Abstract
The discovery and characterization of the opaque endosperm gene provide ideas and resources for the production and application of maize. We found an o213 mutant whose phenotype was opaque and shrunken endosperm with semi-dwarf plant height. The protein, lipid, and starch contents in the o213 endosperm were significantly decreased, while the free amino acid content in the o213 endosperm significantly increased. The aspartic acid, asparagine, and lysine contents were raised in the o213 endosperm by 6.5-, 8.5-, and 1.7-fold, respectively. Genetic analysis showed that this o213 mutant is a recessive single-gene mutation. The position mapping indicated that o213 is located in a 468-kb region that contains 11 protein-encoding genes on the long arm of chromosome 5. The coding sequence analysis of candidate genes between the WT and o213 showed that ZmYSL2 had only a single-base substitution (A–G) in the fifth exon, which caused methionine substitution to valine. Sequence analysis and the allelic test showed that o213 is a new mutant allele of ZmYSL2. The qRT-PCR results indicated that o213 is highly expressed in the stalks and anthers. Subcellular localization studies showed that o213 is a membrane transporter. In the variation analysis of o213, the amplification of 65 inbred lines in GWAS showed that this 3-bp deletion of the first exon of o213 was found only in temperate inbred lines, implying that the gene was artificially affected in the selection process. Our results suggest that o213 is an important endosperm development gene and may serve as a genetic resource.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Maize is an important food crop for humans and livestock (Janick 2021). The kernels of maize are composed of the endosperm, embryo, and seed coat. Maize pollen splits into two sperms, one fused with the egg cell to form a diploid embryo (Olsen 2020), and the other merged with the two polar nuclei of the central cell to form a triploid endosperm (Kiesselbach 1980). The seed coat is mainly composed of cellulose, and its surface is smooth, accounting for approximately 6–8% of the total weight of the seed (Wu and Messing 2014). During seed development, the endosperm supplies nutrients for embryo development (Sabelli and Larkins 2009), embryo absorption, and the transport of nutrients (Ester and Ang 2021). Maize kernel contains approximately 72% starch, 10% protein, and other nutrients. However, the levels of these nutrients are either increased or decreased to different degrees in different opaque endosperm mutants (Segal et al. 2003). Maize quality traits are a guide for evaluating inbred lines and hybrids (Letchworth and Lambert 1998) and a breeding standard for quality protein maize (QPM). The development and adoption of QPM can improve the nutritional value of foods and feed generated from maize products (Bhatnagar et al. 2004). Therefore, the discovery of opaque endosperm mutants is of great significance for analyzing the anabolism of nutrients and improving the nutritional value of maize kernels.
To date, many opaque endosperm mutants have been reported. For example, opaque1 (o1), opaque2 (o2), opaque5 (o5), opaque7 (o7), opaque10 (o10), and opaque11 (o11) have been identified (Wang et al. 2012, 2011; Schmidt et al. 1987; Myers et al. 2011; Feng et al. 2018; Yao et al. 2016). The o1 encodes a myosin XI motor protein, and a 2-bp loss of o1 in exon 14 leads to a frameshift and premature termination of the reading frame (Wang et al. 2012). The o1 mutants have an opaque endosperm phenotype due to abnormal endoplasmic reticulum movement and hampered protein body formation. A study found that the total protein content of o1 was no different from that of wild-type kernels, and there was no difference in the content of the human essential amino acid-lysine (Segal et al. 2003). o2 encodes a basic leucine zipper (bZIP) transcriptional activator (Mertz et al. 1964). A study found that the total protein content of o2 was reduced by 15% compared to wild-type kernels, and the non-zein of o2 increased by 70% (Segal et al. 2003). The o2 mutant can alter amino acid enzyme composition and protein composition changes, resulting in a substantial increase in lysine content (Schmidt et al. 1987). The o5 encodes the monogalactosyldiacylglycerol synthase. Mutation of o5 affects the synthesis of galactolipids necessary for amyloplast and chloroplast function, and exhibits an opaque endosperm with white seeding phenotype (Myers et al. 2011). The o7 encodes an acyl-activating enzyme-like protein that inhibits the synthesis of 19-kD α-zein after mutation, thereby forming the opaque endosperm phenotype (Wang et al. 2011). The maize o10 encodes a cereal-specific protein, and mutation of o10 affects the distribution of zein in the endosperm protein bodies, thus exhibiting an opaque endosperm phenotype (Yao et al. 2016). The o11 encodes a basic helix-loop-helix (bHLH) transcription factor, and mutations in o11 affect the regulatory network of maize endosperm development and nutritional metabolism, resulting in an opaque endosperm phenotype (Feng et al. 2018). The total protein content of o11 is not different from that of wild-type kernels, but the lysine content is 1.8 times that of wild-type kernels. There are many reports on the regulatory mechanism of maize endosperm development, but there are more genes to be discovered and mined.
