Abstract
Cotton fibers are specialized single-cell trichomes derived from epidermal cells, similar to root hairs and trichomes in Arabidopsis. While the MYB-bHLH-WD40 (MBW) complex has been shown to regulate initiation of both root hairs and trichomes in Arabidopsis, the role of their homologous gene in cotton fiber initiation remains unknown. In this study, we identified a R2R3 MYB transcription factor (TF), GhWER, which exhibited a significant increase in expression within the outer integument of ovule at -1.5 DPA (days post anthesis). Its expression peaked at -1 DPA and then gradually decreased. Knockout of GhWER using CRISPR technology inhibited the initiation and early elongation of fiber initials, resulting in the shorter mature fiber length. Additionally, GhWER interacted with two bHLH TF, GhDEL65 and GhbHLH121, suggesting a potential regulatory complex for fiber development. RNA-seq analysis of the outer integument of the ovule at -1.5 DPA revealed that the signal transduction pathways of ethylene, auxin and gibberellin were affected in the GhWER knockout lines. Further examination demonstrated that GhWER directly activated ethylene signaling genes, including ACS1 and ETR2. These findings highlighted the biological function of GhWER in regulating cotton fiber initiation and early elongation, which has practical significance for improving fiber quality and yield.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cotton is a valuable cash crop and a major source of natural fiber for the global textile industry. The cotton fibers, specialized trichomes derived from ovule epidermal cells (Stewart 1975), have been a focus of cotton breeding efforts that prioritize high yield (Naoumkina et al. 2020). However, only 20–30% of ovule epidermal cells can differentiate into fiber cells (Applequist et al. 2001). Therefore, substantial potential exists for enhancing the yield of mature fiber by increasing the number of fiber cells.
Both cotton fibers and Arabidopsis root hairs, as unicellular, unbranched, and elongated specialized trichomes originating from epidermal cells, may share similar mechanisms in cell-fate determination(Lee et al. 2007). The cell-fate determination of Arabidopsis has been studied for several decades, culminating in the identification of a conservative MYB-bHLH-WD40 (MBW) ternary complex controlling root hairs and trichomes formation. In Arabidopsis, the R2R3 MYB transcription factor (TF) WEREWOLF (WER) exerts negative regulation on root hair development, whereas GLABRA1 (GL1), another MYB TF, acts as a positive regulator for trichome initiation, as identified in previous research (Lee and Schiefelbein 1999; Oppenheimer et al. 1991). These proteins, together with the bHLH TF GLABRA 3 / ENHANCER OF GLABRA 3 (GL3/EGL3) and the WD repeat protein TRANSPARENT TESTA GLABRA 1 (TTG1), form the MBW complex that regulates the expression of HD-ZIP TF GLABRA 2 (GL2), thus determining epidermal cell differentiation (Payne et al. 2000; Zhang et al. 2003; Walker et al. 1999; Rerie et al. 1994). Studying the functions of homologous genes to the MBW complex may provide valuable insights into the regulatory mechanisms of cotton fiber initiation.
Recent research has highlighted the importance of pivotal TFs such as the R2R3 MYB TF GhMYB25-like and GhMYB25 in cotton fiber initiation, emphasizing their significant roles (Machado et al. 2009; Walford et al. 2011; Qin et al. 2022). Evidence indicates that silencing the expression of GhMYB25-like in cotton using an RNAi approach results in the production of only a few fibers on seeds, while CRISPR-mediated knockout of GhMYB25-like_At/Dt leads to a complete absence of fibers on seeds (Qin et al. 2022; Walford et al. 2011). Additionally, MYB2 and GhMYB109 exhibit high sequence similarity with GL1 and WER, and have been reported to be involved in fiber development (Guan et al. 2014; Suo et al. 2003).While ectopically expressing GaMYB2/GbMYB2/GhMYB2 in Arabidopsis could complement the phenotype of glabrous mutants gl1, the functions of these homologous genes of MBW complex in cotton fiber initiation has been scarcely documented (Guan et al. 2014; Huang et al. 2013; Wang et al. 2004). The involvement of GhMYB109 has been reported in fiber initiation and elongation, but the details of its mechanisms remain undiscovered (Pu et al. 2008). The function of GL1/WER homologous genes in the regulation of cotton fiber initiation is thus not fully understood.
Plant hormones such as auxin, gibberellin acid (GA), and ethylene play pivotal roles in the initiation and elongation of cotton fiber. An increase in cytokinin (CTK) levels enhances fiber yield without compromising fiber quality (Zhao et al. 2015). Auxin, considered as the most crucial hormone for fiber initiation, significantly increases the IAA content in cotton ovules when epidermal-specific promoter (Floral Binding Protein7, FBP7) drives the expression of IAA biosynthetic genes iaaM, resulting in a 15% increase in lint fiber yield (Zhang et al.2011). In addition to auxin biosynthesis, genes involved in transport and signaling pathway, such as the auxin efflux carrier genes PINs and auxin response factors GhARF2/GhARF18, are also implicated in cotton initiation (Zhang et al. 2017; Xiao et al. 2018). Another hormone, GA, positively regulates fiber initiation and elongation. For instance, the overexpression of the GA 20-oxidase gene GhGA20ox in cotton significantly increases the fiber initials and promotes the elongation of fiber cells (Xiao et al. 2010). Moreover, research has revealed that the crosstalk between auxin and GA significantly affect fiber initiation and elongation, with overexpressing of GhARF18 leading to increased GA content that promotes fiber growth through the upregulation of GA 3-beta-hydroxylase gene GA3OX and the GA 20-oxidase gene GA20OX (Zhu et al. 2022). Ethylene, another vital hormone, promotes cell wall extension and thus fiber elongation (Shi et al. 2006). Despite the established importance of these plant hormones in fiber development, the regulatory mechanisms of their action during fiber initiation and early elongation remain unclear.
This study aims to identify the gene function of WER homologous gene, GhWER, in cotton fiber initiation and early elongation. By integrating gene expression analysis with CRISPR mutants of GhWER, we propose that GhWER is predominantly expressed during fiber initiation and early elongation and contributes positively to the length of mature fibers by influencing the timing of fiber initiation. Additionally, our findings indicate that GhWER interacts with the bHLH TF GhDEL65 and GhbHLH121 as shown by yeast two-hybrid assays. We also reveal that GhWER modulates fiber initiation and early elongation through the regulation of the ethylene signaling pathway. These results offer new insights into the mechanisms of fiber initiation and early elongation, with potential implications for improving cotton fiber quality.
Materials and methods
Plant Materials
The receptor for transformation was Gossypium hirsutum acc. Jin668. Both the wildtype Jin668 and GhWER transgenic lines were cultivated in the experimental field at Huazhong Agricultural University in Wuhan, Hubei, China following standard agricultural practices. The tobacco (Nicotiana benthamiana) used for the LUC assay was grown in a climate-controlled growth chamber at 25℃ with 16 h of light and 8 h of dark.
