Abstract
WNK kinases are a unique class of serine/threonine protein kinases that lack a conserved catalytic lysine residue in the kinase domain, hence the name WNK (with no K, i.e., lysine). WNK kinases are involved in various physiological processes in plants, such as circadian rhythm, flowering time, and stress responses. In this study, we identified 26 WNK genes in soybean and analyzed their phylogenetic relationships, gene structures, chromosomal distribution, cis-regulatory elements, expression patterns, and conserved protein motifs. The soybean WNK genes were unevenly distributed on 15 chromosomes and underwent 21 segmental duplication events during evolution. We detected 14 types of cis-regulatory elements in the promoters of the WNK genes, indicating their potential involvement in different signaling pathways. The transcriptome database revealed tissue-specific and salt stress-responsive expression of WNK genes in soybean, the second of which was confirmed by salt treatments and qRT-PCR analysis. We found that most WNK genes were significantly up-regulated by salt stress within 3 h in both roots and leaves, except for WNK5, which showed a distinct expression pattern. Our findings provide valuable insights into the molecular characteristics and evolutionary history of the soybean WNK gene family and lay a foundation for further analysis of WNK gene functions in soybean.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Protein kinases play pivotal roles in catalyzing protein phosphorylation, which often serves as a regulatory switch. Protein kinases facilitate the transfer of the γ-phosphate group of adenosine triphosphate (ATP) to other proteins at specific residues, including serine, threonine, and tyrosine, and this phosphorylation of substrate proteins alters protein function and modulates cellular processes (Hanks and Hunter 1995; Kumar et al. 2020).
A unique feature of WNK [with no lysine (K)] subfamily is marked by the absence of catalytic lysine (K) residue in kinase subdomain II, which is essential for the coordination of ATP in the active center and conserved among all other kinases. In most kinases, this lysine residue is critical for maintaining protein kinase activity, as it interacts with the α- and β-phosphate groups of ATP, facilitating the binding of ATP for substrate phosphorylation and kinase activation (Nakamichi et al. 2002). In animal WNK kinases, this conserved lysine residue is replaced by cysteine or asparagine, while in plant WNK kinases, it is substituted with a asparagine/serine/glycine residue (Manuka et al. 2015). Despite these amino acid substitutions, WNK kinases are functional and are known to regulate plant growth, hormone responses, and adaptations to biotic and abiotic stressors (Manuka et al. 2019; Wang et al. 2008). This distinctive sequence sets WNK kinases apart as a unique class of protein kinases (Veríssimo and Jordan 2001; Xu et al. 2000).
In plants, the WNK kinase family includes members involved in diverse physiological processes (Kahle et al. 2006; Uchida et al. 2014). In the model plant Arabidopsis thaliana, eleven genes encoding WNK kinases have been identified, with extensive research focused on elucidating the functions of AtWNK1 and AtWNK8 (Huang et al. 2007; Wang et al. 2008). AtWNK1 phosphorylates the clock component APRR3 in vitro and actively participates in regulation of the circadian rhythm (Murakami-Kojima et al. 2002). AtWNK1, AtWNK2, AtWNK4, and AtWNK6 exhibit overlapping functions and contribute to the regulation of the circadian rhythm (Nakamichi et al. 2002). Notably, AtWNK8 physically interacts with Arabidopsis Enhanced Downy Mildew 2 (EDM2), a key regulator of flowering time that modulates the expression of Flowering Locus C (FLC, At5g10140) (Tsuchiya and Eulgem 2010). AtWNK8 demonstrates both autophosphorylation and phosphorylation of multiple sites within the vacuolar H+-ATPase C subunit, thereby participating in the regulation of ion transport in plants (Hong-Hermesdorf et al. 2006). AtWNK8 can also phosphorylate RACK1 to negatively influence stability and function of the RACK1 protein (Urano et al. 2015). In addition to these well-studied WNK genes, AtWNK9 has a positive regulatory role in abscisic acid (ABA) signal transduction and enhances drought tolerance in transgenic Arabidopsis plants (Xie et al. 2014). Collectively, these discoveries highlight the multifaceted roles played by distinct members of the WNK kinase family, shedding light on their diverse functions within plant systems.
Soybean, a globally significant crop cultivated for human and livestock diets, encounters considerable challenges in the field, including its vulnerability to salt stress. Salt stress impedes plant growth and diminishes crop productivity; therefore, it is necessary to thoroughly understand the mechanisms underlying the plant response to salt stress (Papiernik et al. 2005). Members of the WNK gene family are known to be involved in plant responses to stress, including ionic stresses. For instance, a substantial shift in the expression levels of rice WNK genes was observed when subjected to potassium (K +) deficiency (Ma et al. 2012). The nine WNK members in rice (OsWNKs) showed distinct transcript regulation patterns in various tissues as well as in response to diverse abiotic stresses such as drought, salt, heat, and cold. The continuous overexpression of rice OsWNK9 in Arabidopsis increased the tolerance to both salt and drought stress. This enhanced resilience was attributed to an ABA-dependent pathway (Manuka et al. 2019; Manuka et al. 2018).