Many oligopeptide transporters in plants transport nutrients for plant growth and development and participate in plant tissue transport (Delrot et al. 2000). Plants obtain small peptides through protein degradation pathways and biospecific synthesis. Small peptides are hydrolyzed by peptidases in plants and eventually become a source of amino acids, carbon, and nitrogen (Williams and Miller 2001). Therefore, oligopeptide transporters play an essential role in plant growth and development (Stacey et al. 2006). The oligopeptide transporter (OPT) superfamily has two subfamilies: yellow stripe (YS) and peptide transport (PT) (Lubkowitz et al. 1997; Curie et al. 2001; Yen et al. 2001). The YS subfamily is mainly found in plants, fungi, bacteria, and archaea, while the PT subfamily exists in plants and fungi (Lubkowitz 2006). Research has shown that small peptides are involved in different functions, including embryo development (Stacey et al. 2002) and glutathione transport (Bogs et al. 2003; Zhang et al. 2004). The YS subfamily has identified the ZmYS1 and ZmYSL2 (Schaaf et al. 2004; Zang et al. 2020). Early research suggests that the zmys1 mutation affects the Fe3+-phytosiderophore (Fe3+-PS) and Zn-dimethyl acetamide (Zn-DMA) chelate absorption systems defects in the plant (von Wiren et al. 1994). The zmys1 mutation cannot transport the Fe3+-PS complex (von Wiren et al. 1996), leading to yellow of leaf veins and the leaves to appear yellow stripe (Curie et al. 2001; von Wirén et al. 1999). ZmYSL2 is a gene that transports metal ions in plants, and mutations of ZmYSL2 affect kernel development and endosperm texture (Zang et al. 2020). In addition, Sh4 is a classic shrunken kernel mutant, which was confirmed to be the allele of ZmYSL2 through Map-based cloning and genetic transformation, and the Sh4 mutation affected the aleurone layer and starch synthesis of maize kernels (He et al. 2021). Therefore, the discovery of transporters can provide new ideas for the regulatory network of maize kernel development.
This study identified an opaque, shrunken endosperm and semi-dwarf mutant opaque213 (o213). The position mapping result shows that o213 is located in a 468-kb region on the long arm of chromosome 5. o213 encodes a metal nicotinamide transporter, and the results of the allelic test indicated that o213 is a new mutant allele of ZmYSL2. Biochemical analysis results showed that the o213 mutation affected the synthesis of nutrients. In addition, we analyzed the metal ion concentration data and found that zinc, manganese, and copper concentrations were affected. The variation analysis of o213 in the GWAS population indicated that genotype 1 with a 3-bp deletion is conducive to molecular breeding. Collectively, our results provide new insights into the regulatory mechanism of the opaque endosperm.
Materials and methods
Plant materials and phenotypic investigation
Both G213 and o213 mutants were provided by the Maize Research Institute of Sichuan Agricultural University in 2013. G213 is the wild type (WT). Since the mutant kernels show an opaque endosperm phenotype, we named it opaque213 (o213). The o213 mutant originated from the segregation of the WT, and the mutant phenotype can be inherited stably after self-pollination. The WT and o213 mutant (21DAP) were used for cytological observation and biochemical analysis. For the map-based cloning of o213, we constructed the F2 population produced by crossing the o213 mutant with Mo17. In the allelic test, the ZmYSL2 mutant was provided by Jie Zang (University of the Chinese Academy of Sciences). The leaves, taproots, lateral roots, adventitious roots, anthers, stalks, ears, tassel, husks, silks, seeds, embryos, and endosperm samples of o213 mutant and WT were snap-frozen with liquid nitrogen. Leaves, taproots, lateral roots, and adventitious root samples of o213 mutant and WT were collected at the 6-leaf stage. The endosperm of o213 mutant and WT was collected at the filling stage (9DAP, 12DAP, 15DAP, 18DAP, 21DAP, 24DAP). Measure the kernel length, kernel width, 100-kernel weight, plant height, ear height, and the number of internodes of WT and mutants at maturity. Plant materials were grown at the modern base of Sichuan Agricultural University in Chongzhou, China (103° 07'E, 30° 30'N), and Sanya, China (18° 09'N, 108° 56'E). All samples were collected in three biological replicates. A t-test was used for significance analysis.
