Abstract
Low temperature is a major environmental constraint on the production of apples worldwide. An apple rootstock with high cold tolerance was selected to identify genes related to stress tolerance. The transcriptional profiles of the genes in the leaves were examined after various intervals of exposure to cold stress. We obtained three libraries of 14,219, 11,176 and 16,116 tag-mapped predicted coding sequences at three time points (0, 1 and 6 h) during cold stress. In the two time periods, which were from 0 to 1 h and from 1 to 6 h, 139 and 1,085 genes were upregulated, and 1,499 and 381 genes were downregulated, respectively. These groups included a large number of unknown genes. The distribution of genes indicated cold adaptation in the plant. Most of the differential expression genes (DEGs) had temporal specificity and significantly different expression changes at different time points. The classification of DEGs by GO category and KEGG pathway analysis revealed that the DEGs are involved in numerous biological pathways, including metabolism, plant–pathogen interaction and signal transduction. Eleven randomly selected tag-mapped genes were examined by qRT-PCR. The results of the qRT-PCR were in accordance with the transcriptional profiles. The most upregulated gene (MDP0000198054) from 0 to 1 h encodes a dehydration-responsive element-binding protein/C-repeat factor (DREB/CBF). In this study, MDP0000198054 and related genes involved in the cold stress response were discussed. These results could provide new insights into the stress tolerance mechanisms of apple rootstocks.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The apple (Malus × domestica) is one of the most important fruit crops worldwide [1], and China is the leading producer of apples. Low temperature is a major environmental factor that affects apple productivity. Most of the apple-growing regions in China are in mainland monsoon climates characterised by unstable spring temperatures. Early spring chilling below 10 °C and late spring frosts for short time that occur during the fast growth stage of apple trees can severely damage the plants [2, 3]. Identifying cold-resistance genes in apple varieties could lead to a better understanding of the molecular mechanisms and genetic basis of stress tolerance.
With the increasing availability of sequence data, expression profiling has recently been used to identify the genes involved in adaptive stress responses [4–10]. The expressed sequence tag (EST) approach has been used to identify stress tolerance genes and to determine the expression pattern induced by stress in plants [11]. Microarray analysis is another useful technology for assaying the transcriptional responses of plants to stress. High-throughput screening techniques (high-throughput Illumina sequencing technology) with low cost and the capacity to generate large volumes of sequence data have been developed in recent years and have been adapted to monitor the expression of genes in response to abiotic stress. This technology, also referred to as digital gene expression (DGE) tag profiling, can be used to identify millions of short RNAs and differentially expressed genes (DEGs) without prior annotations [12]. However, few studies have used the high-throughput Illumina sequencing technology to analyse the transcriptional differences of the genes that are expressed in response to abiotic stress in the apple tree.
In this study, we selected a cold resistant variety of apple tree, M.26, by comparing it to another common variety, Gala. M.26 is an excellent apple rootstock widely used for grafting in many regions worldwide because it supports the apple tree with strong growth potential, cold and drought hardiness, and wind resistance. We present genome-wide analysis of gene expression changes under cold stress using tag sequencing on a Solexa Illumina platform. A large quantity of novel and DEGs were identified to be involved in cold stress. This study was intended to increase significantly our understanding of the molecular mechanisms and the genetic basis of cold stress tolerance in apple.
Materials and methods
Plant material and experimental treatments
In Chinese main production area of apple, Mailing 26 (M.26) is the most common apple dwarf rootstock and Gala is the most common graft. Tissue-cultured plantlets have the same genetic background with field grown plants, though there are difference at some degree, so we select tissue-cultured plantlets of M.26 and Gala used as experimental samples in this investigation. Besides, experimental conditions could be controlled accurately and convinced results could be obtained.
Tissue-cultured plantlets of M.26 and Gala were grown in MS solid medium containing 3 % sucrose, 0.1 mgl−1 1-Naphthaleneacetic acid (NAA) and 0.6 mgl−1 6-benzyladenine (6-BA). The plantlets were cultured under 80 μmol m−2 s−1 photosynthetic photon flux density with a 12 h illumination and 12 h dark cycle at a constant temperature of 25 °C. After 3 weeks culture, plants with uniform growth were exposed at chilling conditions (4 °C) for 0, 1, 3, 6 and 10 h to simulate the influence of spring chilling. Three independent experiments were performed, and each leaf sample was taken from at least five plantlets.