These and other studies prompted us to identify the WNK genes in soybean. A homology-based alignment against the soybean reference genome identified 26 WNK genes within the soybean genome. Subsequently, we conducted a comprehensive analysis encompassing the physicochemical properties, conserved motifs, gene structures, phylogenetic relationships, and expression patterns of the soybean WNK gene family under salt stress conditions. Our primary objective was to identify the specific WNK genes involved in the soybean response to salt stress. The identification of these genes is valuable genetic resources for the cloning of salt tolerance genes and for future molecular breeding strategies in soybean.
Materials and methods
Plant materials and growth conditions
The soybean variety “Williams 82” was cultivated in a greenhouse maintained at a temperature of 25 °C, with a relative humidity of 70% and a photoperiod of 16 h of light followed by 8 h of darkness. Clean soybean seeds were germinated in vermiculite for a period of 5 days, after which they were transferred to culture tanks containing Hoagland nutrient solution. The plants were grown in these tanks until they reached the V1 developmental stage. To induce stress, 200 mM NaCl was added, while the control group remained in a NaCl-free nutrient solution. Leaves and roots were harvested at 0, 3, 12, and 24 h after the salt stress treatment was initiated. They were wrapped in aluminum foil, submerged in liquid nitrogen, and preserved in a refrigerator at − 80 °C. RNA samples were isolated from young leaves and roots in three biological replicates. Throughout the entire cultivation process, continuous aeration was maintained to ensure adequate oxygen supply to the solution and facilitate root respiration.
Identification and annotation of WNK genes in soybean
To acquire the WNK protein sequences in Glycine max (Wm82.a2.v1), the BLASTP algorithm on the Phytozome 13 website was used with the Arabidopsis thaliana WNK protein sequences as reference templates. Stringent criteria were set, requiring a minimum sequence homology of 50% and an E-value ≤ 1.0e−20. To expand the dataset, WNK sequences from Arabidopsis (TAIR 10), rice (Oryza sativa v7.0), and maize (Zea mays RefGen_V4) were collected from various plant genome databases, including TAIR (http://www.arabidopsis.org/), NCBI (www.ncbi.nlm.nih.gov/), and Phytozome (http://www.phytozome.net/). All gene names are listed in Table S1. All retrieved WNK sequences underwent alignment using MEGA X, and candidate sequences lacking complete homologous and isoform-specific domains were eliminated. The protein structures were analyzed using online tools such as Pfam (http://pfam.xfam.org/), NCBI CDD (https://www.ncbi.nlm.nih.gov/cdd/), and SMART (http://smart.embl.de/). These tools facilitated the identification of sequences with characteristic domains associated with WNK proteins and aided in filtering out incomplete or divergent sequences.
Prediction of physicochemical properties of soybean WNK proteins
To obtain data such as the number of amino acids, molecular weight, isoelectric point, and average hydrophobicity index of the WNK proteins in soybean, a physicochemical property analysis of members of the WNK protein gene family was conducted using Prot Param in the online software ExPASy (https://web.expasy.org/protparam/). TBtools (https://github.com/CJ-Chen/TBtools/releases) was utilized for gene structure analysis to predict the positions and number of exons and introns of each WNK gene (Chen et al. 2020). The WNK protein subcellular localization was predicted by WoLF PSORT (https://psort.hgc.jp/).
Phylogenetic analysis of the WNK genes in soybean
Systematic phylogenetic analysis of the soybean WNK genes was performed to investigate their evolutionary relationships. Multiple sequence alignment of the soybean WNK protein sequences was conducted using the Clustal W algorithm (Thompson et al. 1994). The aligned sequences were then subjected to phylogenetic analysis using the maximum likelihood method implemented in MEGA X software (Liu et al. 2022a). The reliability of the phylogenetic tree was assessed by bootstrap analysis with 1000 replicates.
Gene structure and conserved motifs of WNK genes
Gene structure diagrams of the 26 members of the soybean WNK gene family were constructed using TBtools software based on the soybean gene feature format (GFF) file. Furthermore, the protein sequences of the WNK genes were extracted, and the conserved motifs were predicted and analyzed using the MEME online tool. The maximum number of motifs was set to 10, with other parameters set to their default values. The resulting motif information, including sequence logos and consensus sequences, was obtained. TBtools was used to visualize the MEME results (Chen et al. 2020).
Chromosomal localization and synteny analysis
Phytozome was used to locate the physical positions of the 26 genes on their chromosomes. The chromosomal locations of the gene family members were visualized by importing the genome and gene position files into TBtools. The gene positions were plotted using the chromosome distribution module in TBtools (Pei et al. 2021). Gene duplication events for the GmWNK genes were analyzed by the One Step MCScanX plugin in TBtools.