Determination of protein, starch, lipid, and amino acid contents
To detect the changes in starch, lipid, protein, and amino acid contents in the WT and mutant endosperm, we collected WT and o213 mutant kernels at the mature stage and used blades to remove the seed coats and embryos. The Nanjing Convinced Technology Testing Center analyzed starch, lipid, protein, and amino acid content in the endosperm. Total starch, amylose analysis, and amylopectin analysis methods were based on dual-wavelength colorimetry (Sene et al. 1997). The determination of total lipids was based on the GB 5009.6–2016 protocols developed by the Ministry of Agriculture, People’s Republic of China. The determination of total protein was based on the NY/T 2017–2011 protocols developed by the Ministry of Agriculture, People’s Republic of China. The continuous shaking extraction method determined zein and non-zein content (Wang and Kovacs 2002). Three biological replicates were required for each sample.
Metal ion concentration determination
Immature kernels, endosperms, and embryos were collected at 18DAP and 27DAP. First, we dry the samples and weigh 0.2 g of each piece. Secondly, add the 0.2-g sample and 10 ml of nitric acid to the microwave digestion tube. Next, after the digestion, use EHD-24 equipment (Beijing Donghang Scien-tech Instrument Co., Ltd.) to deacidify at 190 °C for 45 min. Finally, after waiting for the microwave digestion tube to cool, add pure water and pour it into a 10-ml centrifuge tube. The standard solution was measured and analyzed by ICP–MS. Three biological replicates were required for each sample.
Scanning electron microscopy (SEM) and transmission electron microscopy (TEM) analysis
Observe changes in the cytological level of WT and o213 mutant kernels. Immature WT and o213 kernels (21DAP) were excised with a razor blade, immediately placed in 2.5% glutaraldehyde, and sent to Qingdao Sci-tech Innovation Quality Testing Co., Ltd. The samples were observed by scanning electron microscopy (JEOL-JSM-840) and transmission electron microscopy (Japan-JEM1200).
Gene mapping and candidate gene analysis
Polymorphism markers were screened using 1059 SSR markers distributed on ten maize chromosomes. For the preliminary position, kernels from forty-six mutants in the F2 ear were used for preliminary determination linkage markers. Xiangge Zhang (Sichuan Agricultural University) provided InDel markers, and new markers were developed using SSR-Hunter software for fine positioning. Candidate gene prediction was based on the V4 version of the maize reference genome B73 (https://www.maizegdb.org/genome/assembly/Zm-B73-REFERENCE-GRAMENE-4.0).
Phyre2 was used for O213 protein structure prediction (http://www.sbg.bio.ic.ac.uk/phyre2). The gene mapping of o213 and the primer used for sequence analysis of the WT and o213 mutant are shown in Table S1.
Phylogenetic and variation analysis
A BLASTP search was performed in the NCBI database using the O213 protein sequence and related sequences were identified (National Center for Biotechnology Information (nih.gov)). Molecular Evolutionary Genetics Analysis (MEGA) 5.1 software and iTOL: Interactive Tree Of Life (embl.de) constructed a phylogenetic tree of homologous sequences. Select the protein sequence of O213 and other species in the phylogenetic tree, and analyze the putative transmembrane domain of O213 with BioEdit Sequence Alignment Editor.
The variation analysis material was 65 randomly selected from the GWAS (genome-wide association studies) population (Zhang et al. 2016), including 35 temperate materials and 30 tropical materials. To determine the mutation of the o213 exon, the sequence of the o213 coding region in these materials was amplified. MEGA 5.1 software and iTOL: Interactive Tree Of Life (embl.de) were used to construct a phylogenetic tree of genotype. Phyre2 was used to predict the protein structure of four genotypes (http://www.sbg.bio.ic.ac.uk/phyre2) and analyze the putative transmembrane domain of four genotypes with BioEdit Sequence Alignment Editor. The primers used for evolution analysis are shown in Table S1.
Subcellular localization
The full-length o213 and WT coding region sequences without stop codons were cloned into the expression vector pHB-YFP to construct pHB-YFP-O213 and pHB-YFP-WT. The fusion plasmid was transformed into Agrobacterium tumefaciens GV3101 and injected into tobacco leaves. TaWPI6-mCherry (Dangol et al. 2017) was used as a plasma membrane marker. The tobacco leaves after Agrobacterium injection were cut into small pieces and imaged using a confocal laser scanning microscope (Nikon A1; Tokyo, Japan). Excitation and emission wavelengths for yellow fluorescent protein (YFP) were collected from 514 to 527 nm, and excitation and emission wavelengths for green fluorescent protein (GFP) were collected from 488 to 507 nm. The primers used for subcellular localization are shown in Table S1.
RNA extraction, reverse transcription, and qRT-PCR
Total RNA was extracted by grinding in a mortar and adding a TRIzol™ reagent. Reverse transcription of total RNA was performed using the TaKaRa PrimeScript™ II 1st strand cDNA synthesis kit (TaKaRa Biotech, Japan).