Identification of MYB TFs in cotton and construction of phylogenetic tree
The gene and protein sequences of cotton were retrieved from CottonFGD (https://cottonfgd.net), and those of Arabidopsis were download from TAIR (https://www.arabidopsis.org). Sequence alignment was performed using ClustalX software and ESPript (https://espript.ibcp.fr). The aligned sequences were then imported into MEGA-X software for phylogenetic tree construction. The phylogenetic tree was annotated using the iTOL tool (https://itol.embl.de) while domain and gene structure analysis were conducted using the online platform GSDS (http://gsds.gao-lab.org).
qRT-PCR
Tissue samples used for qRT-PCR analysis were collected from G. hirsutum acc. Jin668. Root, stem, and leaf samples were obtained from hydroponic plant, and ovule samples at -1.5 DPA (days post anthesis), -1 DPA, -0.5 DPA, 0 DPA, 3 DPA, and 5 DPA, along with fiber samples at 5 DPA, 10 DPA, 15 DPA, and 20 DPA, were collected from field-grown plants. Three biological replicates were processed for each sample. Subsequently, total RNA from these tissues was extracted using the RNAprep Pure Plant Plus Kit (Tiangen, DP441), and reverse transcription was performed using Reverse Transcriptase M-MLV (Takara, 2641Q). The cotton gene GhUB7 was used as reference gene (Hao et al. 2012). Primer sequences for qRT-PCR were designed referencing the qRT Primer Database (qPrimerDB, https://biodb.swu.edu.cn/qprimerdb) and can be found in Table S3. Gene expression was detected using the ABI 7500 Real-Time PCR system.
Vector construction and transformation
Two sgRNAs targeting GhWER CDS were designed using CRISPR-P (http://cbi.hzau.edu.cn/crispr) and cloned into the pRGEB32-GhU6.9 vector (Wang et al. 2018). The primers used for vector construction can be found in Table S3. The recombinant vector was then transformed into Agrobacterium tumefaciens strain EHA105, which was used to infect the hypocotyl of the Jin668 receptor for transformation (Jin et al. 2006a, 2006b).
Detection of gene editing efficiency
To detect the genotype of target sites in T2 transgenic plants, genomic DNA was extracted from young leaves using the CTAB method. The DNA sequence surrounding the target sites was then determined using high-throughput tracking (HiTOM) (Liu et al. 2019). The process included the following steps: (1) the first round of PCR amplified the target site and its upstream and downstream sequences in a 96-well PCR plate; (2) the second round of PCR added a barcode to the first round of PCR products; (3) the PCR products from the 96-well PCR plate were combined and purified with a DNA purification kit (Tiangen, DP204); (4) the purified DNA were sequenced using high-throughput sequencing from Novogene, and the obtained sequences were analyzed through the Hi-TOM online website (http://www.hi-tom.net/hi-tom) to determine the final gene editing efficiency. The primers for PCR can be found in Table S3.
Ovule observation by scanning electron microscopy
Three to four cotton bolls from WT and three GhWER knockout lines were collected from similar position on the plants at 0 DPA and 1 DPA. The ovules were then stripped from these bolls and fixed in a 2.5% (v/v) glutaraldehyde solution at 4℃. The specific steps for sample preparation, including dehydration using graded ethanol, isoamyl acetate treatment and sample drying, were carried out following procedures outlined in our previous research (Hu et al. 2018). Finally, the dried ovules were observed and photographed using a scanning electron microscopy (JSM-6390/LV).
Measurement of fiber quality
The mature cotton bolls from WT and three GhWER knockout lines were collected from the middle part of plants simultaneously. Three biological replicates were harvested for each line. To measure the lint percentage, lint index, and seed index, all fiber attached to 100 seeds were removed, and the weight of fiber attached to 100 seeds was recorded as the lint index, while the weight of 100 seed was recorded as the seed index. The lint percentage was calculated as the weight of fiber attached to 100 seeds divided by the total weight of 100 seeds and their attached fiber. Subsequently, 15 seeds attached with fiber from the middle part of bolls were selected for the measurement of mature fiber length using the hand-combing method. Finally, 10 g of fiber was selected to measure fiber quality using a semi-automatic high volume cotton tester (PREMIER HFT), including upper-half mean length (UHMI), micronaire value, strength, uniformity index, and elongation.
Interacted protein screening by yeast two hybrid (Y2H)
The Matchmaker Gold Yeast Two-Hybrid System (Clontech, Cat. no. 630489) was employed to identify proteins interacting with GhWER. The CDS of GhWER and its truncated sequence were cloned into the bait vector pGBKT7 and transformed into yeast strains Y2HGold. The CDS of highly expressed TFs in ovules during fiber initiation were cloned into the prey vector pGADT7 and transformed into yeast strains Y187 to construct AD library. Yeast strains Y2HGold harboring pGBKT7 vectors were then plated onto SD/-Trp medium supplemented with X-α-Gal to check for self-activation of GhWER. The Y2HGold strains without self-activation were selected and individually mated with the AD library, followed by incubation at 30℃ with sharking (180 rpm) for 22h. The resulting zygotes were restreaked on SD/-Leu/-Trp medium and SD/-Ade/-His/-Leu/-Trp medium supplemented with X-α-Gal, and then incubated at 30℃ for 3–5 days. AD plasmids from positive colonies were isolated and the cDNA inserts were sequenced to identify candidate proteins that interacted with GhWER.
To detect protein–protein interactions between GhWER and the candidate proteins, the prey vector pGADT7, fused with the CDS of GhDEL65 and GhbHLH65, were transformed into yeast strains Y187. Subsequently, Y2HGold strains carrying GhWER and the Y187 strains carrying GhDEL65 or GhbHLH65 were mated, separately. Finally, the mating yeasts were plated on SD/-Leu/-Trp medium and SD/-Ade/-His/-Leu/-Trp medium supplemented with X-α-Gal, and incubated at 30℃ for 3—5 days to verify the authenticity of protein–protein interaction. All the primers used for Y2H experiments can be found in Table S3.
RNA-seq
Flower buds of both the WT and three GhWER knockout lines were collected at -1.5 DPA, and the ovules were then stripped. Then, the outer integument of the ovules was peeled off in the RNAlater solution (Sigma-Aldrich, R0901) for total RNA extraction. Three biological replicates were established for each transgenic line. BGI was responsible for the cDNA library construction and eukaryote transcriptome sequencing. The raw data obtained from the sequencer was filtered, then the clean data was mapped to the G. hirsutum reference genome of TM-1 using the STAR software (Wang et al. 2019). Next, the FPKM value of each gene was calculated using RSEM software. The differentially expressed genes (DEGs) were identified using the edgeR software on the omicshare online platform (https://www.omicshare.com/tools) with the criteria of a fold change greater than 2 and a P value less than 0.05.