Analysis of cis-acting regulatory elements
The upstream sequences (2 Kb) of each WNK gene were obtained from the genome database for promoter analysis. The PlantCARE program (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was employed to analyze the retrieved sequences and identify putative cis-regulatory elements. The identified cis-regulatory elements were visualized using TBtools, providing a comprehensive view of the types of elements and their distribution within the upstream regions of the WNK genes (Chen et al. 2020).
Expression analysis of WNK genes in different tissues
The expression data for the soybean WNK genes were obtained from various tissues, including flowers, roots, stems, seeds, shoot apical meristems, and pods. This data was sourced from the Phytozome 13 database (https://data.jgi.doe.gov/refine-download/phytozome?organism=Gmax&expanded=508). The FPKM values of the WNK genes of soybean were used to develop a heat map using TBtools (Chen et al. 2020).
Analysis of WNK gene expression in soybean under salt stress
The FastPure Universal Plant Total RNA Isolation kit (Vazyme) was employed for RNA extraction. HiScript III qRT SuperMix for qPCR (Vazyme) was used for reverse transcription to synthesize cDNA, which was subsequently diluted tenfold and stored at − 80 ℃. Specific primers were designed for the 20 WNK genes, and their specificity was confirmed by separating the qRT-PCR products on agarose gels. These primers were then utilized for qRT-PCR analysis. The qRT-PCR reactions were performed using the SYBR Green master mix (ChamQ SYBR qPCR Master Mix, Vazyme). GmACTIN11 was selected as the housekeeping gene. The relative change in expression for each WNK gene in response to salt stress was quantified using the 2−ΔΔCT method (Livak and Schmittgen 2001). Three independent replicates were performed for each treatment. The primers used are described in Table S2.
Statistical analysis
The data were assessed by Duncan’s test (P < 0.05). The data were presented as means accompanied by the standard deviation (SD), derived from three biologically independent replicates for each measured parameter. Significant differences in the data are shown by lowercase letters above the bars. The letters only represent a significant difference within a tissue and not between tissues. The column diagram was performed using GraphPad Prism v8.
Results
Identification of the soybean WNK genes
An extensive BLAST search based on known WNK genes from Arabidopsis was carried out to identify all candidate WNK family proteins encoded within the soybean genome. We further validated the accuracy of these protein predictions using the Pfam database (http://pfam.xfam.org/search). Ultimately, we identified 26 high-confidence genes encoding proteins with serine/threonine protein kinase domains. Following the nomenclature conventions of rice and Arabidopsis, we named the soybean WNK family proteins as GmWNKs, according to the order of the family members on the chromosomes (Table S3). We analyzed the physicochemical properties of the predicted GmWNK protein sequences and estimated that these proteins ranged in length from 297 to 738 amino acids, with molecular weights varying from 33.91 to 84.54 kDa. The predicted isoelectric points of the proteins ranged from 4.84 to 6.58. The estimated instability index of the proteins ranged from 35.33 to 53.28. The predicted aliphatic index of the proteins ranged from 66.44 to 87.99. Using the online tool WoLF PSORT (https://www.genscript.com/wolf-psort.html?src=leftbar), we predicted subcellular localization and found that 13 GmWNK proteins should be localized in the nucleus, six in the chloroplasts, six in the cytoplasm, and one in the cytoskeleton. These results indicated that these WNK genes may have different functional roles.
To study the phylogenetic relationships of the WNK proteins in soybean and other species, 54 WNK protein sequences from soybean (G. max), Arabidopsis, rice (Oryza sativa) and maize (Zea mays) were selected to construct an evolutionary tree (Fig. 1). Based on the topology of the phylogenetic tree, the WNK proteins in plants can be divided into four clades, namely clade I, II, III, and IV. In addition, clade IV was divided into clades IVA and IVB (Fig. 1). In each clade, the WNK genes from monocots and dicots were divided into two sub-branches, with high bootstrap values. For instance, in Clade IVB, the proteins from the dicots—GmWNK13, 26, 14, 25, 23, 6 and AtWNK1, 9, 2—were separate from the proteins from the monocots: OsWNK1, 3 and ZmWNK3, 5 (Fig. 1). The phylogenetic analysis provided insights into the potential functional conservation or diversification of the soybean WNK genes.
The phylogenetic tree of WNK proteins from dicotyledonous and monocotyledonous plants. The phylogenetic tree was constructed using WNK protein sequences from Arabidopsis thaliana (At; black triangles), Glycine max (Gm; red stars), Oryza sativa (Os; green squares), and Zea mays (Zm; blue circles) using the maximum likelihood method with 1000 bootstrap replications in MEGA X. Clades I–IV are distinguished by different colors
Distribution and colinearity analysis of soybean WNK genes
The physical locations of the 26 WNK genes in the soybean genome were unevenly distributed on 15 chromosomes (Fig. 2). There is only one WNK gene located on chromosomes Chr01, Chr04, Chr07, Chr08, Chr09, Chr13, Chr16, and Chr19. There are two WNK genes on chromosomes Chr03, Chr06, and Chr10. There are three WNK genes on chromosomes Chr02, Chr14, Chr18, and Chr20. No genes were mapped to chromosomes Chr05, Chr11, Chr12, or Chr15 (Fig. 2). Most of the genes were located near the end of a chromosome, while a few were located in the middle region of the chromosome. The number of WNK genes on each chromosome is also different.
