted using Molecular Evolutionary Genetic Analysis software (Mega5) [28]. Genetic relationships were inferred from neighbor-joining amino acid alignment after 1,000 bootstrap replicates, using pairwise deletion and amino acid p-distance parameters.
The prediction of the LEA signal peptide was performed using the Signal P 3.0 online program (http://www.cbs.dtu.dk/services/SignalP/). The TMHMM ver.2.0 (http://www.cbs.dtu.dk/services/TMHMM/) program was used for the prediction of the transmembrane regions. Subcellular localization prediction of the deduced amino acids was performed using TargetP (http://www.psort.nibb.ac.jp/form.html).
Exon–intron structures and motif analyses
The diagrams of exon–intron structures were obtained using the online Gene Structure Display Server (GSDS) bioinformatic tools (http://gsds.cbi.pku.edu.cn/) from both coding sequence (CDS) and genomic sequences.
Protein motif analyses were performed using the MEME suite tool (http://meme.sdsc.edu/meme) with the following parameters: distribution of motifs: zero or one per sequence; maximum number of motifs: 5; minimum number of sites: 2; maximum number of sites: 5; minimum motif width: 6 and maximum motif width: 50.
Chromosome localization of StLEA genes
The chromosomal localization of StLEA genes was retrieved from phytozome and MapChart 2.2 [29] to visualize chromosome linkage on the twelve potato chromosomes. CDS of genes in individual duplicate blocks were aligned using Clustal W software. A codon-based Z-test was performed for each block in MEGA 5.05 using the Pamilo-Bainchi-Li [30, 31] substitution model, the bootstrap variance estimation method (1,000 replicates), and pairwise deletion. Only blocks that show significant results (P value < 0.05) were considered.
Physical and chemical characteristics of predicted LEA proteins in potato
The basic physical and chemical characteristics of potato LEA proteins were calculated using the ProtParam online tool (http://www.expasy.org/tools/protparam.html), including the number of amino acids, molecular weight, theoretical isoelectric point (pI), aliphatic index and grand average of hydropathicity (GRAVY). The protein folding state was predicted by the FoldIndex program (http://bioportal.weizmann.ac.il/fldbin/findex).
In silico gene expression analyses
The relative gene expression Fragments Per Kilobase of transcript per Million fragments mapped (FPKM) values of the 29 LEA genes were obtained from Xu et al. [32]. The expression values were clustered using the quality threshold (QT) clustering tool in the Multiple Array Viewer [33].
Plant material, in vitro culture and stress treatments
Potato plants cv. Claustar were multiplied under in vitro conditions in Murashige and Skoog medium (MS) [34] supplemented with vitamins [35] and under a 16 h photoperiod (70 ± 5 µmol m−2 s−1 photosynthetic photon flux density) at 25 ± 2 °C and 50–60 % relative humidity.
Three weeks old plantlets were then submitted to NaCl (100 mM) or polyethylene glycol 8,000 (PEG, 10 %) treatments for 3, 6, 9 and 24 h. The plantlets were harvested. Plants grown on MS basal medium at 25 °C served as control. All material was frozen in liquid nitrogen for subsequent use. For each condition three biological repetitions were carried out.
RNA isolation and DNase treatment
Total RNA was isolated from 100 mg of leaves, shoots and roots according to Vaewoerd et al. [36] modified by Saïdi et al. [37]. All samples were treated with RNase-free DNase (Promega) and the absence of DNA was verified by PCR using EF1α (Elongation factor 1α) specific primers (EF1αF GATGCTACCACCCCGAAGTA and EF1αR ACGAAGGGGTTTGTCTGTTG). After this control, RNA concentration was quantified spectrophotometrically at 260 nm.