Electrolyte conductivity
Electrolyte conductivity content was measured as described previously [13]. Five leaf discs (5 mm in diameter) of the five different treatment groups were punched and rinsed with distilled water. These discs were incubated in distilled water at room temperature and shaken for 4 h. The initial electrolyte conductivities of the solutions were measured with a conductivity detector (DDS SJ-308A; China). The solutions were boiled and then were used to measure the absolute conductivity. Electrolyte leakage was calculated using the initial and absolute conductivity [13].
MDA content
MDA content was measured as described previously [14]. Leaf samples (0.5 g) from the five treatment groups were homogenised in 5 ml of 10 % trichloroacetic acid (TCA) and centrifuged at 12,000 rpm for 10 min. Then, 2 ml of 0.6 % thiobarbituric acid (TBA) in 10 % TCA was added to 2 ml of the supernatant. The mixture was heated in boiling water for 15 min and then quickly cooled in the mixture of ice and water. Following centrifugation at 12,000 rpm for 10 min, the absorbance of the supernatant at 450, 532 and 600 nm was measured spectrophotometrically to calculate MDA content.
Sample preparation and total RNA extraction
The leaf samples from ten individual plantlets with cold treatments (4 °C) for 0, 1 and 6 h were harvested and mixed for gene expression analysis. The collected tissue samples were immediately frozen in liquid nitrogen and stored at −60 °C until further use. Total RNA was isolated from the leaves using the cetyltrimethylammonium bromide (CTAB) procedure [15]. The quality of RNA (ratios of A260/A280 ranging from 1.9 to 2.1) and the concentration were measured using a NanoDrop Spectrometer (ND-1000 Spectrophotometer, Peqlab). The integrity of the RNA samples was examined using an Agilent 2100 Bioanalyser (RNA Nano Chip, Agilent, Santa Clara, CA, USA) [15].
Library construction and deep sequencing
20 μg total RNA samples of three libraries were prepared for Illumina sequencing. Magnetic beads with attached poly T oligos were used to purify the mRNA from the total RNA. The first- and second-strand cDNA was synthesised using a poly T oligo primer. The bead-bound cDNA was digested with NlaIII to generate small fragments from the most 3′ CATG to the poly A-tail. Subsequently, Illumina adapter 1 was ligated to the free 5′ end, and the sample was digested with MmeI, which cut the cDNA 17 bp downstream of the CATG site and detached the fragments from the beads. After dephosphorylation and phenol extraction, Illumina adapter 2 was ligated to the 3′ end of the tag. Finally, the short cDNA fragments were prepared for Illumina sequencing on an Illumina Genome Analyzer (San Diego, CA, USA) using the manufacturer’s protocol and the reagents of the Illumina Gene Expression Sample Prep Kit and a Solexa Sequencing Chip (flow cell).
Data processing
All data were minimum information about a microarray experiment (MIAME)-compliant. The raw data were filtered to remove low-quality tags containing short tags (<21 nt), empty reads, and singletons (tags that occurred only once). The remaining dataset consisting of clean tags was subsequently classified according to the copy number in the library. The percentage of total clean tags was calculated, and the saturation of the library was analysed [16]. The clean tags were mapped to the apple genome. Tags that mapped to more than one transcript were excluded from our analysis. When multiple tag types aligned to different positions of the same gene, the gene expression levels were represented by a summation of all tags. Subsequently, the matched genes were assigned to broad functional categories based on similarity comparison to the Genbank Non-Redundant protein database using BLASTx.
Differential expression of genes
Tag libraries were constructed for the two stressed samples and one control sample. A statistical analysis of the frequency of each tag in the different cDNA libraries was performed to compare the differential expression of genes in both stressed and control conditions.
To compare the differential expression of genes across the samples, all clean tags were mapped to the reference sequences, and only 1-bp mismatches were considered. The clean tags that mapped to reference sequences from multiple genes were filtered. The remaining clean tags were designed to be unambiguous. The number of unambiguous clean tags for each gene was calculated and normalised to the TPM (the number of transcripts per million clean tags) [17]. A rigorous algorithm [18] was used to identify DEGs between the three samples. The P value corresponds to the differential gene expression test. The false discovery rate (FDR) is a method that determines the threshold of the P value in multiple tests and analyses, and the P value is obtained by manipulating the FDR value. We used FDR ≤ 0.001 and the absolute value of \(\log_{2} ratio\; \ge \;1\) as the threshold to judge the significance of the gene expression differences. For gene expression profiling, gene ontology (GO) enrichment analysis of functional significance was applied using the hypergeometric test to map all DEGs to terms in the BGI WEGO. The focus was on finding significantly enriched GO terms in the DEGs and comparing these GO terms to the transcriptome database.