The distribution of soybean WNK genes across the chromosomes. Each WNK was mapped to its chromosomal position by its physical positions on the soybean genome. The chromosome number is labeled next to each chromosome. The scale bar is in mega bases (Mb). The color on the chromosome indicates the density of genes, with red indicating the highest density and blue indicating the lowest density
Tandem and segmental duplications are key mechanisms underlying the expansion of gene families (Cannon et al. 2004). Typically, tandem duplications are characterized by the occurrence of two adjacent genes on the same chromosome, usually separated by no more than five genes (Wang et al. 2023). Segmental duplications lead to the presence of large repetitive chromosomal blocks in the genome and are often associated with chromosomal rearrangements and polyploid events (Lallemand et al. 2020). Colinearity analysis revealed the presence of 21 segmental duplication events associated with 26 WNK genes throughout the soybean genome and the absence of tandem duplications (Fig. 3). These results suggested that the expansion of the WNK gene family in soybean can be attributed to segmental duplications.
Chromosomal distribution and inter-chromosomal relationship of WNK genes. Red curves connect segmentally duplicated gene pairs. The outer section illustrates the positions of these genes, with yellow boxes corresponding to different chromosomes (Chr1–Chr20). A red zigzag line highlights the gene density on each chromosome. The inner boxes of the diagram further emphasize gene density within the chromosomes, with red denoting the highest density and blue indicating the lowest density
WNK gene structures and predicted protein motifs
To investigate the conservation and diversity of the gene structures within the soybean WNK gene family, the exons and introns of the 26 WNK genes were predicted based on their coding sequences and genomic information. The number of exons varied among the WNK genes, with most members containing 6–8 exons, while the six members in Clade I had only 2 exons (Fig. 4C).
Structure of GmWNK members. A Phylogenetic relationships among GmWNK members and distribution of conserved protein motifs. A total of 10 motifs were identified (Fig. S1). The bottom scale represents protein length. B Predicted conserved protein domains of GmWNK proteins. The gray bars represent the length of each protein sequence, and the conserved protein kinase domains are depicted with colored boxes. C Exon–intron structures of WNK genes. Yellow boxes represent exons (CDS), black lines represent introns, and green boxes represent 5′ and 3′ UTR regions
Analysis of the conserved protein motifs further supported the phylogenetic relationships and classification of the soybean WNK gene family. Ten conserved motifs were identified among the 26 GmWNK proteins (Fig. 4A). All WNK proteins possessed essential STKc_WNK or PKc_like superfamily domains, which included those motifs labeled 1, 2, 3, 4, 5, 7, and 10 (Fig. 4B). The proteins in Clades II, III, and IVB, except for WNK9, also contained an additional OSR1_C superfamily domain. Apart from the proteins in Clade I, the other GmWNK members carried motifs 8 and 6 (Fig. S1). These results provide insights into the structural characteristics of the soybean WNK gene family, and the encoded conserved motifs provide clues about potential functions of the GmWNK proteins.
Analysis of cis-elements in the soybean WNK gene promoters
Cis-regulatory elements play crucial roles in the transcriptional regulation of gene expression. In this study, the upstream 2000-bp sequences of the identified WNK genes were extracted from the soybean genome, and PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was employed to search for cis-regulatory elements. A total of 14 types of cis-regulatory elements were identified in the promoter regions of the WNK genes (Fig. 5), with the greatest number of elements associated with hormones, including auxin response, salicylic acid response, gibberellin response, abscisic acid response, and MeJA response (Fig. 5 and Table S4). Additionally, numerous light-responsive elements and circadian rhythm control elements were present in the promoters, indicating that WNK genes may be involved in light signaling (Fig. 5 and Table S4). Other identified elements in the promoters were related to endosperm- and meristem-specific expression, such as zein metabolism. The large number and wide distribution of elements associated with defense, stress, drought, anaerobic, and low-temperature responses suggested that WNK genes are involved in various biological processes and exhibit different responses to abiotic stresses (Fig. 5 and Fig. S2).