Semi-quantitative RT-PCR analysis of gene expression
The investigation of transcript expression profiles was performed by semi-quantitative RT-PCR analysis as described [38]. All the tests were performed in triplicates. Total RNA (2 µg) were used for first strand cDNA synthesis for both control and the treated samples using 200 units of MoMLV (Moloney Murine Leukemia Virus) reverse transcriptase (Invitrogen) in a final volume of 20 µl. One microliter of each cDNA was used as template for sqRT-PCR with 2 units of Taq DNA polymerase (Invitrogen). The following PCR amplification conditions were used: heating at 95 °C for 5 min, then 25 cycles of (95 °C for 30 s, 60 °C for 30 s, and 72 °C for 5 min). A final extension at 72 °C for 10 min was performed. For the LEA gene expression level analysis, specific primers were designed using the Primer3 v.0.4.0 software (http://frodo.wi.mit.edu/). Primers used for amplification of the StLEA genes and their annealing temperatures are presented in Table 1.
The cDNA product standardization for semi-quantitative RT-PCR was performed using EF1α primers with hybridization temperature of 60 °C and for 25 cycles. The RT-PCR amplified products were analyzed and quantified on ethidium bromide stained agarose gels using the Gel DocXR Gel Documentation System (BioRad). The calculated average band densities were recorded and used in graphical analyses. Error bar was determined from two separate biological replicates.
Results and discussion
Sequence identification and phylogenetic analysis
To identify the LEA genes in potato, a series of basic local alignment search tool (BLAST) searches were first performed by using LEA proteins of Arabidopsis as query sequences and then a second round of BLASTp on the potato genome was made. Sequences which did not present start or stop codons were re-predicted using the fgenesh software (http://linux1.softberry.com/berry.phtml).
A total of 29 StLEA gene models were selected (Table 2). Phylogenetic analyses with LEA sequences from Arabidopsis and rice (Supplementary material 1) using the neighbor-joining methods (Fig. 1) revealed that potato LEA proteins can be divided into seven major phylogenetic groups (Table 2; Fig. 1) as described [16, 18]. Indeed, LEA_4 group (also known as group 3 or D-7) is the most dominant, containing six members (StLEA5, StLEA8, StLEA19, StLEA23, StLEA24 and StLEA26). StLEA5 was also identified as a co-ortholog of AtLEA19 and AtLEA36, while StLEA23 is an ortholog of AtLEA25.
Phylogenetic relationship between LEA protein in rice (OsLEA), Arabidopsis (AtLEA) and potato (StLEA).The tree was constructed using the neighbor-joining algorithm. Scale bar corresponds to 0.1 amino acid substitution per residue. The different LEA groups are indicated by different colors. (Color figure online)
The second important group is known as the dehydrin group (group 2 or D-11) which contains five potato proteins (StLEA6, StLEA7, StLEA13, StLEA18 and StLEA27). They are homologous to rice and A. thaliana LEA proteins. Based on the phylogenetic tree, we can suggest that StLEA7 is a co-ortholog of AtLEA34 (Xero1) and AtLEA51 (RAB18), in Arabidopsis. Xero1 transcripts were detected in seeds [19]. Furthermore, in this cluster, StLEA27 was identified as an ortholog of AtLEA14. It seems also that StLEA13 is an ortholog of OsLEA23.
The LEA_5 group includes StLEA12 as an ortholog of OsLEA21/EMP1. This latter was shown previously to be up-regulated by drought stress [39]. Two StLEA genes (StLEA21 and StLEA25) belonged to the SMP group (Fig. 1).
Analysis of potato LEA gene structure
A comparison of the full-length cDNA sequences with the potato genomic DNA allowed the determination of the exons and introns content of each StLEA gene (Fig. 2) using GSDS. The data showed that the CDS of all the StLEA genes contain one or two introns. The number and position of the introns are relatively conserved in each family; thus, the StLEA gene in the LEA_1, LEA_3 and dehydrin families contained a single intron and two exons. The members of the seed maturation protein (SMP) family have two introns and three exons.