Quantitative real-time PCR (qRT-PCR) analysis
The expression levels of the selected candidate genes were determined using quantitative real-time-PCR (qRT-PCR). Eleven candidate genes from the differential expression libraries were validated by qRT-PCR using the same RNA samples that were used for the DGE library construction. Tissue samples were removed from the −60 °C freezer. The first-strand cDNA fragments were synthesised from total RNA using a TransScript™ One-step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China). Eleven gene-specific primer pairs were designed based on the target gene sequences using the Beacon Designers 8 software. The reactions were performed with a Stratagene Mx3000P instrument (Agilent) in final volumes of 20 μl. Each reaction contained 0.8 μl of cDNA, 10 μl 2 × SYBR premix Ex Taq™ (Takara, Shiga, Japan) and 10 μM of the forward and reverse primers. The thermal cycling conditions were as follows: 44 cycles of 95 °C denaturation for 15 s, 55 °C annealing for 30 s and extension for 15 s. The apple actin gene was used as an internal control. The relative expression levels were calculated as 2−(ΔCt of treatment−ΔCt of control) [19].
Results
M.26 has strong cold tolerance
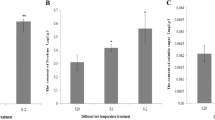
To determine the time from cold treatment to remarkable gene differential expression, we tested the physiological indices of electrolyte conductivity and malondialdehyde (MDA) content in the apple rootstocks M.26 and the Gala cultivation variety. Electrolyte leakage occurs after membrane damage and is a typical symptom of cold injury [20]. Figure 1a indicates that the relative electrolyte leakage levels in M.26 and Gala increased significantly within 1 h of cold stress. The electrolyte leakage in Gala plateaued between 1 and 6 h of cold treatment and later continued to increase between 6 and 10 h. In M.26, the relative electrolyte leakage peaked after 1 h of cold treatment and subsequently decreased to the initial level by 10 h.
Relative electrolyte leakage and MDA content analyses of M.26 and Gala plantlets at 4 °C. Relative electrolyte leakage (a) and MDA content (b) were measured at 0, 1, 3, 6, and 10 h treatment at 4 °C. The values of electrolyte leakage of M.26 and Gala at 0 h were treated as 100 %. Values are the means of three experiments ±SD
The amount of MDA in cells is commonly used as a lipid peroxidation indicator, and the MDA levels increased during cold treatments [14]. Figure 1b shows that the MDA content increased in both M.26 and Gala apples. However, after 6 h of cold stress, a more significant increase was observed in Gala than in M.26. After 10 h of cold stress, the MDA content in Gala was approximately 1.4 times higher than in M.26. These results suggest that M.26 has more cold hardiness than Gala and that 1 and 6 h are two important cold exposure time points; thus, our study focused on the three time points of 0, 1 and 6 h to investigate the cold adaptation mechanism of M.26.
Illumina sequencing and gene annotation
To examine the transcriptome globally after cold stress in M.26, a Solexa Genome Analyzer was used to perform high-throughput tag sequencing analysis on poly (A)-enriched RNAs from the three libraries of 0, 1 and 6 h of cold treatment. The total number of tags per library was approximately 4.6 million. After removing low-quality tags, the proportion of clean tags were more than 92 % (Table 1). The distribution of total and distinct tag counts exhibited similar tendencies for all libraries (Fig. 2). Among the distinct tags, less than 6.5 % had copy numbers higher than 100; more than 22 % of the tags had copy numbers between 5 and 50 copies; and more than 56 % had copy numbers lower than 5. A total of 146,031, 122,171 and 288,569 distinct clean tags corresponding to the three libraries were generated (Table 1).
To annotate sequences by mapping the tags to genes is an important step, which can reveal the molecular events behind the observed gene expression. In this study, more than 10 % of the distinct tags were unambiguously mapped to the predicted coding sequences (CDSs). The tags that mapped to the database of the Malus × domestica genome (http://genomics.research.iasma.it/) generated 14,219, 11,176 and 16,116 tag-mapped predicted CDSs for the three libraries described above, respectively. The tag-mapped predicted CDSs were annotated in GenBank Non-Redundant protein databases using BLASTx.