Cis-element analysis of WNK promoters. The cis-elements in the 2000-bp upstream promoter regions are mapped on each gene, with the different types of cis-elements represented by different colors, as identified on the right. The length of the promoter sequence is indicated by scale bars at the bottom of the figure
Expression patterns of WNK genes in soybean
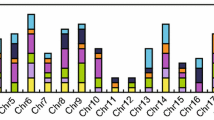
To explore the potential functions of the WNK genes in soybean, we downloaded the gene expression data package from the Phytozome database and obtained the FPKM values of WNK genes in various tissues, including pods, leaves, roots, nodules, seeds, shoot apical meristem (SAM), stems, and flowers (Fig. 6 and Table S5). The majority of the genes showed low expression levels across most, if not all, tissues, while WNK9 and WNK15 exhibited relatively high constitutive expression levels in all tissues. However, several WNK genes displayed high expression levels in specific tissues. For instance, WNK5 was highly expressed in pods, WNK20 in leaves and flowers, WNK3 and WNK5 in roots, WNK5 in root nodules, WNK9 in seeds, WNK25 in SAM, and WNK15 in stems (Fig. 6 and Table S5). These findings suggested that different WNK genes may play specific roles in particular tissues, indicating tissue-specific functions of members of the GmWNK gene family.
Response of WNK genes under salt stress treatments
To investigate if any of the WNK genes showed transcriptional changes under salt stress, we subjected soybean plants to salt treatment (0 h, 3 h, 12 h, and 24 h) and then collected leaves and roots for RNA extraction. qRT-PCR analysis was used to analyze the expression levels of 20 WNK genes, which were then compared between the 0-h treatment and the three other time points. The expression of these 20 WNK genes was induced by 200 mM salt stress in both roots and leaves, indicating that soybean WNK genes were responsive to salt stress (Fig. 7). The expression levels of several genes, including WNK1, WNK10, WNK15, WNK17, WNK19, WNK20, and WNK23, were significantly up-regulated after 3 h or 12 h of exposure to 200 mM NaCl, followed by down-regulation (Fig. 7). These genes exhibited similar expression responses to salt stress in both leaves and roots, implying that they may share common upstream signaling components and possibly the similar function of conferring salt tolerance in aerial and subterranean tissues. Interestingly, WNK5 displayed a distinct expression pattern under salt stress in roots, with a marked decrease in expression level after 3 h, a marked increase at 12 h, and a marked decrease again at 24 h.
Analysis of WNK gene expression under salt treatment.
Transcript levels were determined in roots and leaves using qRT-PCR. The differential expression analysis was conducted based on the 2−∆∆ct method. Data was the most representative of three biological replicates, and the GmActin11 gene was chosen as the internal reference. Following analysis of variance, the significant differences, as identified by Duncan’s test (P < 0.05) using SPSS v.25, are represented by different letters.
Discussion
WNK proteins are serine/threonine protein kinases (STKs) that can be activated by upstream signals through phosphorylation to regulate the activity of downstream target substrates, serving as important regulators in cellular physiology. Research on WNK kinases has primarily focused on humans and animals. In the human genome, the WNK gene family consists of four genes: WNK1, WNK2, WNK3, and WNK4 (Veríssimo and Jordan 2001; Xu et al. 2000). Lower organisms such as yeast and bacteria do not possess WNK kinases, and all four WNK kinases are found almost exclusively in mammals (Tang 2016). WNK kinases are relatively recent proteins in biological evolution and are present in complex eukaryotic organisms. In comparison to animals, plants have a greater number of WNK genes (Wang et al. 2008). Currently, there is limited research on the WNK protein family in plants, mainly focused on model plants such as Arabidopsis and rice. Eleven WNK genes have been reported in Arabidopsis, and nine WNK genes have been reported in rice (Manuka et al. 2015; Wang et al. 2008). There is little information regarding WNK family members in soybean, and the evolutionary relationships between soybean WNK genes and other plant WNK genes remains unknown. Therefore, further research in this area is warranted.
Gene duplication is not only a major source of evolutionary innovation but also a primary cause of gene family expansion (Schmutz et al. 2010). In soybean, a total of 26 WNK genes have been identified (Table S1), which is significantly higher than in Arabidopsis (11), rice (9), or maize (9) and which is consistent with the idea that soybean has undergone two rounds of large-scale whole-genome duplication (WGD) events, resulting in a highly duplicated genome and an increased number of many genes compared to other diploid species (Gill et al. 2009). A phylogenetic tree of WNK proteins from soybean, Arabidopsis, rice, and maize showed that the WNK family can be divided into four clades and that the orthologs cluster together with WNK members fro20.00 m other plant species, indicating that they may have undergone similar evolutionary diversification (Fig. 1). Based on phylogenetic clustering and sequence similarity, paired GmWNKs, such as GmWNK1/5, GmWNK15/24, GmWNK4/16, GmWNK3/17, and GmWNK14/25, show high sequence similarity (> 90%) and high internal node bootstrap values (99%) (Fig. 1), suggesting that these genes may have parallel relationships and functional redundancy may have arose from gene duplication events.