Motif display of predicted StLEA proteins in potato
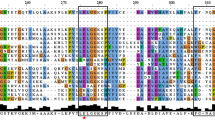
The MEME (Multiple EM for Motif Elicitation) web server was used to analyze the motif distribution of potato dehydrin proteins (Fig. 3; Supplementary material 2). These proteins are characterized by K, S and Y segments [13]. Five genes belonging to the dehydrin group were identified (group 2; Fig. 1) in the potato genome. Each protein contains the conserved K segment (Fig. 3), in addition to a cluster of serines (S segment) present in all these dehydrins. A third domain known as Y segment (Fig. 3) was also found at the N-terminal end of three potato dehydrins (StLEA6, StLEA7 and StLEA27). These dehydrins were divided into five subgroups [40, 41] based on the presence and the organization of the different motifs. The KYS classification for all dehydrin proteins is given in Supplementary material 3. Since, the K and S segments are present in all dehydrins, they were considered as distinguishing traits of this group, while the Y segment is present only in three proteins. It was reported that the phosphorylation of the S segment, leads to calcium binding activity in some dehydrins [21, 42]. However, other S segment containing dehydrins didn’t exhibit such function [41].
Motif distribution of dehydrin proteins in potato using the MEME web server. Each colored box represents the DHN domain and conserved motifs, as indicated in the legends below the trees. The length of the blue line shows the length of a sequence relative to the other sequences; the position of a block shows where a motif has matched the sequence. The amino acid sequences of the conserved motifs are summarized in Supplementary material 2. (Color figure online)
Characteristics of predicted LEA genes in potato
In silico analyses allowed to determine the molecular weight average of the LEA proteins that ranged from 10 to 30 kDa (Supplementary material 4). Few of them are very small LEA proteins (<10 kDa). Some LEA proteins that belong to the LEA_4 phylogenetic group showed high MWs (~45 kDa).
Based on their theoretical pI value, the 29 LEA proteins can be divided into two subgroups. Thirteen LEA proteins sequence are rich in alkaline amino acids while 16 possess more acidic amino acids. However in the LEA_2 and SMP groups, all the amino acids are acidic. The LEA_3 group exhibit a predominance of alkaline amino acids. The important differences in most GRAVY values indicate that the LEA proteins of all groups fall into hydrophilic space on the hydropathy plot. LEA_5 seems to be the most hydrophilic and LEA_2 group the most hydrophobic. The dehydrin and LEA_4 groups showed a wide range of GRAVY values that are all hydrophilic. One consistent property of the major LEA protein groups is the hydrophilicity that seems to be responsible for their lack of secondary structure in the hydrated state, and their solubility at high temperatures [17]. The folding states of the potato LEA protein family showed that the majority of the StLEAs identified were predicted to be unfolded proteins (62 %) under physiological conditions (Supplementary material 4).
Transmembrane helix predictions using TMHMM v.2 [43] suggested that LEA proteins are expressed in subcellular compartments.
The Target P software was used to investigate the subcellular localization of StLEAs proteins. The results indicate that the majority of StLEA proteins are predicted to be in ambiguous sub-cellular localizations, but in the LEA_4 group, two proteins are predicted to be involved in the secretory pathway and one member (StLEA26) is probably targeted to chloroplasts. In addition, in the LEA_3 group, one member (StLEA28) is also predicted to be involved in the secretory pathway.
Genomic organization of the potato LEA genes
The 29 StLEA genes are distributed in the entire potato genome. Indeed, chromosomes I and II contain the highest number of LEA with five genes for each (Fig. 4). Most of the genes are sparsely located. On the other hand, six genes are considered as duplicated among the potato chromosomes (Fig. 4) which could be the result of the polyploidy event of this genome. However, the phylogenetic relationships (Fig. 1) suggest that they are closely related to each other. Therefore, StLEA20 and StLEA28, StLEA10 and StLEA17and StLEA2 and StLEA22 are considered as duplicated genes (Fig. 4). Moreover, StLEA1/StLEA2 on chromosome I and StLEA15/StLEA16 on chromosome X appear as tandem duplications defined by pairs of genes belonging to the same phylogenetic group within a 100 kb distance. They may result from duplications of small parts of a chromosome. These pairs of genes have five or less non-homologous genes between them (Supplementary material 5) [44].