The template is used for qRT-PCR after diluting 20 times with 1 μl stock solution. The primers were designed using Beacon Designer 7 software and Primer3Plus (http://www.primer3plus.com/cgi-bin/dev/primer3plus.cgi). Gene fragments were amplified using Fast Start Essential DNA Green Master Mix (Roche Biotech, Germany) on a q TOWER3G fluorescent quantitative PCR instrument (Analytik Jena AG). Two-way ANOVA evaluated gene expression data. The primers used for qRT–PCR are shown in Table S1.
RNA-Seq data analysis
Total maize endosperm RNA was extracted from immature (12DAP) WT and o213 kernels for RNA-Seq analysis. A total amount of 2 µg of RNA per sample was used as the input material for the RNA sample preparations. Sequencing libraries were generated using VAHTS mRNA-seq v2 Library Prep Kit for Illumina, following the manufacturer’s recommendations and adding an index code to identify the sequence of each sample. Differentially expressed genes were defined as those for which the adjusted P value was less than 0.05 and the log2 (fold change) was more significant than 1. The GO and KEGG enrichment analysis of differentially expressed gene sets is implemented in GO sequence R and KOBAS 3.0 packages, respectively. GO terms with an adjusted P-value below 0.05 were significantly enriched by the differentially expressed genes. GO analysis and KEGG pathway analysis were performed using the online GO database (http://www.geneontology.org) and KEGG pathway database (http://www.kegg.jp/), respectively. Three biological replicates were measured for each sample.
Results
The o213 mutant has opaque, shrunken endosperm, and semi-dwarf phenotype
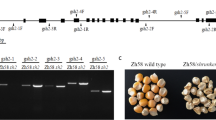
The o213 mutant is a natural variation of the G213. The o213 mutant was crossed with Mo17 to produce F1 and self-crossed to obtain F2. In the offspring of F2, the Mo17 and o213 kernels were present in a ratio of 3:1 (421:135, χ2 = 0.029 < χ20.05 = 3.84), indicating that the o213 mutant is a recessive single-gene inherited mutant (Fig. 1A). During the kernel filling stage, the WT kernel was yellow, while the o213 mutant kernel was white (Fig. 1B). The o213 mutant exhibits opaque, shrunken endosperm (Fig. 1C). In addition, the o213 mutant had a cavity between the seed coat and embryo (Fig. 1D). Compared to the WT, the 100-kernel weight, kernel length, and width of the o213 mutant were significantly reduced (Fig. 1E, F). However, the germination rate of the o213 mutant was hardly affected.
Comparison of the difference between WT and o213 kernel morphology. a F2 ears of Mo17 × o213 mutant. Arrows show o213 kernels (bar = 1 cm); b 15DAP ears of WT and o213 mutant (bar = 1 cm); c observe under the light transmission of WT and o213 endosperm (bar = 1 cm); d longitudinal section of mature WT and o213 maize kernels. Arrows show the gap between the seed coat and embryo (bar = 1 cm); e 100-kernel weight comparison between WT and o213 kernel; f comparison of kernel length and width of WT and o213 kernel. g Seedling stage of WT and o213 mutant (bar = 1 cm). h Mature stage of WT and o213 mutant (bar = 10 cm). i Comparison of plant height, ear height, and number of the internodes of WT and o213 mutant. Student’s t-test, 0.05 < (**) P < 0.01 and (***) P < 0.001
In the vegetative growth stage, the plant height of the o213 mutant from seedling to maturity was weaker than that of the WT (Fig. 1G, H). To measure the growth difference between the WT and o213 mutant, we measured the plant height, ear height, and the number of internodes and found that the plant height, ear height, and the number of internodes of the o213 mutant were significantly lower than those of the WT (Fig. 1I). So, the o213 mutant showed a semi-dwarf phenotype. Collectively, these results indicate that the o213 mutant has a defective opaque, shrunken endosperm, and semi-dwarf phenotype.