The members of the WNK gene family in soybean are predicted to encode proteins ranging from 297 to 738 amino acid in length (Table S3), similar to the lengths of WNK proteins in rice and Arabidopsis (Manuka et al. 2015; Wang et al. 2008). The predicted gene structures of the soybean WNK contain two to eight exons (Fig. 4C), similar to the gene structures in Arabidopsis, with three to nine exons (Liu et al. 2022b; Manuka et al. 2015). The GmWNK proteins were predicted to localize to various cellular compartments, including the nucleus, chloroplast, cytoplasm, and cytoskeleton (Table S3). In contrast, LOCTREE3 predicted that all OsWNK proteins localized to the cytoplasm, while mPLoc predicted their localization in the nucleus (Manuka et al. 2015).
STKs catalyze the transfer of the gamma-phosphoryl group from ATP to serine/threonine residues on protein substrates. WNKs form a subfamily of STKs with a distinctive positioning of the catalytic lysine compared to other protein kinases (Huang et al. 2007; Richardson and Alessi 2008). WNK family members lack a catalytic lysine residue in subdomain II of the N-terminal kinase domain, which is conserved in all other kinases and plays a critical coordinating role in the ATP active site (McCormick and Ellison 2011). A catalytic lysine residue in subdomain I of WNK proteins has been proposed to substitute for the lysine residue in subdomain II as a key phosphorylation site (Wilson et al. 2001). Predicted conserved domains of soybean WNK gene members revealed the presence of N-terminal protein kinase domains; domain I contained the motif of Gly-X-Gly-X-X-Lys-X-Val, and in domain II, the absence of lysine residues was replaced by aspartate/serine residues (Fig. 4B). This altered domain is also found in Arabidopsis and rice WNK proteins (Manuka et al. 2015; Wang, et al. 2008). In Clades II, III, and IVB, all of the GmWNK proteins, except GmWNK9, possessed an additional oxidative-stress-responsive kinase 1 C-terminal domain (OSR1 domain), approximately 40 amino acids in length (Fig. 4B). OSR1 is involved in the signaling cascade that activates the Na/K/2Cl cotransporter during osmotic stress. This domain in the C-terminal domain of OSR1 recognizes a motif (Arg-Phe-Xaa-Val) on the OSR1-activating protein, which is a WNK kinase (Villa et al. 2007).
Cis-regulatory elements regulate responses to hormones and various stresses during plant growth and development (Nishimura et al. 2010). Analysis of the promoter regions of all WNK genes revealed the presence of diverse cis-regulatory elements, including those involved in light response, plant hormone signaling, and stress resistance (Fig. 5). In Arabidopsis, the WNK gene family regulates plant flowering time through the photoperiod pathway (Wang et al. 2008). It is noteworthy that light-responsive cis-elements frequently occur in WNK gene promoters, suggesting their potential involvement in soybean photoperiodic response or circadian rhythm (Nakamichi et al. 2002). Jasmonic acid and abscisic acid are well-known signaling molecules involved in biotic and abiotic stress responses, inducing plant resistance to these stressors (Sah et al. 2016; Verma et al. 2016; Wang et al. 2020). The frequent occurrence of abscisic acid- and jasmonic acid-responsive elements in the GmWNK promoters indicates an association between WNK family genes and plant stress (Fig. 6) (Feng et al. 2019; Gómez-Porras et al. 2007).
Soybean, as a globally important food and oil crop with a large impact on the economies of several countries, can be improved through the identification of stress-related genes such as the WNK kinases that may be introduced during the development of new varieties. Plant protein kinases play crucial roles in stress-induced signal transduction (Kumar et al. 2013; Sun et al. 2022). Other WNK genes are known to be involved in responses to abiotic stress. For instance, the PeWNKs of Moso bamboo are involved in salt stress response (Liu et al. 2022b). Disruption of AtWNK8 enhances Arabidopsis tolerance to salt and osmotic stresses by modulating proline content and the activities of catalase and peroxidase (Zhang et al. 2013). In our study, we observed that salt stress rapidly induced the transcription levels of most WNK genes in soybean to significantly increased levels within 3 h in both roots and leaves, with the exception of GmWNK5 (Fig. 7). Interestingly, GmWNK5 exhibited an opposite expression pattern compared to the other WNK genes, with a significant decrease in expression within 3 h in roots (Fig. 7). Similar results have been reported in previous studies, where soybean WNK1 (in this article, it is WNK5) was downregulated after 2 h of NaCl treatment, and overexpression of GmWNK1 in Arabidopsis altered sensitivity to salt and osmotic stress (Wang et al. 2010). These findings suggest that these genes are responsive to salt stress and may be involved in regulation of salt tolerance. These observations provide a foundation for further exploring the molecular mechanisms underlying the specific roles of the WNK family in soybean.
Data availability
All data used in this paper has been released.