Dual-luciferase reporter assays (LUC)
The dual-luciferase reporter system was used to verify the upstream and downstream relationship between GhWER and hormone-related genes. We cloned the promoter fragments of hormone-related genes into the pGreenII 0800-LUC vector, and the CDS of GhWER into the pGreenII 62-SK vector, with the specific primers used for vector construction listed in Table S3. These constructed vectors were then transformed into Agrobacterium tumefaciens strain GV3101, with the empty vectors serving as a negative control. The A. tumefaciens strain harboring the pGreenII 0800-LUC and pGreenII 62-SK vector were activated, and then mixed at a 1:9 ratio. This mixed solution was subsequently injected into N. benthamiana leaves and cultivated at 25℃ for 48-72 h. After cultivation, the leaves were treated with luciferin (Promega, P1041) and observed using a whole-body fluorescent imaging system (Berthold, LB985 NightSHADE).
Results
GhWER shares high homology with WER
In our previous study, we identified a series of TFs that exhibited preferential expression during the initiation of fiber cell in G. hirsutum, including an R2R3-MYB TF GhWER (Ghir_A08G017600) (Hu et al. 2018). As a homologous gene of WER, GhWER appeared to potentially fulfill a similar function to WER in determining the fate of epidermal cells (Fig. 1A). GhWER was a R2R3-MYB TF coding a 230 aa protein. Sequence alignment indicated a conserved sequence of R2 and R3 domain shared by both WER and GhWER (Fig. 1A). In allotetraploid G. hirsutum, GhWER had one homologous copy (Ghir_D08G018460), which shared homology with GaMYB2 (Ga08G1971) in diploid Gossypium arboretum and Gorai_004G196800 in diploid Gossypium raimondii (Fig. 1A). Only 8 different amino acids were identified among these genes in different cotton species (Fig. 1A), further suggesting a relative conservation of GhWER during the evolution of cotton species.
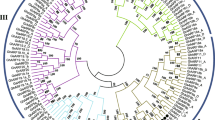
Sequence alignment and evolution analysis of MYB TFs in cotton and Arabidopsis. A. Amino acid sequence alignment of WER proteins in cotton and Arabidopsis. The black lines indicate the R2 motif and R3 motif of the R2R3-MYB domain. B. Gene structure and phylogenetic tree of R2R3-MYB TFs related to cotton fiber initiation or trichomes development of Arabidopsis
To further investigate the function of GhWER, we conducted an evolution analysis of GhWER and other R2R3 MYB TFs associated with fiber/trichome development in cotton and Arabidopsis (Fig. 1B). Gene structure analysis revealed variations in the UTR, intron, and exon regions of these MYB TFs (Fig. 1B). Phylogenetic analysis divided these R2R3 MYB TFs into two clades: the first clade, containing GhWER and GhMYB109 from cotton, as well as WER, GL1 and AtMYB23 from Arabidopsis, had 2—3 exons; while the second clade, comprising GhMYB25 and GhMYB25-like from cotton, as well as AtMYB106, AtMYB16 and AtMYB17 from Arabidopsis, contained 3—4 exons (Fig. 1B). The close phylogenetic relationship between GhWER, GhMYB109 in cotton, and WER in Arabidopsis (Fig. 1B) suggests that GhWER may play an essential role in the early stage of fiber development, similar to GhMYB109.
GhWER is preferentially expressed in ovules during fiber initiation
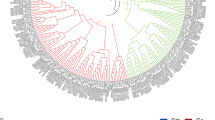
To further determine the biological function of GhWER in fiber development, we constructed a phylogenetic tree that included GhWER and its homologous genes in cotton. By aligning the amino acid sequence of WER, a total 48 R2R3-MYB TFs were identified in cotton. These MYB proteins were categorized into 5 subclasses based on their phylogenetic relationship, and their expression patterns were depicted on the phylogenetic tree. GhWER belonged to subclass II and exhibited preferential expression in 0—3 DPA ovules and 5—10 DPA fibers (Fig. 2A). Additionally, other genes in subclass II, such as Ghir_A05G037220 (GhMYB109), Ghir_A11G000100, and Ghir_A06G008810, also showed predominant expression during fiber initiation and the early stage of fiber elongation (Fig. 2A). These findings suggest that these genes may have functional redundancy with GhWER in controlling fiber initiation.
Expression pattern of GhWER in G. hirsutum. A. Phylogenetic relationship and expression pattern of GhWER and its homologous genes in G. hirsutum. GhWER is marked by the red box. 0 D, 1 D, 3 D represent the ovules at 0 DPA, 1 DPA, and 3 DPA, respectively; 5 DF, 10 DF, and 20 DF represent the fibers at 5 DPA, 10 DPA, and 20 DPA, respectively. B. Expression pattern in different tissues of GhWER in G. hirsutum ‘Jin668’, as detected by qRT-PCR. R, root; S, stem; L, leaf; -1.5 D, -1 D, -0.5 D, 0 D, 1 D, 3 D, 5 D represent ovules at -1.5 DPA, -1 DPA, -0.5 DPA, 0 DPA, 3 DPA, 5 DPA; and 5 DF, 10 DF, 15 DF, 20 DF represent fibers at 5 DPA, 10 DPA, 15 DPA, 20 DPA, respectively. GhUb7 was used as the reference gene
The expression of GhWER was detected in various tissues of G. hirsutum accession Jin668, including roots, stems, leaves, and different developmental stages of ovules and fibers. The qRT-PCR results showed that GhWER was expressed at -1.5 DPA in ovules and its expression peaked at -1 DPA before gradually decreasing (Fig. 2B). In addition, GhWER was expressed at a moderate level in roots, leaves and early development stages of fibers (5 to 10 DPA), and at a low level in stems and later development stage of fibers (15 to 20 DPA) (Fig. 2B). Analysis of the expression pattern of GhWER in the single-cell RNA-seq (scRNA) atlas of the outer integument of ovules showed that GhWER was preferentially expressed in outer pigment layer cell cluster and fiber cell cluster of ovules (Fig. S1)(Qin et al. 2022). This preferential expression of GhWER in the early developmental stages of ovule integument suggests that GhWER may be involved in fiber initiation.
The later initiation timing of fiber initials in GhWER gene editing line result in shorter fibers
To verify the gene function of GhWER in cotton fiber development, knockout lines of GhWER were generated in cotton using CRISPR/Cas9 technology. A vector containing two sgRNAs targeting to the R2R3-MYB domain of GhWER_At/Dt was transformed into the Jin668 receptor line, resulting in the generation of thirteen independent transgenic T0 plants. Three of these plants (CR-1, CR-38 and CR-44) were randomly chosen for this study. The genotypes of the target sites in the T2 plants of these three independent transgenic lines were detected using Hi-Tom, which revealed that the most abundant mutations observed were 1–3 insertions/deletions (Table S1). Additionally, the mutants CR-1–8-1 and CR-1–8-3 showed a 297 bp nucleotides insertion at target 2, which may be induced by DNA repair (Table S1).