o213 mutant affects the synthesis of nutrients in the kernel
To explore the morphological changes of the protein bodies, SEM and TEM were used to observe the immature WT and o213 mutant (21DAP). The results showed that the filling material between the starch grains in the endosperm of the o213 mutant was loose (Fig. 2A). TEM shows a decrease in the number of protein bodies in the endosperm of the mutant (Fig. 2B, C). Biochemical analysis showed that the total starch, amylose, amylopectin, lipid, the total protein, zein, and non-zein contents of the o213 endosperm all decrease (~ 12.8%, ~ 39%, ~ 10.9%, ~ 71.3%, ~ 43.2%, ~ 60.5%, and ~ 8.6%, respectively (Fig. 2D–F)). Most free amino acid content in the o213 mutant endosperm increased significantly. The free asparagine, aspartic acid, and lysine contents in the o213 mutant endosperm were 8.5 times, 6.5 times, and 1.7 times higher than those in the WT, respectively (Fig. 2G–I). Inductively coupled plasma-mass spectrometry (ICP–MS) analysis showed that the zinc concentrations in the embryos, endosperm, and seeds of immature o213 kernels (18DAP, 27DAP) were significantly reduced (Fig. S1). The manganese concentration in the endosperm of immature o213 kernels (18DAP) was significantly increased, and the manganese concentration in the endosperm (27DAP) was significantly decreased. There was no difference between the embryos and seeds (Fig. S1). The copper concentration in the immature kernel o213 embryos (18DAP, 27DAP) was significantly reduced, and there was no difference between the endosperm and seeds (Fig. S1). These results indicate that the nutrients and mineral element content of o213 kernels were affected.
Cytology and nutritional composition differences. a Scanning electron microscopy of the endosperm of immature (21DAP) WT and o213 (bar = 10 µm). b Transmission electron microscopy of the endosperm of immature (21DAP) WT and o213 (bar = 5 µm). c Comparison of the number of protein bodies between WT and o213 endosperm. d–f Comparison of the lipid, starch, and protein between WT and o213 endosperm. g–i Comparison of total free amino acid and free amino acid content between WT and o213 endosperm. Student’s t-test, (*) P < 0.05, 0.05 < (**) P < 0.01 and (***) P < 0.001
Mapping of the causal gene of o213
Using 1059 pairs of markers distributed on 10 maize chromosomes to screen polymorphic markers among Mo17 and o213 mutant, a total of 300 pairs of polymorphic markers were screened for preliminary positioning. The mutation phenotypes of 46 kernels from F2 ears were used for preliminary linkage analysis for preliminary mapping. The preliminary positioning results stated that the o213 gene was located between the markers umc1502 and umc1524 on the long arm of chromosome 5. For fine mapping, 20,000 mutant kernels were used to locate o213 within the 468-kb range between the InDel19029 and InDel435. This interval contains 11 predicted genes (Zm00001d017422, Zm00001d017423, Zm00001d017424, Zm00001d017425, Zm00001d017427, Zm00001d017428, Zm00001d017429, Zm00001d017430, Zm00001d017432, Zm00001d017433, Zm00001d017434 (Fig. 3A)). Subsequently, the ORFs (open reading frames) of 11 candidate genes were sequenced. Only Zm00001d017427 had a single nucleotide polymorphism (SNP) mutation between the WT and o213 mutant. The SNP (A to G) in the fifth exon of the Zm00001d017427 gene causes the amino acid in the o213 coding sequence to convert from methionine to valine (Fig. 3B, C). This gene was previously annotated as ZmYSL2/Sh4, which encodes a metal-nicotianamine transporter (Fig. 3D). The o213 mutant had a similar phenotype to the zmysl2 mutant and sh4 mutant. For the allelic test, the zmysl2 mutant as the female parent and the o213 mutant as the male parent were crossed to produce F1, and all F1 kernels showed a zmysl2 mutant phenotype, indicating that the mutant phenotypes of o213 and zmysl2 were controlled by the same gene (Fig. 3E). These results further illustrate that o213 is a novel mutant allele of zmysl2.
Fine mapping of o213 and the allelic test. a The o213 gene is located in the 468-kb region between InDel19029 and InDel435 on chromosome 5. The number below the corresponding markers indicates recombinants between the markers and o213. b The sequencing results of WT and o213; the shaded part is the mutation site. c The amino acid sequence of WT and o213 mutants; the red font is the mutant amino acid. d Structure of the ZmYSL2 and its mutation sites in the one allele. Nucleotide substitutions in the allele mutants are indicated. The o213 has one nucleotide A1371 substitution in the fifth exon. The solid black boxes are exons, the white solid boxes are UTR, and the black lines are introns. e The allelic test of ZmYSL2 mutant and o213 mutant (bar = 1 cm)
Sequence alignment of O213
To identify the homologous sequences of O213 in other species, we downloaded the homologous sequences of different species related to O213 from the NCBI database and performed phylogenetic tree analysis. Protein sequence alignment showed that O213 has high sequence similarity with other species, including Sorghum bicolor, Dichanthelium oligosanthes, Panicum hallii var. hallii, Setaria viridis, Oryza sativa, Hordeum vulgare, Avena sativa, etc. (Fig. 4A). At the same time, we analyzed the transmembrane domain of O213 and other species in the phylogenetic tree and found that the transmembrane domain of O213 is conserved among different species (Fig. S2). Therefore, the combination of the phylogenetic tree and transmembrane domain analysis shows that the sequence evolution relationship of O213 among other species is highly conserved.