Exploration of novel and differentially expressed genes (DEGs) in response to low temperature stress
In Fig. 3, 1,638 and 1,466 genes were differentially expressed in the 0–1 and 1–6 h cold treatment periods, respectively. Among these DEGs, 139 and 1,085 genes were upregulated, while 1,499 and 381 genes were downregulated in the 0–1 and 1–6 h cold treatment periods, respectively. The results indicate that short time stress (1-h) downregulates the expression of a large number of genes and upregulates the expression of a small number of genes, which suggests that the plants were injured under the stress condition. When the stress time was elongated to 6 h, the number of downregulated genes decreased (from 1,499 to 381), but the number of upregulated genes increased (from 139 to 1,085). To identify the known and unknown genes, all sequences were annotated and classified through alignment with the GenBank and Rfam databases. As a result, 71 unknown and 68 known genes were upregulated, and 508 unknown and 991 known genes were downregulated between 0 and 1 h. While from 1 to 6 h, 510 unknown and 575 known genes were upregulated, and 193 unknown and 188 known genes were downregulated. All of these DEGs were selected for further analysis. We found large divergences in the fold-changes of the known and unknown DEGs. The fold-change of the majority of DEGs ranged from 2 (\(\log_{2} ratio\; = \;1\)) to 32-fold (\(\log_{2} ratio = 5\)); only a small fraction of genes exhibited differences of greater than 32-fold in the two differentially expressed libraries (Table S1 and Table S2). Besides, we found a number of transcripts which have not been reported in the previous studies to be involved in plant stress with the fold-change from 2 to 1,024 fold (\(\log_{2} ratio = 10\)). Some of them were transcription factors (TFs) and some were unknown genes as listed in Table S5. Among them, we found an intriguing gene, encoding Tubby-like protein 7 (TLP7), an obesity-like gene related to animal specific characters.
The distribution of the DEGs was shown in Fig. 4. The red and green regions with dots revealed the genes with markedly different expression, and the blue region showed the genes with no obvious expression diversity (Fig. 4). There were a few upregulated genes (red dots) between 0 and 1 h (Fig. 4a), but there were many more upregulated genes between 1 and 6 h (Fig. 4b). The downregulated genes (green dots) exhibited the opposite trend. In the treatment period between 1 and 6 h, the cold tolerance genes were greatly upregulated (Fig. 4b). This indicates that the cold tolerance processes might be reflected by time-dependent polygenic expression alterations.
The Venn diagram in Fig. 5 depicts the genes that were differentially expressed in response to low temperature in the two different time periods. The 520 overlapped genes were co-expressed, while 1,118 and 946 genes were expressed in the different time periods, indicating that these genes have temporal specificity.
Venn diagram showing overlaps and non-overlaps among DGEs under low temperature stress during the time periods of 0–1 and 1–6 h. The numbers outside the circles denote the total number of genes in each time period. The numbers in one circle denote specific genes, and numbers in the two intersecting circles denote overlapped genes
Gene expression changes can provide important clues about the roles of functionally unknown genes in biologically active processes because the functions of genes sharing the same tendency are likely to be connected. Moreover, comparing the expression changes of stress-regulated genes between different treatment time periods provides important clues regarding the differences and similarities in the response to cold stress during these two time periods.
Functional categorisation and Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis of stress-regulated genes
GO is an international standardised gene functional classification system that describes the properties of genes and gene products in any organism. The DEGs (FDR ≤ 0.001 and \(|\log_{2} ratio|\; \ge \;1\)) were classified into functional categories according to the BGI WEGO (Web Gene Ontology Annotation Plot, http://www.geneontology.org/) database. Comparisons of the two time periods of cold stress yielded similar results in terms of the percentage of genes classified into each category. The differentially expressed genes in both time periods were classified into 16 categories on the basis of cellular components, 10 categories on the basis of molecular function and 13 categories on the basis of biological processes (Fig. 6).