These mutations caused frame shifts and premature termination of translation at different positions of GhWER in six plants from the three T2 knockout lines (Fig. 3A). Specifically, only one type of mutant protein, namely m1, was detected in each plant of knockout lines CR-1–8 and CR-44–7, with early termination of GhWER at 52aa (Fig. 3A). On the other hand, line CR-38–1 was found to be heterozygous, with mutant CR-38–1-8 possessing two types of mutations (m1 and m2) leading to early termination at 78aa, and mutant CR-38–1-9 having a different set of mutations (m2 and m3) resulting in early termination at 123aa (Fig. 3A). In conclusion, these three GhWER knockout lines were identified as loss-of-function mutant.
Knockout of GhWER in cotton causes delayed fiber initiation and elongation. A. Schematic presentation of GhWER and its mutant proteins (right). Types of GhWER mutant proteins in T2 transgenic plants (left). B. SEM images of 0 DPA and 1 DPA ovules in wild type (WT) and GhWER knockout lines. The magnification is 50 times and 500 times. C. Statistics of fiber initials in the middle part of 0 DPA ovules. * P < 0.05. D-F. Lint percentage (D), lint index (E), and seed index (F) of WT and GhWER knockout lines. Data are presented as mean ± SD; n = 15, * P < 0.05, ** P < 0.01. G. Phenotypes of seeds and mature fibers in WT and GhWER knockout lines. Bar = 1cm. H. Fiber length of mature fibers in WT and GhWER knockout lines. Data are presented as mean ± SD; n = 15, * P < 0.05, ** P < 0.01
To better understand the gene function of GhWER in fiber initiation, we observed the fiber initials on 0 DPA and 1 DPA ovules of WT and GhWER knockout lines using a scanning electron microscope (SEM). At a magnification of 500 times, clear observations of fiber initials were made on 0 DPA ovules in both the WT and the three GhWER knockout lines (Fig. 3B). Subsequently, the number of fiber initials was quantified to compare between the WT and the three GhWER knockout lines. It was found that, relative to the WT, the number of initiated fiber cells in the three GhWER knockout lines was slightly reduced, but not significantly at 0 DPA (Fig. 3B and 3C). However, on 1 DPA ovules, the length of fiber initials in the three GhWER knockout lines was shorter than that of the WT (Fig. 3B). These results indicate a slower initiation and early elongation of fiber cells in the absence of GhWER.
To confirm whether the delayed early fiber development of GhWER knockout lines affects the length of mature fiber, the yield and quality trait for GhWER knockout lines of T2 generation was investigated. The lint percentage of the three GhWER knockout lines was found to be 41.27 ± 0.30%, 39.66 ± 0.11%, and 39.08 ± 0.70%, respectively, which were significantly lower than that of the WT (43.05 ± 0.26%) (Fig. 3D). However, there was no significant difference in lint index between the WT and the three GhWER knockout lines (Fig. 3E). Furthermore, although the seed size did not show a significant difference between the WT and knockout lines (Fig. 3G), the seed index of the three GhWER knockout lines (9.03 ± 0.04 g, 9.04 ± 0.14 g, 8.87 ± 0.20 g) was significantly higher than that of the WT (8.00 ± 0.13 g) (Fig. 3F), suggesting that the decreased lint percentage in the knockout lines was due to the increased seed index. Therefore, there was no significant change in lint yield of the knockout lines compared with the WT. Additionally, the fiber quality traits were measured using a semi-automatic high volume cotton tester (PREMIER HFT), revealing no significant difference in the other fiber quality traits (micronaire value, strength, uniformity index, elongation) between the knockout lines and the WT (Table S2). However, the UHMI of the GhWER knockout lines (26.18 ± 0.24 mm for CR-1–8, 26.25 ± 0.10 mm for CR-38–1 and 25.75 ± 0.21 mm for CR-44–7) were significantly shorter than that of the WT (27.40 ± 0.07) (Table S2). The fiber length of the GhWER knockout lines (25.57 ± 0.57 mm for CR-1–8, 25.57 ± 0.44 mm for CR-38–1 and 25.20 ± 0.51 mm for CR-44–7) obtained from hand-combing was also significantly shorter than that of the WT (27.13 ± 0.43 mm) (Fig. 3G and 3H), which was consistent with the UHMI results. In conclusion, knocking out GhWER resulted in the delay of fiber cell initiation and early elongation, ultimately causing shorter mature fibers. This suggests that GhWER positively regulates fiber initiation and elongation.
GhWER interacts with bHLH TFs—GhDEL65 and GhbHLH121
To investigate the regulation mechanism of GhWER, a Y2H assay was employed to identify proteins that interact with GhWER. First, the self-activation of GhWER was examined. The pGBKT7 vector, which contained the CDS sequence of GhWER, was transformed into the yeast strain Y2H and grown on the SD/-Trp medium supplemented with X-α-Gal. The appearance of light blue colonies harboring GhWER indicated weak self-activation (Fig. 4A). To further analyze the self-activation, two N-terminal truncated and two C-terminal truncated GhWER proteins were assessed using the Blue-White Screening method. It was found that two N-terminal truncated GhWER proteins (residues 1–149 and 1–190) did not display self-activation, while two C-terminal truncated GhWER proteins (residues 150–230 and 191–230) exhibited evident self-activation (Fig. 4A).
GhWER interacts with GhDEL65 and GhbHLH121. A. The self-activation of GhWER in the yeast strain Y2HGold. The yeast strains Y2HGold were plated onto SD/-Trp (with X-α-Gal) medium. B. The interaction between GhWER and GhDEL65/GhbHLH121 was verified by Y2H assay. Yeast cells were plated on SD/–Leu/–Trp and SD/–Ade/–His/–Leu/–Trp (with X-α-Gal) media. C. The expression pattern of GhDEL65 (left) and GhbHLH121 (right) in different tissues of G. hirsutum. R, root; S, stem; L, leaf; Pe, petal; An, anther; St, stamen; 0 D, 1 D, 3 D, 10 D, 20 D represent ovules at 0 DPA, 1 DPA, 3 DPA, 10 DPA, 20 D DPA; and 5 DF, 10 DF, 15 DF, 20 DF represent fibers at 5 DPA, 10 DPA, 15 DPA, 20 DPA, respectively
Subsequently, the N-terminal truncated GhWER protein GhWERN2 (residues 1–190) was used as bait to screen for GhWER-interacting proteins in a Y2H library consisting of TFs related to fiber initiation. Two proteins, GhDEL65 (Ghir_D08G020010) and GhbHLH121 (Ghir_A03G022080), were identified to interact with GhWER, and both belong to the bHLH class of TFs. Furthermore, the interaction between GhWER and GhDEL65/GhbHLH121 was also confirmed through point-to-point verification (Fig. 4B). It has been reported that GhDEL65 shares functional similarity with GL3 or EGL3 in trichome development and positively regulated fiber elongation in cotton (Shangguan et al. 2016). Moreover, both GhDEL65 and GhbHLH121 exhibited relatively high expression level at 0—3 DPA ovules (Fig. 4C), indicating that GhWER and GhDEL65/GhbHLH121 may form a complex to regulate fiber initiation and early elongation.