Variation analysis of o213 in GWAS population
We sequenced the coding regions of 65 materials, including 30 tropical and 35 temperate inbred lines, and compared them with the CDS of ZmYSL2 in the B73 reference genome. We found that o213 had a 3-bp AGC deletion at 110–112 of the first exon of temperate inbred lines, while there was no AGC deletion in tropical inbred lines (Table S2). A survey of 65 materials in the GWAS population revealed a clear bias in 3-bp AGC deletions between temperate and tropical inbred lines: ~ 14.2% of temperate inbred lines had 3-bp AGC deletion, but no 3-bpAGC deletions in tropical inbred lines. This result showed that this 3-bp deletion might be an artificial selection for maize genetic improvement. This 3-bp deletion resulted in glutamine (Q) deletion at position 37 in the O213 amino acid sequence without causing premature termination of the amino acid sequence (Fig. S5). Subsequently, we observed the phenotype of temperate inbred lines with a 3-bp deletion and found no difference (Fig. S3). We analyzed the sequence of temperate inbred lines with 3-bp deletion and found 4 genotypes (1, 2, 3, 4) (Fig. 4B).
Then, we analyzed the agronomic traits and quality content data of four genotype temperate inbred lines with 3-bp deletion at three test sites (Chongzhou, China; Hongya, China; Yunnan, China) (Zhang et al. 2016). We found that among these traits, there are 27 significant differences between genotype 1 and genotypes 2, 3, and 4; there are 27 significant differences between genotype 2 and genotypes 1, 3, and 4. There are 20 significant differences between genotype 3 and genotypes 1, 2, and 4; there are 24 significant differences between genotype 4 and genotypes 1, 2, and 3 (Table S3). At the same time, we found that there are significant differences between genotype 1 and genotypes 2 and 4 in ear length (EL), ear diameter (ED), row number per ear (RN), kernel depth (KD), and oil content (OC). In kernel number per row (KN) and kernel width (KW), genotype 1 and genotype 4 are significantly different. In cob weight (CW), genotype 1 differs significantly from genotypes 2 and 3. In kernel thickness (KT), genotype 1 is significantly from genotypes 3 and 4. In kernel radio (KR), starch content (SC), and glutamic acid content (GC), genotype 1 differs significantly from genotypes 2, 3, and 4. In protein content (PC), genotype 1 and genotype 3 are significantly different. In dry matter weight (DM), genotypes 1 and 2 are significantly different (Table S3). In ear length, cob weight, kernel radio, yield (Y), dry matter weight, starch content, and glutamic acid content, genotype 2 is significantly different from genotype 1. In ear diameter, row number per ear, and kernel depth, there is a significant difference between genotype 2 and genotype 1, 3, 4. In oil content, there are significant differences between genotypes 2 and genotypes 1 and 3. In moisture content (MC), genotype 2 differs significantly from genotype 3. In lysine content (LC) and kernel width, genotype 2 and type 4 genotypes are significantly different. In kernel thickness and kernel number per row, genotype 2 is significantly different from genotype 4 (Table S3). In ear diameter, row number per ear, kernel depth, and oil content, genotype 3 is significantly different from genotype 2, 4. There are significant differences between genotype 3 and genotype 1 in cob weight, kernel radio, kernel thickness, and starch content. In moisture content, kernel width, and lysine content, genotype 3 and genotype 2 are significantly different. In protein content and glutamate content, genotype 3 is significantly different from genotypes 1 and 4 (Table S3). There are significant differences between genotype 4 and genotype 1 in ear length, kernel radio, and starch content. There are significant differences between genotype 4 and genotypes 1, 2, and 3 in ear diameter, row number per ear, and kernel depth. In oil content and glutamic acid content, genotype 4 is significantly different from genotypes 1, and 3. In protein content, genotype 4 is significantly different from genotype 3. In kernel number per row, kernel width, and kernel thickness, genotype 4 differs significantly from genotypes 1 and 2. In lysine content, genotype 4 is significantly different from genotype 2 (Table S3). These results indicate that genotypes 1 and 2 significantly impact kernel traits and quality content. Genotypes 3 and 4 have a minor effect on kernel traits and quality content.