Localization of the identified LEA genes on the potato chromosomes. The chromosome number is indicated at the top of each chromosome. The genes marked in red color indicate genes of the dehydrin family. Colored squares indicate potential segmental duplicated LEA gene pairs: StLEA20 and StLEA28 (red triangle), StLEA10 and StLEA17 (blue triangle) and StLEA2 and StLEA22 (gray triangle). The thick lines join tandem repeated genes (StLEA1 and StLEA2). The chromosome size was retrieved from Xu et al. [32]. P values for individual pairs of duplicate pairs are available in supplementary material 5. (Color figure online)
Expression profiling of LEA genes
The expression profile of the 29 StLEA genes was first analyzed via the in silico analyses from vegetative libraries (Fig. 5). Expression analysis was performed on leaves, stems and roots and under different abiotic stress conditions. We used transcriptome data derived from Illumina RNA-Seq reads generated by PGSC [32] and analyzed by Massa et al. [45].
Expression analysis and hierarchical clustering of all LEA genes in S. tuberosum in different organs (a) and under different stress conditions in leaves: b under hormone induction; c abiotic stress; d infection with P. infestans. Color scale at the end of each dendrogram represents relative expression levels: green represents low level and red indicates high level asterisk indicates dehydrin genes. (Color figure online)
High expression levels in the vegetative organs were observed for nine genes in the absence of any stress or growth regulator treatment (Fig. 5). The expression of LEA genes seemed to be enhanced mainly by abiotic stress rather than by infection with Phytophtora infestans. Very low expression levels of the genes were observed under standard conditions in leaves. They remain low in salt-stressed leaves compared to hormonal treatments. Nineteen StLEA genes seemed to be highly upregulated by ABA treatment, suggesting that these StLEA proteins may be related to the ABA signaling pathway. Indeed, this hormone plays important roles in plant response to dehydration [5]. Many reports have claimed that some DHN genes are not induced by the endogenous ABA accumulation, while they respond to ABA [5, 22].
These data suggest that various StLEAs may be associated with different functions as was shown for Arabidopsis orthologs, (StLEA7 as ortholog of AtLEA51; RAB18). For example, Arabidopsis LEA51 was reported to be up regulated by salt and cold stresses and more strongly by drought [19].
Phylogenetic analysis showed that StLEA23 is an ortholog of Arabidopsis AtLEA25 (COR15b) that was shown previously to be highly upregulated by cold and salinity stress [19].
According to RNAseq data, StLEA23 may be induced by ABA and benzo (l, 2, 3) thiadiazole-7-carbothioic acid S-methyl ester (BTH) treatment but not by salt stress (Fig. 5).
Ten StLEA genes seem not to be expressed in potato in any tested condition or organ. They may correspond to pseudogenes or they may be expressed in other particular conditions.
Expression of potato DHN genes
The expression analysis was carried out by semi-quantitative RT-PCR on the five dehydrin family members. Expression analysis was followed under conditions of 24 h of dehydration (PEG8000; 10 %) and of salt stress (100 mM NaCl). The results show that under dehydration stress (Fig. 6), StDHN2a (StLEA18), StDHN1 corresponding to StLEA7 and StDHN25 corresponding to StLEA13 are expressed in all the tissues studied but at different levels. Expression of StDHN1 gene is higher in both stems and roots than in leaves. StDHN1 mRNAs is most abundant in roots. On the contrary, weak StDHN25 levels are detected in roots.
Expression profiles of potato StDHN genes following treatment with PEG. Total RNA was extracted from different organs (leaves, shoots and roots) of S. tuberosum submitted to PEG treatment. Relative intensity levels of dehydrin transcripts were determined using semi-quantitative RT-PCR during 24 h of PEG treatment. SqRT-PCR data were normalized using the potato elongation factor (EF1α) gene. Error bars indicate standard deviation across two biological replicates. Black square leaves, dark gray square stems, light gray square roots
In roots, StDHN2a expression increased in response to the PEG treatment, starting after 3 h of treatment. Maximum expression was observed at 6 h (Fig. 6). However, no expression of TAS14 (an ABA and environmental stress inducible DHN) was detected in roots.