Knocking out of GhWER in cotton affects hormone signaling pathway
Since GhWER plays an important role in fiber cell initiation and early elongation, RNA-seq was performed to identify the regulatory network of GhWER downstream genes in the outer integument of ovules at -1.5 DPA. A total of 241 DEGs were identified between the WT (Jin668) and three knockout lines (CR-38–1-8, CR-38–1-9, CR-44–7-2), with 112 genes being up-regulated and 129 genes being down-regulated in the knockout lines (Fig. S2A and S2B). To understand the regulatory pathways that are involved in these DEGs, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were performed. The GO analysis showed that these DEGs were mainly enriched in response to hormones (especially related to ethylene), response to organic substances, response to endogenous stimuli, and so on (Fig. 5A). The KEGG analysis indicated that these DEGs were mainly enriched in the pathway of plant hormone signal transduction and the MAPK signaling pathway (Fig. 5B). In conclusion, the DEGs were found to be mainly enriched in hormone-related pathways.
DEGs between WT and GhWER knockout lines are enriched in the hormone signaling pathway. A. Top 20 enriched GO terms derived from DEGs between WT and GhWER knockout lines. The size of dots represents the number of DEGs; the color of dots represents the significance (q value) of the GO terms. B. Top 20 enriched KEGG terms derived from DEGs between WT and GhWER knockout lines. The color of bars represents the significance (q value) of KEGG terms. C. Relative expression of DEGs using qRT-PCR in -1.5 DPA ovules of WT and three GhWER knockout lines. Error bars represent ± SD. GhUb7 was used as the reference gene
Further, we focused on the expression of hormone-related DEGs (Fig. S2C), in light of previous studies that have emphasized the crucial role of hormones in fiber development, such as ethylene (Shi et al. 2006), auxin (Zhang et al. 2011) and GA (Xiao et al. 2010). In comparison to the WT, the GhWER knockout lines showed a down-regulation of ethylene synthase gene ACS1 (Ghir_D12G024500), ethylene sensors RAV1 (Ghir_A12G018050), ETR2 (Ghir_D02G004190), and ethylene responsive factor ERF017 (Ghir_D06G008570), as well as the auxin-responsive gene SAUR50 (Ghir_A05G026080), and Gibberellin-regulated genes SN2 (Ghir_A09G017900) and GASA14 (Ghir_D07G023560) (Fig. S2D). These findings were further validated in -1.5 DPA ovules through qRT-PCR (Fig. 5C) and the expression pattern indicated that these hormone-related genes were predominantly expressed during the early development stages of ovules (0 -3 DPA) (Fig. S2D). This suggests that GhWER may facilitate fiber cell initiation and early elongation by regulating the signal transduction of ethylene, auxin and GA.
Moreover, other TFs preferentially expressed during early development stages, such as the ethylene response factor ERF017 (Ghir_D06G008570) and stimulus–response related TFs WRKY70 (Ghir_A06G019420), WRKY33 (Ghir_A04G010400) and bHLH162 (Ghir_A12G014970), were found to be down-regulated in GhWER knockout lines. In contrast, the TFs bHLH094 (Ghir_D02G017450) and MYB21 (Ghir_A12G015320) were up-regulated in GhWER knockout lines, indicating their potential involvement in fiber cell initiation (Fig. S2C and S2D, Fig. 5C).
GhWER regulates the expression of various genes in ethylene signaling pathway
Based on the results of GO and KEGG analysis, which indicated that DEGs between WT and GhWER knockout lines were primarily enriched in hormone response (Fig. 5A), we investigated the relationship between GhWER and these hormone-related DEGs. To do so, we predicted the cis-elements binding sites on the 2 Kb DNA sequence upstream of the start codon of hormone-related genes, including ethylene synthase gene ACS1 and ethylene sensors ETR2, using the PlantRegMap website (http://plantregmap.gao-lab.org). Our findings revealed at least one MYB cis-elements on their promoter (Fig. 6A), suggesting the potential for GhWER to bind the promoter of ACS1 and ETR2.
GhWER protein activates transcription of the ethylene signaling pathway genes ACS1 and ETR2. A. Description of MYB related cis-elements on the promoter of ACS1 and ETR2. The black solid lines represent the promoters; the black rectangle represents the gene body; the blue ovals on the promoter represent cis-elements, and the grey solid line represents the promoters used for the LUC assay. B. Schematic diagram of reporters and effectors. C-D. Analysis of GhWER activation on the promoter of ACS1 (C) and ETR2 (D) using LUC assay. The reporters and effectors were injected into tobacco leaves. Bar = 2 cm
To verify whether GhWER is capable of activating the transcriptional activation of ACS1 and ETR2, we conducted the LUC assay in tobacco leaves. GhWER was fused with 35S promoter to act as the effector, while the promoter fragments of ACS1 and ETR2, which contained MYB cis-elements, drove the expression of luciferase as the reporter (Fig. 6B). The empty vectors were used as negative controls (Fig. 6B). The tobacco leaves co-transformed with the effector (GhWER) and reporter (proACS1 or proETR2) displayed increased fluorescence intensity compared to the negative control (Fig. 6C and 6D), suggesting that GhWER directly activates the transcriptional activation of ACS1 and ETR2. In conclusion, we inferred that GhWER might regulate fiber initiation and early elongation by directly controlling the expression of genes related to ethylene signaling transduction. The down-regulation of the ethylene pathway in GhWER knockout lines could potentially impede the initiation and early development of fibers.
Discussion
Our research dedicated significant effort to investigate the function of GhWER, which was primarily expressed in fiber initiation and early elongation (Fig. 2B). Gene sequence alignment and phylogenetic analysis revealed that GhWER shared the highest homology with WER of the MBW complex in Arabidopsis (Fig. 1A), suggesting its role in determining the cell fate of ovule epidermal cells in cotton. The MBW complex is critical for multiple biological functions, such as trichome development (regulated by GL1-GL3/EGL3-TTGl), root hair development (controlled by WER-GL3/EGL3-TTG1), anthocyanin biosynthesis (governed by PAP1/PAP2-TT8/GL3/EGL3-TTG1), and seed-coat mucilage production (regulated by MYB61-TT8/EGL3-TTG1) in Arabidopsis (Ramsay and Glover 2005; Lepiniec et al. 2006; Xu et al. 2015). The bHLH and WD40 components within these MBW complexes exhibit a high degree of conservation, while their diverse functionalities are governed by distinct MYB TFs. Furthermore, although reports on MBW complexes may be lacking across different species, various MYB genes have been confirmed to participate in the formation of epidermal hairs and anthocyanin biosynthesis. For example, AaMIXTA1 regulated glandular trichomes initiation in Artemisia annua (Shi et al. 2018), CsMYB6 regulated fruit trichome initiation in cucumber (Cucumis sativus), and two PAP2 homologous genes, BnaPAP2.C6a and BnaPAP2.A7b, controlled stem and flower color in oilseed rape (Brassica napus) (Chen et al. 2023). In soybean, research on seed color identified 13 QTLs (quantitative trait loci), with MYB TFs emerging as crucial candidate genes (Song et al. 2023). All these studies collectively demonstrate the significant role of MYB TFs in plant development.