References
Cannon SB, Mitra A, Baumgarten A, Young ND, May G (2004) The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4:10
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13:1194–1202
Feng R, Ren M, Lu L, Peng M, Guan X, Zhou D, Zhang M, Qi D, Li K, Tang W, Yun T, Chen Y, Wang F, Zhang D, Shen Q, Liang P, Zhang Y, Xie J (2019) Involvement of abscisic acid-responsive element-binding factors in cassava (Manihot esculenta) dehydration stress response. Sci Rep 9:12661
Gill N, Findley S, Walling JG, Hans C, Ma J, Doyle J, Stacey G, Jackson SA (2009) Molecular and chromosomal evidence for allopolyploidy in soybean. Plant Physiol 151:1167–1174
Gómez-Porras JL, Riaño-Pachón DM, Dreyer I, Mayer JE, Mueller-Roeber B (2007) Genome-wide analysis of ABA-responsive elements ABRE and CE3 reveals divergent patterns in Arabidopsis and rice. BMC Genom 8:260
Hanks SK, Hunter T (1995) Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. Faseb j 9:576–596
Hong-Hermesdorf A, Brüx A, Grüber A, Grüber G, Schumacher K (2006) A WNK kinase binds and phosphorylates V-ATPase subunit C. FEBS Lett 580:932–939
Huang CL, Cha SK, Wang HR, Xie J, Cobb MH (2007) WNKs: protein kinases with a unique kinase domain. Exp Mol Med 39:565–573
Kahle KT, Rinehart J, Ring A, Gimenez I, Gamba G, Hebert SC, Lifton RP (2006) WNK protein kinases modulate cellular Cl- flux by altering the phosphorylation state of the Na-K-Cl and K-Cl cotransporters. Physiology (bethesda) 21:326–335
Kumar K, Kumar M, Kim SR, Ryu H, Cho YG (2013) Insights into genomics of salt stress response in rice. Rice 6:27
Kumar K, Raina SK, Sultan SM (2020) Arabidopsis MAPK signaling pathways and their cross talks in abiotic stress response. J Plant Biochem BioT 29:700–714
Lallemand T, Leduc M, Landès C, Rizzon C, Lerat E (2020) An overview of duplicated gene detection methods: why the duplication mechanism has to be accounted for in their choice. Genes (basel) 11:1046
Liu H, Liu X, Zhao Y, Nie J, Yao X, Lv L, Yang J, Ma N, Guo Y, Li Y, Yang X, Lin T, Sui X (2022a) Alkaline α-galactosidase 2 (CsAGA2) plays a pivotal role in mediating source-sink communication in cucumber. Plant Physiol 189:1501–1518
Liu R, Vasupalli N, Hou D, Stalin A, Wei H, Zhang H, Lin X (2022b) Genome-wide identification and evolution of WNK kinases in Bambusoideae and transcriptional profiling during abiotic stress in Phyllostachys edulis. PeerJ 10:e12718
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Ma TL, Wu WH, Wang Y (2012) Transcriptome analysis of rice root responses to potassium deficiency. BMC Plant Biol 12:161
Manuka R, Saddhe AA, Kumar K (2015) Genome-wide identification and expression analysis of WNK kinase gene family in rice. Comput Biol Chem 59:56–66 (Pt A)
Manuka R, Saddhe AA, Kumar K (2018) Expression of OsWNK9 in Arabidopsis conferred tolerance to salt and drought stress. Plant Sci 270:58–71
Manuka R, Karle SB, Kumar K (2019) OsWNK9 mitigates salt and drought stress effects through induced antioxidant systems in Arabidopsis. Plant Physiol 24:168–181
McCormick JA, Ellison DH (2011) The WNKs: atypical protein kinases with pleiotropic actions. Physiol Rev 91:177–219
Murakami-Kojima M, Nakamichi N, Yamashino T, Mizuno T (2002) The APRR3 component of the clock-associated APRR1/TOC1 quintet is phosphorylated by a novel protein kinase belonging to the WNK family, the gene for which is also transcribed rhythmically in Arabidopsis thaliana. Plant Cell Physiol 43:675–683
Nakamichi N, Murakami-Kojima M, Sato E, Kishi Y, Yamashino T, Mizuno T (2002) Compilation and characterization of a novel WNK family of protein kinases in Arabidopsis thaliana with reference to circadian rhythms. Biosci Biotechnol Biochem 66:2429–2436
Nishimura N, Sarkeshik A, Nito K, Park SY, Wang A, Carvalho PC, Lee S, Caddell DF, Cutler SR, Chory J, Yates JR, Schroeder JI (2010) PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J 61:290–299
Papiernik SK, Grieve CM, Lesch SM, Yates SR (2005) Effects of salinity, imazethapyr, and chlorimuron application on soybean growth and yield. Commun Soil Sci Plan 36:951–967
Pei X, Wang X, Fu G, Chen B, Nazir MF, Pan Z, He S, Du X (2021) Identification and functional analysis of 9-cis-epoxy carotenoid dioxygenase (NCED) homologs in G. hirsutum. Int J Biol Macromol 182:298–310
Richardson C, Alessi DR (2008) The regulation of salt transport and blood pressure by the WNK-SPAK/OSR1 signalling pathway. J Cell Sci 121:3293–3304