Semi-quantitative RT-PCR showed that StLEA6 (TAS14) was weakly expressed in leaves and stems at 9 and 24 h of PEG treatment.
Similarly to its ortholog in A. thaliana (Fig. 1; 19), the expression levels of the StDHN1 gene were very low in drought-stressed leaves compared to control leaves. Moreover, StLEA27 was not expressed in any tissue under normal growth conditions or dehydration stress suggesting that this DHN may not be related to drought stress response. In contrast, Wang et al. [22] reported that wheat DHN from the YSK2 group were mostly induced in dehydrated leaves.
Salt stress (Fig. 7) led to the induction of almost all StDHNs within the leaves. StDHN1 and StDHN25 expression was detected in all organs. However, almost no expression of StDHN2a and TAS14 was detected in stems and roots.
Relative intensity levels of dehydrin genes under exposure to NaCl treatment in potato. Total RNA was extracted from leaves, shoots and roots and used in semi-quantitative RT-PCR analyses. Ef1α constitutive gene expression was used as control to normalize the amount of templates in PCR samples. Each value represents an average of at least two independent replicates. Error bars indicate the standard deviations. Black square leaves, dark gray square stems, light gray square roots
Under salt stress the expression of StDHN2a was detected only in leaves starting at 3 h, reaching a maximum after 6 h and then gradually decreased. This treatment seemed also to induce the expression of StLEA27 in leaves after 6 h. Expression of the TAS14 gene was induced at 3 and 9 h post-treatment in leaves and roots respectively.
Moreover, salt treatment induced the expression of StDHN25 in leaves after 3 h. This expression seems to be maintained at a high level for 24 h.
Thus, three of these genes (StDHN2a, TAS14 and StLEA27) were expressed only under salt and/or drought stress conditions, suggesting that they play specific roles under stress conditions. However, the StDHN1 (YSK2) and StDHN25 (SK3) genes were expressed in all tissues in standard conditions, suggesting that LEA genes also are involved in normal plant growth and development. Similarly to StDHN25, expression of the wheat SK3 type TaDHN2 was induced in dehydration and salt stressed leaves and roots [22].
The RT-PCR analyses are in agreement with the in silico predictions and confirmed that the expression of potato DHN are related to abiotic stress response in a tissue specific pathway. These data corroborate also several others studies on LEA proteins [4, 5, 8, 10, 25].
Conclusion
This report describes a detailed analysis of the LEA gene family in Solanum tuberosum. A wide range of sequence diversity was revealed from the in silico analyses. The noticed distinct phylogenetic tree topology and expression patterns were confirmed by experimental data.
These results can provide additional information for the understanding of physiological properties and roles of LEA proteins in the annotated potato genome.
This will provide important information for gene-cloning and function analyses to better understand the role of LEA proteins in abiotic stress responses.