The above genes were further categorised according to the number of genes expressed during different time periods. The molecular function of genes with altered expression between 0 and 1 h (Fig. 6a) were mainly involved in binding (31.7 % upregulated and 38.2 % downregulated) and catalytic activity (26.6 % upregulated and 37.1 % downregulated). In terms of cellular components, low temperature stress affected genes associated with the cell plastid (15.1 % upregulated and 13.2 % downregulated) and cytoplasm (18.0 % upregulated and 22.8 % downregulated). In terms of biological processes, low temperature stress primarily affected metabolic processes (30.2 % upregulated and 38.1 % downregulated) and stress responses (10.1 % upregulated and 8.1 % downregulated). The genes with altered expression between 1 and 6 h (Fig. 6b) showed similar results of categorisation to the genes with altered expression between 0 and 1 h treatment (Fig. 6a). These differentially expressed genes were related to catalytic reactions, transcriptional regulation, and transport activity. All of these events are often observed in plant tolerance responses.
The DEGs were classified according to KEGG function annotations to identify the pathways that were involved in the cold stress response. The KEGG pathway annotation showed that the DEGs in both time periods were annotated for 116 biological processes with P < 1, and most of the DEGs were involved in cellular metabolism, plant–pathogen interaction and signal transduction. The most enriched annotated pathways were the metabolic pathways, with 239 and 205 DEGs representing 22.91 and 23.98 % in the two time periods, respectively, followed by plant–pathogen interaction (6.81 and 9.01 %) and plant hormone signal transduction (5.27 and 5.15 %). The KEGG pathway annotations are presented in Table S3 and Table S4.
Validation of DGE data by qRT-PCR
To validate the reliability of the sequencing data, eleven candidate genes were selected at random for qRT-PCR assays to analyse the expression levels. The qRT-PCR expressions relative to the reference gene, actin, were compared with the high-throughput Illumina sequencing analysis results (Fig. 7). The change tendencies showed similar results between the qRT-PCR and the sequencing methods, suggesting that the sequencing methods were valid.
Discussion
Plant tolerance to biotic and abiotic stresses, such as low or high temperature, drought, salt and disease factors, is a defence response involving multiple genes. Plant species are highly variable with respect to the optimum environment; therefore, a harsh environmental condition for one plant species might not be stressful for another [21, 22]. This variability is reflected in a multitude of different stress-response mechanisms. Identifying specific genes that affect complex traits (i.e., biological traits affected by several genetic and environmental factors is a difficult task for geneticists [23]. Global analysis of the transcriptome in specific species will facilitate the identification of systemic gene expression levels and regulatory mechanisms for the development of tolerance to low temperatures.
In this study, we determined that the degree of damage caused by cold temperatures is much smaller in M.26 than in Gala (Fig. 1). For the apple rootstock M.26, 1 h is a representative time point of chilling damage, and 6 h is a representative time point of cold adaptation (Fig. 1). A comprehensive analysis of the transcriptome changes of M.26 in response to cold stress was performed using tag sequencing on an Illumina sequencing platform. More than 11,000 tag-mapped genes were identified for each library (Table 1). Mapping the tags to genes to annotate sequences can reveal the molecular events behind the gene expression. This set of tag-mapped genes covers almost 40 % of the total predicted CDSs in the CDS database of the Malus × domestica genome, which suggests that these genes adequately define the transcriptome. Analysis of these transcriptomes is helpful for studying the cold stress response molecular mechanism in the apple rootstock M.26.
In the present study, a total of 2,584 DEGs in two differentially expressed libraries (Fig. 5), including a large number of unknown genes, were found to be involved in the cold stress response. These genes encode cellular components, molecular functions and biological processes. The expression changes of numerous genes expressed during the two time periods were significantly different (Fig. S1). The DEGs observed in this study provided useful information for understanding how M.26 responds to cold stress and how M.26 seedlings can adapt to stressful conditions. There were 520 genes that were differentially expressed in the 0–1 and 1–6 h cold treatments (Fig. 5). Additionally, 1,118 genes were differentially expressed only in the 0–1 h time period, and 946 genes were differentially expressed only in the 1–6 h time period. The progressive adjustment of the metabolism with sustained, transient, early- and late-responsive metabolic alterations by DEGs might be the mechanism of cold adaptation.