To date, the molecular mechanism of cotton fiber initiation has been well-documented. The MYB subgroup 9 is a specific clade for Malvaceae, with its members playing a crucial role in fiber initiation (Paterson et al. 2012; Zhang et al. 2015). Among these members, GhMYB25-like has been identified as playing the most important role in fiber initiation, as its absence resulted in the failure of fiber initiation (Walford et al. 2011; Qin et al. 2022). Additionally, GhMYB25, its homologous gene, has been found to promote the formation of fiber initials (Machado et al. 2009). In contrast, GhWER belong to MYB subgroup 15, which is distinct from MYB subgroup 9. In this study, GhWER knockout lines showed a delay in the early elongation of fiber initials and shorter mature fiber length compared to WT (Fig. 2). Similar expression patterns and sequence similarities between GhMYB109 and GhWER (Fig. 1B and Fig. 2A) suggest a shared function, as suppression of GhMYB109 mirrored the phenotypic effects seen in GhWER knockout lines (Suo et al. 2003). However, the functional parallels between GhWER/GhMYB109 and WER/GL1 from Arabidopsis in fiber initiation are less pronounced than their roles in Arabidopsis trichome/root hair development. Based on the above findings, it can be inferred that GhWER is not directly responsible for fiber initiation, in contrast to its involvement in trichome development in Arabidopsis.
Through Y2H library screening, we found that two bHLH TF, GhDEL65 and GhbHLH121, interacted with the R2R3 MYB TF GhWER (Fig. 3). This interaction mirrored the dynamics observed between bHLH TF GL3/EGL3 and MYB TF GL1/WER in Arabidopsis during the formation of trichomes and root hairs (Payne et al. 2000; Bernhardt et al. 2003). It has been noted that in Arabidopsis, the MBW complex consisting of homodimers or heterodimers of GL3/EGL3 acts as a bridge linking MYB TF GL1/WER and the WD40 protein TTG1 (Payne et al. 2000). Moreover, similar to GL3/EGL3’s role in Arabidopsis, GhDEL65 appears to influence trichome development (Shangguan et al. 2016). However, unlike the trichome development in Arabidopsis, researches reveal no evidence of a functional WD-repeat protein equivalent to TTG1 in cotton, leading us to infer that GhWER may not form a typical MBW complex during the early stage of fiber development.
Plant hormones play a crucial role in regulating various aspects of plant growth and development, including fiber initiation in cotton and trichome development in Arabidopsis. In Arabidopsis, the hormones GA, CTK and jasmonic acid (JA) synergistically control trichome development. Two modes of action of plant hormones were exhibited in Arabidopsis trichome development. One mode of action involves the integration of GA and CTK signaling, which activate the expression of C2H2 zinc finger proteins (GIS, GIS3, ZFP5 and ZFP6). These proteins then act on the MBW complex to regulate trichome development in the inflorescence organs of Arabidopsis (Gan et al. 2007b, 2007a; Zhou et al. 2013; Sun et al. 2015). Another mode of action involves JA and GA relieving the inhibition of JAZs and DELLAs to the MBW complex by inducing the promoting degradation of JAZs and DELLAs (Qi et al. 2014, 2011). In both modes of action, the effective functioning of hormone signals relies on their interaction with the MBW complex. However, in this study, RNA-seq analysis of the outer integument of ovule between WT and GhWER knockout lines revealed downregulation of hormone signaling components in GhWER knockout lines, especially in the ethylene, auxin and GA signaling pathway (Fig. 5). The LUC assay demonstrated that GhWER acted as the direct upstream to regulate the expression of components of hormone signaling (Fig. 6). These results indicate that GhWER operates at different levels within the regulatory pathway of fiber initiation and epidermal development.
Data availability
The RNA-seq data have been deposited in the NCBI SRA database (https://www.ncbi.nlm.nih.gov/bioproject/) with BioProject number PRJNA1064389. For any other data inquiries, interested parties can contact the corresponding authors and request access.
References
Applequist W, Cronn R, Wendel J (2001) Comparative development of fiber in wild and cultivated cotton. Evol Dev 3(1):3–17. https://doi.org/10.1046/j.1525-142X.2001.00079.x
Bernhardt C, Lee MM, Gonzalez A, Zhang F, Lloyd A, Schiefelbein J (2003) The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130(26):6431–6439. https://doi.org/10.1242/dev.00880
Chen D, Jin Q, Pan J, Liu Y, Tang Y, E Y, Xu L, Yang T, Qiu J, Chen X, Wang J, Gong D, Ge X, Li Z, Cui C (2023) Fine mapping of genes controlling pigment accumulation in oilseed rape (Brassica napus L.). Mol Breed 43(3). https://doi.org/10.1007/s11032-023-01365-5
Gan Y, Liu C, Yu H, Broun P (2007a) Integration of cytokinin and gibberellin signalling by Arabidopsis transcription factors GIS, ZFP8 and GIS2 in the regulation of epidermal cell fate. Development 134(11):2073–2081. https://doi.org/10.1242/dev.005017
Gan Y, Yu H, Peng J, Broun P (2007b) Genetic and molecular regulation by DELLA proteins of trichome development in Arabidopsis. Plant Physiol 145(3):1031–1042. https://doi.org/10.1104/pp.107.104794
Guan X, Pang M, Nah G, Shi X, Ye W, Stelly DM, Chen ZJ (2014) miR828 and miR858 regulate homoeologous MYB2 gene functions in Arabidopsis trichome and cotton fibre development. Nat Commun 5:3050. https://doi.org/10.1038/ncomms4050
Hao J, Tu L, Hu H, Tan J, Deng F, Tang W, Nie Y, Zhang X (2012) GbTCP, a cotton TCP transcription factor, confers fibre elongation and root hair development by a complex regulating system. J Exp Bot 63(17):6267–6281. https://doi.org/10.1093/jxb/ers278