References
Shih MD, Hsieh TY, Lin TP, Hsing YC, Hoekstra FA (2010) Characterization of two soybeans (Glycine max L.) LEA IV proteins by circular dichroism and fourier transform infrared spectrometry. Plant Cell Physiol 51:395–407
Du D, Zhang Q, Cheng T, Pan H, Yang W, Sun L (2012) Genome-wide identification and analysis of late embryogenesis abundant (LEA) genes in Prunus mume. Mol Biol Rep. doi:10.1007/s11033-012-2250-3
Lan T, Gao J, Zeng QY (2013) Genome-wide analysis of the LEA (late embryogenesis abundant) protein gene family in Populus trichocarpa. Tree Genet Genomes 9:253–264
Dure L (1993) Structural motifs in LEA proteins. In: Close TJ, Bray EA (eds) Plant responses to cellular dehydration during environmental stress. American Society of Plant Physiologists, Rockville, pp 91–103
Hanin M, Brini F, Ebel C, Toda Y, Takeda S, Masmoudi K (2011) Plant dehydrins and stress tolerance. Plant signal behav 6:1503–1509
Brini F, Hanin M, Lumbreras V, Irar S, Pagès M, Masmoudi K (2007) Functional characterization of DHN5, a dehydrin showing a differential phosphorylation pattern in two Tunisian durum wheat (Triticum durum Desf.) varieties with marked differences in salt and drought tolerance. Plant Sci 172:20–28
Finkelstein RR, Rock C (2002) Abscisic acid biosynthesis and response. In: Meyerowitz EM, Somerville CR (eds) The Arabidopsis book http://www.aspb.org/publications/arabidopsis
Bies-Ethève N, Gaubier-Comella P, Debures A, Lasserre E, Jobet E, Raynal M, Cooke R, Delseny M (2008) Inventory, evolution and expression profiling diversity of the LEA (late embryogenesis abundant) protein gene family in Arabidopsis thaliana. Plant Mol Biol 67:107–124
Filiz E, Ozyigit II, Tombuloglu H, Koc I (2013) In silico comparative analysis of LEA (Late embryogenesis abundant) proteins in Brachypodium distachyon L. Plant Omics 6:433–440
Close TJ (1997) Dehydrins: a commonality in the response of plants to dehydration and low temperature. Physiol Plant 100:291–296
Battista JR, Park MJ, McLemore AE (2001) Inactivation of two homologues of proteins presumed to be involved in the desiccation tolerance of plants sensitizes Deinococcus radiodurans R1 to desiccation. Cryobiology 43:133–139
Goyal K, Pinelli C, Maslen SL, Rastogi RK, Stephens E, Tunnacliffe A (2005) Dehydration-regulated processing of late embryogenesis abundant protein in a desiccation-tolerant nematode. FEBS Lett 579:4093–4098
Close TJ (1996) Dehydrins: emergence of a biochemical role of a family of plant dehydration proteins. Physiol Plant 97:795–803
Reyes JL, Rodrigo MJ, Colmenero-Flores JM, Gil JV, Garay-Arroyo A, Campos F, Salamini F, Bartels D, Covarrubias AA (2005) Hydrophilins from distant organisms can protect enzymatic activities from water limitation effects in vitro. Plant, Cell Environ 28:709–718
Irar SEO, Pagès M, Goday A (2006) Towards the identification of late-embryogenic-abundant phosphoproteome in Arabidopsis by 2-DE and MS. Proteomics 6:175–185
Bray EA (1993) Molecular responses to water deficit. Plant Physiol 103:1035–1040
Tunnacliffe A, Wise M (2007) The continuing conundrum of the LEA proteins. Naturwissenschaften. doi:10.1007/s00114-007-0254-y
Wise MJ (2003) LEAping to conclusions: a computational reanalysis of late embryogenesis abundant proteins and their possible roles. BMC Bioinformatics 4:52
Hundertmark M, Hincha DK (2008) LEA (Late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9:118. doi:10.1186/1471-2164-9-118
Espelund M, Saeboe-Larssen S, Hughes DW, Galau GA, Larsen F, Jakobsen KS (1992) Late embryogenesis-abundant genes encoding proteins with different numbers of hydrophilic repeats are regulated differentially by abscisic acid and osmotic stress. Plant J2:241–252
Arumingtyas EL, Savitri ES, Purwoningrahayu RD (2013) Protein profiles and dehydrin accumulation in some soybean varieties (Glycine max L. Merr) in drought stress conditions. Am J Plant Sci 4:134–141
Wang Y, Xu H, Zhu H, Tao Y, Zhang G, Zhang L, Zhang C, Zhang Z, Ma Z (2014) Classification and expression diversification of wheat dehydrin genes. Plant Sci 214:113–120