The genes that were upregulated or downregulated in the two time periods of cold stress (Fig. 4, Table S1 and Table S2) are likely part of distinct cold stress response mechanisms in M.26. The unknown genes accounted for 35 and 48 % (Fig. 4) of the 0–1 and 1–6 h time periods, respectively. There are over half of the genes identified in this study could be placed into a potential functional category based on similarity to previously characterized genes. The known genes could be involved in several mechanisms of the cold stress response. These results show that the high-throughput sequencing data were highly enriched for DEGs under cold stress. TFs and signalling regulators are the most important categories of the genes involved in regulating the expression of downstream cold process-responsive genes. TFs involved in entire stress-response pathways are important regulators and are therefore key targets for genetic engineering to enhance the stress tolerance in valuable crop plants [24].
The highest upregulated gene (MDP0000198054), which had an expression change of more than 1,000-fold (\(\log_{2} ratio = 10.13\)) during the first hour and more than fivefold (\(\log_{2} ratio = 2.40\)) during the next 5 h, encodes a dehydration-responsive element-binding protein (DREB/CBF) of Malus × domestica. DREB/CBF of Malus × domestica and DREB1A of Arabidopsis both belong to the A-1 subgroup of the DREB/CBF family [25]. Members of the DREB/CBF protein family play a critical role in improving the abiotic-stress tolerance of plants. The ICE1–CBF transcriptional cascade is important for cold adaptation in many plant species (Fig. 8). The ICE1 gene encodes a MYC-like basic helix-loop-helix (bHLH) transcription factor that can bind to MYC-binding motifs in the DREB1A/CBF3 promoter. The over-expression of ICE1 in plants enhances the expression of the CBF regulon in cold and freezing tolerance [26], suggesting the activation of the ICE1 protein in response to low-temperature signals. The stability of ICE1, the expression of DREB1/CBF genes and the freezing tolerance are negatively regulated via ubiquitination of ICE1 by the RING finger E3 ligase HOS1 [27, 28]. Miura et al. [29] found that sumoylation of ICE1 by SIZ1, a SUMO E3 ligase, inhibits polyubiquitination of ICE1. The repressed cold-inducible expression of DREB1A and the reduced freezing tolerance in siz1 mutants indicates the importance of sumoylation for the function of ICE1 and freezing tolerance of Arabidopsis [28, 29]. Researchers [30, 31] found that the promoter regions of the cold-inducible DREB1/CBF genes have several evening elements (EEs; AAAATATCT) [30] and circadian clock associated 1 (CCA1) binding sites (AATCT) [31]. These sites are target sequences for the two MYB-related TFs, CCA1 and late elongated hypocotyl (LHY), which are components of the central oscillator [32, 33]. CCA1 has been detected to bind to DREB1 promoter regions. Furthermore, the genes induced by DREB1 under cold stress were dramatically reduced in the cca1-11 lhy-21 double mutant. Therefore, CCA1 and LHY are positive regulators of DREB1 cold induction. Many of these DREB family genes control related gene expression differences in the transcriptome of M.26. The transcriptional regulation of cold-inducible DREB1A in apple rootstocks is shown in Fig. 8. Zhao et al. [25] suggested that MDP0000147009 in subgroup A-2 could be sensitive to cold temperatures in another apple rootstock (Malus prunifolia); however, we found no difference in the expression of MDP0000147009 in the transcriptome of M.26. Therefore, we propose that although the DREBs play an important role in the response of apples to low temperatures, there are differences among apple varieties regarding DREB involvement.
Transcriptional regulation of cold-inducible DREB1A in apple rootstock. The activity of DREB1A is mainly regulated at the transcriptional level. The transcription of DREB1A is under the control of low-temperature signals. Expressed DREB1A proteins bind to DRE/CRT and activate the transcription of target cold-inducible genes, including transcription factors, and thus induce transcriptional changes that eventually cause cold stress responses. Expression change extents of DREB1/CBF-related genes in cold treatment were displayed with colour bars (0 vs. 1 and 1 vs. 6 h). Colour saturation reflects the magnitude of \(\log_{2} ratio\) for each gene
Compared with previous studies, we listed some genes which may be involved in plant stress (Table S5) [34–38]. Among them, TLP7, a gene related to animal specific characters, was largely up-regulated with 512-fold (\(\log_{2} ratio = 9\)). In mammals, genetic mutation of tubby or other TLPs can result in certain disease phenotypes, for example, obesity, retinal degeneration, hearing loss. We have successfully verified the stress-resistant function of TLP7 in prokaryotic cell and identified the β-barrel as its functional region [38].