Hu H, Wang M, Ding Y, Zhu S, Zhao G, Tu L, Zhang X (2018) Transcriptomic repertoires depict the initiation of lint and fuzz fibres in cotton (Gossypium hirsutum L.). Plant Biotechnol J 16(5):1002–1012. https://doi.org/10.1111/pbi.12844
Huang Y, Liu X, Tang K, Zuo K (2013) Functional analysis of the seed coat-specific gene GbMYB2 from cotton. Plant Physiol Biochem 73:16–22. https://doi.org/10.1016/j.plaphy.2013.08.004
Jin S, Zhang X, Nie Y, Guo X, Liang S, Zhu H (2006a) Identification of a novel elite genotype for in vitro culture and genetic transformation of cotton. Biol Plantarum 50(4):519–524. https://doi.org/10.1007/s10535-006-0082-5
Jin SX, Liang SG, Zhang XL, Nie YC, Guo XP (2006b) An efficient grafting system for transgenic plant recovery in cotton (Gossypium hirsutum L.). Plant Cell Tiss Org 85(2):181–185. https://doi.org/10.1007/s11240-005-9068-9
Lee MM, Schiefelbein J (1999) WEREWOLF, a MYB-related protein in arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99(5):473–483. https://doi.org/10.1016/S0092-8674(00)81536-6
Lee JJ, Woodward AW, Chen ZJ (2007) Gene Expression Changes and Early Events in Cotton Fibre Development. Ann Bot 100(7):1391–1401. https://doi.org/10.1093/aob/mcm232
Lepiniec L, Debeaujon I, Routaboul J-M, Baudry A, Pourcel L, Nesi N, Caboche M (2006) Genetics and Biochemistry of Seed Flavonoids. Annu Rev Plant Biol 57(1):405–430. https://doi.org/10.1146/annurev.arplant.57.032905.105252
Liu Q, Wang C, Jiao XZ, Zhang HW, Song LL, Li YX, Gao CX, Wang KJ (2019) Hi-TOM: a platform for high-throughput tracking of mutations induced by CRISPR/Cas systems. Sci China Life Sci 62(1):1–7. https://doi.org/10.1007/s11427-018-9402-9
Machado A, Wu Y, Yang Y, Llewellyn DJ, Dennis ES (2009) The MYB transcription factor GhMYB25 regulates early fibre and trichome development. Plant J 59(1):52–62. https://doi.org/10.1111/j.1365-313X.2009.03847.x
Naoumkina M, Zeng L, Fang DD, Wang M, Thyssen GN, Florane CB, Li P, Delhom CD (2020) Mapping and validation of a fiber length QTL on chromosome D11 using two independent F2 populations of upland cotton. Molecular Breeding 40(3). https://doi.org/10.1007/s11032-020-01111-1
Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD (1991) A Myb Gene Required for Leaf Trichome Differentiation in Arabidopsis Is Expressed in Stipules. Cell 67(3):483–493. https://doi.org/10.1016/0092-8674(91)90523-2
Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, Jin D, Llewellyn D, Showmaker KC, Shu S, Udall J, Yoo MJ, Byers R, Chen W, Doron-Faigenboim A, Duke MV, Gong L, Grimwood J, Grover C, Grupp K, Hu G, Lee TH, Li J, Lin L, Liu T, Marler BS, Page JT, Roberts AW, Romanel E, Sanders WS, Szadkowski E, Tan X, Tang H, Xu C, Wang J, Wang Z, Zhang D, Zhang L, Ashrafi H, Bedon F, Bowers JE, Brubaker CL, Chee PW, Das S, Gingle AR, Haigler CH, Harker D, Hoffmann LV, Hovav R, Jones DC, Lemke C, Mansoor S, Ur Rahman M, Rainville LN, Rambani A, Reddy UK, Rong JK, Saranga Y, Scheffler BE, Scheffler JA, Stelly DM, Triplett BA, Van Deynze A, Vaslin MF, Waghmare VN, Walford SA, Wright RJ, Zaki EA, Zhang T, Dennis ES, Mayer KF, Peterson DG, Rokhsar DS, Wang X, Schmutz J (2012) Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492(7429):423–427. https://doi.org/10.1038/nature11798
Payne C, Zhang F, Lloyd A (2000) GL3 Encodes a bHLH Protein That Regulates Trichome Development in Arabidopsis Through Interaction With GL1 and TTG1. Genetics 156:1349–1362
Pu L, Li Q, Fan X, Yang W, Xue Y (2008) The R2R3 MYB transcription factor GhMYB109 is required for cotton fiber development. Genetics 180(2):811–820. https://doi.org/10.1534/genetics.108.093070
Qi TC, Song SS, Ren QC, Wu DW, Huang H, Chen Y, Fan M, Peng W, Ren CM, Xie DX (2011) The Jasmonate-ZIM-Domain Proteins Interact with the WD-Repeat/bHLH/MYB Complexes to Regulate Jasmonate-Mediated Anthocyanin Accumulation and Trichome Initiation in Arabidopsis thaliana. Plant Cell 23(5):1795–1814. https://doi.org/10.1105/tpc.111.083261
Qi T, Huang H, Wu D, Yan J, Qi Y, Song S, Xie D (2014) Arabidopsis DELLA and JAZ proteins bind the WD-repeat/bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy. Plant Cell 26(3):1118–1133. https://doi.org/10.1105/tpc.113.121731
Qin Y, Sun M, Li W, Xu M, Shao L, Liu Y, Zhao G, Liu Z, Xu Z, You J, Ye Z, Xu J, Yang X, Wang M, Lindsey K, Zhang X, Tu L (2022) Single-cell RNA-seq reveals fate determination control of an individual fibre cell initiation in cotton (Gossypium hirsutum). Plant Biotechnol J 20(12):2372–2388. https://doi.org/10.1111/pbi.13918
Ramsay NA, Glover BJ (2005) MYB–bHLH–WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci 10(2):63–70. https://doi.org/10.1016/j.tplants.2004.12.011
Rerie WG, Feldmann KA, Marks MD (1994) The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev 8(12):1388–1399. https://doi.org/10.1101/gad.8.12.1388
Shangguan XX, Yang CQ, Zhang XF, Wang LJ (2016) Functional characterization of a basic helix-loop-helix (bHLH) transcription factor GhDEL65 from cotton (Gossypium hirsutum). Physiol Plant 158(2):200–212. https://doi.org/10.1111/ppl.12450
Shi YH, Zhu SW, Mao XZ, Feng JX, Qin YM, Zhang L, Cheng J, Wei LP, Wang ZY, Zhu YX (2006) Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 18(3):651–664. https://doi.org/10.1105/tpc.105.040303
Shi P, Fu X, Shen Q, Liu M, Pan Q, Tang Y, Jiang W, Lv Z, Yan T, Ma Y, Chen M, Hao X, Liu P, Li L, Sun X, Tang K (2018) The roles of AaMIXTA1 in regulating the initiation of glandular trichomes and cuticle biosynthesis in Artemisia annua. New Phytol 217(1):261–276. https://doi.org/10.1111/nph.14789