Sah SK, Reddy KR, Li J (2016) Abscisic acid and abiotic stress tolerance in crop plants. Front Plant Sci 7:571
Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu S, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, Du J, Tian Z, Zhu L, Gill N, Joshi T, Libault M, Sethuraman A, Zhang XC, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht J, Grimwood J, Rokhsar D, Stacey G, Shoemaker RC, Jackson SA (2010) Genome sequence of the palaeopolyploid soybean. Nature 463:178–183
Sun Z, Feng Z, Ding Y, Qi Y, Jiang S, Li Z, Wang Y, Qi J, Song C, Yang S, Gong Z (2022) RAF22, ABI1 and OST1 form a dynamic interactive network that optimizes plant growth and responses to drought stress in Arabidopsis. Mol Plant 15:1192–1210
Tang BL (2016) (WNK)ing at death: with-no-lysine (Wnk) kinases in neuropathies and neuronal survival. Brain Res Bull 125:92–98
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tsuchiya T, Eulgem T (2010) The Arabidopsis defense component EDM2 affects the floral transition in an FLC-dependent manner. Plant J 62:518–528
Uchida S, Sohara E, Rai T, Sasaki S (2014) Regulation of with-no-lysine kinase signaling by Kelch-like proteins. Biol Cell 106:45–56
Urano D, Czarnecki O, Wang X, Jones AM, Chen JG (2015) Arabidopsis receptor of activated C kinase1 phosphorylation by with no lysine8 kinase. Plant Physiol 167:507–516
Veríssimo F, Jordan P (2001) WNK kinases, a novel protein kinase subfamily in multi-cellular organisms. Oncogene 20:5562–5569
Verma V, Ravindran P, Kumar PP (2016) Plant hormone-mediated regulation of stress responses. BMC Plant Biol 16:1–10
Villa F, Goebel J, Rafiqi FH, Deak M, Thastrup J, Alessi DR, van Aalten DM (2007) Structural insights into the recognition of substrates and activators by the OSR1 kinase. EMBO Rep 8:839–845
Wang Y, Liu K, Liao H, Zhuang C, Ma H, Yan X (2008) The plant WNK gene family and regulation of flowering time in Arabidopsis. Plant Biol (stuttg) 10:548–562
Wang Y, Suo H, Zheng Y, Liu K, Zhuang C, Kahle KT, Ma H, Yan X (2010) The soybean root-specific protein kinase GmWNK1 regulates stress-responsive ABA signaling on the root system architecture. Plant J 64:230–242
Wang J, Song L, Gong X, Xu J, Li M (2020) Functions of jasmonic acid in plant regulation and response to abiotic stress. Int J Mol Sci 21:1446
Wang J, Sun Z, Liu H, Yue L, Wang F, Liu S, Su B, Liu B, Kong F, Fang C (2023) Genome-wide identification and characterization of the soybean Snf2 gene family and expression response to rhizobia. Int J Mol Sci 24:7250
Wilson FH, Disse-Nicodème S, Choate KA, Ishikawa K, Nelson-Williams C, Desitter I, Gunel M, Milford DV, Lipkin GW, Achard JM, Feely MP, Dussol B, Berland Y, Unwin RJ, Mayan H, Simon DB, Farfel Z, Jeunemaitre X, Lifton RP (2001) Human hypertension caused by mutations in WNK kinases. Science 293:1107–1112
Xie M, Wu D, Duan G, Wang L, He R, Li X, Tang D, Zhao X, Liu X (2014) AtWNK9 is regulated by ABA and dehydration and is involved in drought tolerance in Arabidopsis. Plant Physiol Biochem 77:73–83
Xu B, English JM, Wilsbacher JL, Stippec S, Goldsmith EJ, Cobb MH (2000) WNK1, a novel mammalian serine/threonine protein kinase lacking the catalytic lysine in subdomain II. J Biol Chem 275:16795–16801
Zhang B, Liu K, Zheng Y, Wang Y, Wang J, Liao H (2013) Disruption of AtWNK8 enhances tolerance of Arabidopsis to salt and osmotic stresses via modulating proline content and activities of catalase and peroxidase. Int J Mol Sci 14:7032–7047
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 32301822), the China Postdoctoral Science Foundation (Grant No. 2022M720882), the Innovation Research Project of Coarse Cereals Specialty in Guizhou Province [2019[4012]], and the Regional First-class Discipline of Ecology in Guizhou Province (XKTJ[2020]22).
Author information
Authors and Affiliations
Contributions
BS and ZS conceived and designed the research. BS and ZS performed the experiments and collected the data. JW and YZ supervised the experiments. BS wrote the manuscript. FK and BL modified manuscript. TG polished the manuscript. FW and TF polished the images. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Yes.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Su, B., Ge, T., Zhang, Y. et al. Genome-wide identification and expression analysis of the WNK kinase gene family in soybean. Mol Breeding 44, 16 (2024). https://doi.org/10.1007/s11032-024-01440-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-024-01440-5