The comprehensive transcriptome profiling analysis of an apple rootstock under cold stress would enrich the knowledge of the stress tolerance of apple rootstocks at the molecular level and provide possible genetic engineering targets for plant improvement.
Abbreviations
- CBF:
-
C-repeat factor
- CCA1:
-
Circadian clock associated 1
- CDS:
-
Coding sequence
- DREB:
-
Dehydration-responsive-element-binding
- DEGs:
-
Differential expression genes
- DGE:
-
Digital gene expression
- FDR:
-
False discovery rate
- GO:
-
Gene ontology
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- LHY:
-
Late elongated hypocotyl
- M.26:
-
Mailing 26
- MDA:
-
Malondialdehyde
- qRT-PCR:
-
Quantitative real-time PCR
- TFs:
-
Transcription factors
- TLP:
-
Tubby-like protein
- BGI WEGO:
-
Web gene ontology annotation plot
References
Flachowsky H, Peil A, Sopanen T, Elo A, Hanke V (2007) Overexpression of BpMADS4 from silver birch (Betula pendula Roth.) induces early-flowering in apple (Malus × domestica Borkh.). Plant Breed 126(2):137–145. doi:10.1111/j.1439-0523.2007.01344.x
Manuchehr F, Mohammad R, Gholam Ali K, Theodoros M (2009) Modelling apple tree bud burst time and frost risk in Iran. Meteorol Appl 9999(9999):45–52. doi:10.1002/met.159
Vestal CK (1971) First and last occurrences of low temperatures during the cold season. Mon Weather Rev 99(8):650–652. doi:10.1175/1520-0493(1971)099<0650:falool>2.3.co;2
Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA (2005) LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci USA 102(29):10387–10392. doi:10.1073/pnas.0503029102
Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14(8):1675–1690
Kawasaki S, Borchert C, Deyholos M, Wang H, Brazille S, Kawai K, Galbraith D, Bohnert HJ (2001) Gene expression profiles during the initial phase of salt stress in rice. Plant Cell 13(4):889–905
Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130(4):2129–2141. doi:10.1104/pp.008532
Oono Y, Seki M, Satou M, Iida K, Akiyama K, Sakurai T, Fujita M, Yamaguchi-Shinozaki K, Shinozaki K (2006) Monitoring expression profiles of Arabidopsis genes during cold acclimation and deacclimation using DNA microarrays. Funct Integr Genomics 6(3):212–234. doi:10.1007/s10142-005-0014-z
Rensink WA, Buell CR (2005) Microarray expression profiling resources for plant genomics. Trends Plant Sci 10(12):603–609. doi:10.1016/j.tplants.2005.10.003
Oztur ZN, Talame V, Deyholos M, Michalowski CB, Galbraith DW, Gozukirmizi N, Tuberosa R, Bohnert HJ (2002) Monitoring large-scale changes in transcript abundance in drought- and salt-stressed barley. Plant Mol Biol 48(5–6):551–573
Fan XD, Wang JQ, Yang N, Dong YY, Liu L, Wang FW, Wang N, Chen H, Liu WC, Sun YP, Wu JY, Li HY (2013) Gene expression profiling of soybean leaves and roots under salt, saline-alkali and drought stress by high-throughput Illumina sequencing. Gene 512(2):392–402. doi:10.1016/j.gene.2012.09.100
Xiao S, Mo D, Wang Q, Jia J, Qin L, Yu X, Niu Y, Zhao X, Liu X, Chen Y (2010) Aberrant host immune response induced by highly virulent PRRSV identified by digital gene expression tag profiling. BMC Genomics 11:544. doi:10.1186/1471-2164-11-544
Knepper C, Savory EA, Day B (2011) Arabidopsis NDR1 is an integrin-like protein with a role in fluid loss and plasma membrane-cell wall adhesion. Plant Physiol 156(1):286–300. doi:10.1104/pp.110.169656
Hong Z, Lakkineni K, Zhang Z, Verma DP (2000) Removal of feedback inhibition of delta(1)-pyrroline-5-carboxylate synthetase results in increased proline accumulation and protection of plants from osmotic stress. Plant Physiol 122(4):1129–1136
Gasic K, Hernandez A, Korban S (2004) RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol Biol Rep 22(4):437–438. doi:10.1007/bf02772687
t Hoen PA, Ariyurek Y, Thygesen HH, Vreugdenhil E, Vossen RH, de Menezes RX, Boer JM, van Ommen GJ, den Dunnen JT (2008) Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res 36(21):e141. doi:10.1093/nar/gkn705