Expression patterns of o213 and subcellular location
To detect the expression patterns of o213 and WT, we analyzed the qRT-PCR results of o213 and WT. The results showed that o213 is expressed in various tissues (Fig. 5A). Compared with the WT, the expression of o213 peaked in the endosperm at 18DAP and began to decrease at 21DAP. The expression of o213 increased significantly in the mutant’s taproots, leaves, stalks, ears, and anthers. The expression of o213 significantly reduced in the seeds of the mutant. To explore the subcellular location of o213. We fused the full-length coding regions of o213 and WT without stop codons with 2 × 35S: pHB-YFP, and expressed them transiently in tobacco leaves. WT-YFP and O213-YFP fluorescence were co-localized with the fluorescence of the plasma membrane marker OsPIP2.1-mCherry, and GFP was used as a control (Fig. 5B). Collectively, o213 is a membrane protein, and o213 is expressed in various tissues.
Expression pattern of the o213 gene in WT and mutant and subcellular localization. a The expression pattern of the o213 gene in different tissues in WT and mutant. Data are means ± SD of three biological replicates. (**) P < 0.01, as determined by the paired t-test. b Subcellular localization of o213. The O213-YFP fusion and WT-YFP fusion were transiently expressed in tobacco epidermal cells, and observe the YFP signal with the confocal microscope, GFP as a control. The plasma membrane marker was OsPIP2.1-mCherry. Capture images on day 2 of infiltration (bars = 50 µm)
Altered expression of nutritional quality and kernel development-related genes in the o213 mutant
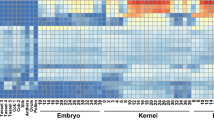
To explore the effects of o213 on endosperm development and nutritional quality. Immature WT and o213 mutant endosperm (12DAP) were used for RNA-Seq analysis. The sequencing results showed a total of 3924 significantly differentially expressed genes (DEGs) after filtering. There were 1942 upregulated expressed genes among these differentially expressed genes and 1982 downregulated expressed genes (Fig. 6A, B). Gene ontology (GO) enrichment analysis showed that the downregulated genes included lipid storage, seed maturation, and dormancy. The upregulated genes included zinc II ion transport and iron ion homeostasis (Fig. 6C). In the lipid storage term, 5 DEGs were downregulated. In the seed dormancy and maturation terms, 2 DEGs were downregulated in the mutant kernels (Dwarf8 (D8), Zm00001d033680; DELLA protein SLN1-like, Zm00001d044065). In the zinc II ion transport term, 10 DEGs were upregulated in the mutant kernels (ZIP4, Zm00001d049375; zinc transporter 2, Zm00001d029497; zinc transporter 5, Zm00001d038437). The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of DEGs shows that o213 is involved in the synthesis of nutrients such as starches, sugars, lipids, and amino acids were involved, as well as hormone synthesis and signal transduction (Fig. 6D, E). The upregulated DEGs were related to the tricarboxylic acid cycle (TCA) cycle (aconitase3, Zm00001d048358 and succinate dehydrogenase4, Zm00001d007966), starch and sucrose metabolism (fructokinase1, Zm00001d042536), fatty acid metabolism (acetyl-CoA carboxylase2 (ACC1), Zm00001d004125), and amino acid metabolism (beta-alanine synthase1 (bas1), Zm00001d021506; 4-hydroxyphenylpyruvate dioxygenase 1 (hppd1), Zm00001d015356). The downregulated DEGs were related to plant hormone biosynthesis and signal transduction including (cis-zeatin O-glucosyltransferase1 (czog1), Zm00001d000237; cytokinin oxidase10 (CKX10), Zm00001d032046; auxin import carrier1 (aic1), Zm00001d030310; D8, Zm00001d033680; bZIP-transcription factor 92 (bZIP92), Zm00001d022550; and Aux/IAA-transcription factor 8(iaa8), Zm00001d039624).
Transcriptome data analysis of gene expression in o213 mutant. a Heatmap of DEGs between o213 mutant and WT. b Volcano of DEGs between in o213 mutant and WT. c GO term enrichment of the DEGs; GO terms contain biological processes (BP), molecular function (MF), and cellular component (CC) categories. d KEGG pathway enrichment of upregulated genes and downregulated genes. e KEGG pathway enrichment of upregulated genes and downregulated genes
Discussion
Opaque endosperm mutants are essential materials for studying starch biosynthesis, endosperm development mechanisms, and quality protein breeding. This study characterized a novel mutant allele of ZmYSL2, o213. Besides, we found a reported mutant allele of ZmYSL2, Sh4. The sh4 mutant contains a 1-bp insertion in the ZmYSL2 gene, resulting in a frameshift and premature stoppage of translation (He et al. 2021). The phenotype of the o213 mutant is similar to that of the previously reported sh4 mutant and zmysl2 mutant, but the o213 mutation site is new. The mutation site of o213 is located in the sixth predicted transmembrane domain (Fig. S2). The subcellular localization of o213 is situated on the membrane (Fig. 5B). We speculate that the replacement of this single base at the front end of the sixth transmembrane domain may not affect the positioning of the membrane (Fig. S2). Both methionine and valine are non-polar, hydrophobic amino acids in terms of amino acids. The mutation of methionine in O213 to valine may cause changes in the protein structure, affecting the function of O213 (Fig. S4) (Albenne et al. 2009). The qRT-PCR results showed that o213 was expressed in different tissues of WT and mutants (Fig. 5A), but o213 was highly expressed in the stalk and anther tissues of mutants, which is different from that previously reported (Zang et al. 2020).