Xu H, Yang Y, Xie L, Li X, Feng C, Chen J, Xu C (2014) Involvement of multiple types of dehydrins in the freezing response in loquat (Eriobotrya japonica). PLoS One. doi:10.1371/journal.pone.0087575
Yang Y, He M, Zhu Z, Li S, Xu Y, Zhang C, Singer SD, Wang Y (2012) Identification of the dehydrin gene family from grapevine species and analysis of their responsiveness to various forms of abiotic and biotic stress. BMC Plant Biol 12:140
Garay-Arroyo A, Colmenero-Flores JM, Garciarrubio A, Covarrubias AA (2000) Highly hydrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit. J Biol Chem 275:5668–5674
Galau GA, Wang HYC, Hughes DW (1993) Cotton Lea5 and Lea14 encode atypical late embryogenesis-abundant proteins. Plant Physiol 101:695–696
Higgins D, Thompson J, Gibson T (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi:10.1093/nar/22.22.4673
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol Biol Evol 28:2731e9
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Li WH (1993) Unbiased estimation of the rates of synonymous and non synonymous substitution. J Mol Evol 36:96–99
Pamilo P, Bianchi NO (1993) Evolution of the Zfx and Zfy genes: rates and interdependence between the genes. Mol Biol Evol 10:271–281
Xu X, Pan S, Cheng S (2011) Genome sequence and analysis of the tuber crop potato. Nature 475:189–195
Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M et al (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374–378
Murachige T, skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Morel G, Wetmore RH (1951) Fern callus tissue culture. Am J Bot 38:141–143
Vaewoerd TC, Dekker BMM, Hoekema A (1989) A small scale procedure for the rapid isolation of plant RNAs. Nucleic Acid Res 17:23–62
Saidi MN, Gargouri-Bouzid R, Rayanni M, Drira N (2009) Optimization of RNA isolation from brittle leaf disease affected date palm leaves and construction of a subtractive cDNA library. Mol Biotechnol 41:63–68
Degenhardt J, Al-MasriA N, Kurkcuoglu S, Szankowski I, Gau AE (2005) Characterization by suppression subtractive hybridization of transcripts that are differentially expressed in leaves of apple scab-resistant and susceptible cultivars of Malus domestica. Mol Genet Genomics 273:326–335
Wang XS, Zhu HB, Jin GL, Liu HL, Wu WR, Zhu J (2007) Genome-scale identification and analysis of LEA genes in rice (Oryza sativa L.). Plant Sci 172:414–420
Campbell SA, Close TJ (1997) Dehydrins: genes, proteins, and associations with phenotypic traits. New Phytol 137:61–74
Alsheikh MK, Svensson JT, Randall SK (2005) Phosphorylation regulated ion-binding is a property shared by the acidic subclass dehydrins. Plant, Cell Environ 28:1114–1122
Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg E, Man O, Beckmann JS, Silman I, Sussman JL (2005) FoldIndex©: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics 21:3435–3438
Krogh A, Larsson B, Von Heijne G, Sonnhammer ELL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580
Hanada K, Zou C, Lehti-Shiu MD, Shinozaki K, Shiu SH (2008) Importance of lineage specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol 148:993–1003
Massa AN, Childs KL, Lin H, Bryan GJ, Giuliano G, Buell CR (2011) The transcriptome of the reference potato genome Solanum tuberosum Group Phureja clone DM1-3 516R44. PLoS ONE. doi:10.1371/journal.pone.002680
Acknowledgments
This work was financed by the Tunisian Ministry of High Education and Scientific Research. The authors are grateful to Anne-Lise Haenni from the Institute Jacques Monod (France) for reading and improving the English.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Charfeddine, S., Saïdi, M.N., Charfeddine, M. et al. Genome-wide identification and expression profiling of the late embryogenesis abundant genes in potato with emphasis on dehydrins. Mol Biol Rep 42, 1163–1174 (2015). https://doi.org/10.1007/s11033-015-3853-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-015-3853-2