Morrissy AS, Morin RD, Delaney A, Zeng T, McDonald H, Jones S, Zhao Y, Hirst M, Marra MA (2009) Next-generation tag sequencing for cancer gene expression profiling. Genome Res 19(10):1825–1835. doi:10.1101/gr.094482.109
Audic S, Claverie JM (1997) The significance of digital gene expression profiles. Genome Res 7(10):986–995
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25(4):402–408. doi:10.1006/meth.2001.1262
Puhakainen T, Hess MW, Makela P, Svensson J, Heino P, Palva ET (2004) Overexpression of multiple dehydrin genes enhances tolerance to freezing stress in Arabidopsis. Plant Mol Biol 54(5):743–753. doi:10.1023/B:PLAN.0000040903.66496.a4
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681. doi:10.1146/annurev.arplant.59.032607.092911
Poorter H, Larcher W (2004) Physiological plant ecology. 4th edition. Ann Bot 93(5):616–617. doi:10.1093/aob/mch084
Risch NJ (2000) Searching for genetic determinants in the new millennium. Nature 405(6788):847–856. doi:10.1038/35015718
Century K, Reuber TL, Ratcliffe OJ (2008) Regulating the regulators: the future prospects for transcription-factor-based agricultural biotechnology products. Plant Physiol 147(1):20–29. doi:10.1104/pp.108.117887
Zhao T, Liang D, Wang P, Liu J, Ma F (2012) Genome-wide analysis and expression profiling of the DREB transcription factor gene family in Malus under abiotic stress. Mol Genet Genomics 287(5):423–436. doi:10.1007/s00438-012-0687-7
Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17(8):1043–1054. doi:10.1101/gad.1077503
Dong CH, Agarwal M, Zhang Y, Xie Q, Zhu JK (2006) The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc Natl Acad Sci USA 103(21):8281–8286. doi:10.1073/pnas.0602874103
Ishitani M, Xiong L, Lee H, Stevenson B, Zhu JK (1998) HOS1, a genetic locus involved in cold-responsive gene expression in arabidopsis. Plant Cell 10(7):1151–1161
Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM (2007) SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19(4):1403–1414. doi:10.1105/tpc.106.048397
Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290(5499):2110–2113
Wang ZY, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM (1997) A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9(4):491–507. doi:10.1105/tpc.9.4.491
Yanovsky MJ, Kay SA (2001) Signaling networks in the plant circadian system. Curr Opin Plant Biol 4(5):429–435 S1369-5266
Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293(5531):880–883. doi:10.1126/science.1061320
Iyer LM, Aravind L (2002) The catalytic domains of thiamine triphosphatase and CyaB-like adenylyl cyclase define a novel superfamily of domains that bind organic phosphates. BMC Genomics 3(1):33
Briknarova K, Gehrmann M, Banyai L, Tordai H, Patthy L, Llinas M (2001) Gelatin-binding region of human matrix metalloproteinase-2: solution structure, dynamics, and function of the COL-23 two-domain construct. J Biol Chem 276(29):27613–27621. doi:10.1074/jbc.M101105200
Aplan PD, Nakahara K, Orkin SH, Kirsch IR (1992) The SCL gene product: a positive regulator of erythroid differentiation. EMBO J 11(11):4073–4081
Cubas P, Lauter N, Doebley J, Coen E (1999) The TCP domain: a motif found in proteins regulating plant growth and development. Plant J 18(2):215–222
Du F, Xu JN, Zhan CY, Yu ZB, Wang XY (2014) An obesity-like gene MdTLP7 from apple (Malusxdomestica) enhances abiotic stress tolerance. Biochem Biophys Res Commun 445(2):394–397. doi:10.1016/j.bbrc.2014.02.005
Acknowledgments
This work was supported by the Special Research Fund of Public Welfare of China Agricultural Ministry (201303093).
Author information
Authors and Affiliations
Corresponding author
Additional information
Fan Du and Jia-Ning Xu have contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Du, F., Xu, JN., Li, D. et al. The identification of novel and differentially expressed apple-tree genes under low-temperature stress using high-throughput Illumina sequencing. Mol Biol Rep 42, 569–580 (2015). https://doi.org/10.1007/s11033-014-3802-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3802-5