Song J, Xu R, Guo Q, Wu C, Li Y, Wang X, Wang J, Qiu L-J (2023) An omics strategy increasingly improves the discovery of genetic loci and genes for seed-coat color formation in soybean. Mol Breed 43 (9). https://doi.org/10.1007/s11032-023-01414-z
Stewart J (1975) Fiber initiation on the cotton ovule (Gossypium hirsutum). Am J Bot 62(7):723–730. https://doi.org/10.1002/j.1537-2197.1975.tb14105.x
Sun LL, Zhang AD, Zhou ZJ, Zhao YQ, Yan A, Bao SJ, Yu H, Gan YB (2015) GLABROUS INFLORESCENCE STEMS3 (GIS3) regulates trichome initiation and development in Arabidopsis. New Phytol 206(1):220–230. https://doi.org/10.1111/nph.13218
Suo J, Liang X, Pu L, Zhang Y, Xue Y (2003) Identification of GhMYB109 encoding a R2R3 MYB transcription factor that expressed specifically in fiber initials and elongating fibers of cotton (Gossypium hirsutum L.). Biochim Biophys Acta 1630(1):25–34. https://doi.org/10.1016/j.bbaexp.2003.08.009
Walford SA, Wu Y, Llewellyn DJ, Dennis ES (2011) GhMYB25-like: a key factor in early cotton fibre development. Plant J 65(5):785–797. https://doi.org/10.1111/j.1365-313X.2010.04464.x
Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC (1999) The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11(7):1337–1349. https://doi.org/10.1105/tpc.11.7.1337
Wang S, Wang JW, Yu N, Li CH, Luo B, Gou JY, Wang LJ, Chen XY (2004) Control of plant trichome development by a cotton fiber MYB gene. Plant Cell 16(9):2323–2334. https://doi.org/10.1105/tpc.104.024844
Wang P, Zhang J, Sun L, Ma Y, Xu J, Liang S, Deng J, Tan J, Zhang Q, Tu L, Daniell H, Jin S, Zhang X (2018) High efficient multisites genome editing in allotetraploid cotton (Gossypium hirsutum) using CRISPR/Cas9 system. Plant Biotechnol J 16(1):137–150. https://doi.org/10.1111/pbi.12755
Wang M, Tu L, Yuan D, Zhu SC, Li J, Liu F, Pei L, Wang P, Zhao G, Ye Z, Huang H, Yan F, Ma Y, Zhang L, Liu M, You J, Yang Y, Liu Z, Huang F, Li B, Qiu P, Zhang Q, Zhu L, Jin S, Yang X, Min L, Li G, Chen LL, Zheng H, Lindsey K, Lin Z, Udall JA, Zhang X (2019) Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat Genet 51(2):224–229. https://doi.org/10.1038/s41588-018-0282-x
Xiao YH, Li DM, Yin MH, Li XB, Zhang M, Wang YJ, Dong J, Zhao J, Luo M, Luo XY, Hou L, Hu L, Pei Y (2010) Gibberellin 20-oxidase promotes initiation and elongation of cotton fibers by regulating gibberellin synthesis. J Plant Physiol 167(10):829–837. https://doi.org/10.1016/j.jplph.2010.01.003
Xiao G, He P, Zhao P, Liu H, Zhang L, Pang C, Yu J (2018) Genome-wide identification of the GhARF gene family reveals that GhARF2 and GhARF18 are involved in cotton fibre cell initiation. J Exp Bot 69(18):4323–4337. https://doi.org/10.1093/jxb/ery219
Xu W, Dubos C, Lepiniec L (2015) Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci 20(3):176–185. https://doi.org/10.1016/j.tplants.2014.12.001
Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130(20):4859–4869. https://doi.org/10.1242/dev.00681
Zhang M, Zheng X, Song S, Zeng Q, Hou L, Li D, Zhao J, Wei Y, Li X, Luo M, Xiao Y, Luo X, Zhang J, Xiang C, Pei Y (2011) Spatiotemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fiber yield and quality. Nat Biotechnol 29(5):453–458. https://doi.org/10.1038/nbt.1843
Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, Zhang J, Saski CA, Scheffler BE, Stelly DM, Hulse-Kemp AM, Wan Q, Liu B, Liu C, Wang S, Pan M, Wang Y, Wang D, Ye W, Chang L, Zhang W, Song Q, Kirkbride RC, Chen X, Dennis E, Llewellyn DJ, Peterson DG, Thaxton P, Jones DC, Wang Q, Xu X, Zhang H, Wu H, Zhou L, Mei G, Chen S, Tian Y, Xiang D, Li X, Ding J, Zuo Q, Tao L, Liu Y, Li J, Lin Y, Hui Y, Cao Z, Cai C, Zhu X, Jiang Z, Zhou B, Guo W, Li R, Chen ZJ (2015) Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol 33(5):531–537. https://doi.org/10.1038/nbt.3207
Zhang M, Zeng JY, Long H, Xiao YH, Yan XY, Pei Y (2017) Auxin Regulates Cotton Fiber Initiation via GhPIN-Mediated Auxin Transport. Plant Cell Physiol 58(2):385–397. https://doi.org/10.1093/pcp/pcw203
Zhao J, Bai W, Zeng Q, Song S, Zhang M, Li X, Hou L, Xiao Y, Luo M, Li D, Luo X, Pei Y (2015) Moderately enhancing cytokinin level by down-regulation of GhCKX expression in cotton concurrently increases fiber and seed yield. Mol Breed 35(2). https://doi.org/10.1007/s11032-015-0232-6
Zhou Z, Sun L, Zhao Y, An L, Yan A, Meng X, Gan Y (2013) Zinc Finger Protein 6 (ZFP6) regulates trichome initiation by integrating gibberellin and cytokinin signaling in Arabidopsis thaliana. New Phytol 198(3):699–708. https://doi.org/10.1111/nph.12211
Zhu L, Jiang B, Zhu J, Xiao G (2022) Auxin promotes fiber elongation by enhancing gibberellic acid biosynthesis in cotton. Plant Biotechnol J 20(3):423–425. https://doi.org/10.1111/pbi.13771
Funding
This work was supported by grants from the National Natural Science Foundation of China (projects No. 32172071 to lilitu) and National Key R&D Program of China (projects No. 2022YFF1001400 to lilitu).
Author information
Authors and Affiliations
Contributions
L.T. conceived the project and designed the experiments. G.Z., W.L. contributed to the vector construction. W.L., L.S., M.X., M.S. and G.Z. performed the transgenic experiments and phenotype investigation. W.L., L.S., M.X. and G.Z. performed the RNA-seq and qRT-PCR. G.Z. performed the LUC assay. G.Z. and L.T. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, G., Li, W., Xu, M. et al. GhWER controls fiber initiation and early elongation by regulating ethylene signaling pathway in cotton (Gossypium hirsutum). Mol Breeding 44, 38 (2024). https://doi.org/10.1007/s11032-024-01477-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-024-01477-6