The nutritional contents of the endosperm are essential for the development of maize kernels. Endosperm mutant affects the lysine content in the kernel. Lysine is a vital amino acid in maize kernels, but it is present in low amounts (Massey 2003). Therefore, a change in lysine content has research value in maize endosperm mutants. In addition, lysine content is divided into three categories in maize endosperm mutants (Segal et al. 2003). First, the lysine content in the o2 mutant is 2 times higher than that in the WT. Second, the lysine content of the o11 mutant is 1.8 times higher than that in the WT. Third, the lysine content in the o5 mutant and o9 mutant is 1.4 times higher than that of the WT (Segal et al. 2003). The lysine content in the endosperm of the o213 mutant is different from that of the zmysl2 mutant and sh4 mutant. The lysine content of the o213 mutant was 1.7 times higher than that of the WT, indicating that the o213 mutant belongs to the second classification category of lysine content. In other words, the increase of lysine content in o213 endosperm can improve kernels’ nutritional quality and value. The advent of quality protein maize has shown more advantages than ordinary maize in yield, agronomic characteristics, kernel texture, lysine and tryptophan contents (Vasal 2000). Therefore, the increase in lysine contents in the o213 endosperm may provide more possibilities for QPM breeding. According to reports, zinc is vital for seed development and germination (Ozturk et al. 2006; Boi et al. 2020; Ullah et al. 2020; Tunes et al. 2015). In previous reports, the zinc concentration of the zmysl2 mutant was significantly reduced in the kernels and embryos, but there was no difference in the endosperm (Zang et al. 2020). The zinc concentration of the sh4 mutant is significantly reduced in the kernel, endosperm, and embryo (He et al. 2021). The zinc concentration in the o213 mutant was similar to the zinc concentration in the sh4 mutant, showing significant decreases in the kernels, embryos, and endosperm (Fig. S1). This reduction in zinc concentration will hinder protein synthesis and cause a significant accumulation of amino acids (Falchuk et al. 1978; Snehi Dwivedi and Takkar 1974).
Gene selection is a process of natural evolution. We amplified the coding region of the o213 gene in 65 materials and found a 3-bp AGC deletion in temperate inbred lines but not in tropical inbred lines. Our results found that temperate inbred lines with 3-bp deletion have four genotypes (Fig. 4B). Then, we explored the transmembrane domains and protein structures of the four genotypes, with ZmYSL2 as a control. We found that the 3-bp deletion in the 4 genotypes was not in the transmembrane domain (Fig. S5). However, we found that genotype 1 is a type of protein structure, and genotypes 2, 3, and 4 are a type of protein structure, compared with the protein structure of ZmYSL2 (Fig. S6). We speculate that the 3-bp deletion may not affect the protein function of O213, and other amino acid mutations may cause the difference in the protein structure of the four genotypes. We performed genotype analysis on temperate materials with a 3-bp deletion and found that genotype 1 has higher lysine content, glutamic acid content, and oil content than genotypes 2, 3, and 4. Currently, molecular markers have been widely used in molecular breeding. In quality protein maize breeding, o2 was used as a molecular marker transferred to two early-maturing inbred lines. Early-maturing QPM and Vivek QPM were produced through hybridization, and their tryptophan contents increased by 30% and 40%, respectively (Gupta et al. 2009). In addition, molecular markers can screen for alleles (Chen et al. 2010) and restorer lines that are highly resistant to bacterial blight (Deng et al. 2007). Therefore, our results reveal that genotype 1 with 3-bp deletion is conducive to molecular marker-assisted selection (MAS) breeding (Zhang et al. 2005).
Many genes play a role in the kernel filling stage (Jin et al. 2013). Transcriptome sequencing showed that DEGs were mainly involved in nutritional metabolism, hormone synthesis, and zinc II ion transport (Fig. 6E). Our results found that the expression levels of differentially expressed genes related to nutrient metabolism and the contents of biochemical indicators had the same trend. Therefore, we speculate that the kernel quality of the o213 mutant may be regulated by the expression of genes related to kernel development. In addition, zinc is mainly stored in maize kernels and combined with phytic acid in endosperm protein bodies (Mazzolini et al. 1985). Zinc binds tightly with phytic acid to form a stable protein-zinc-phytic acid complex. When seeds germinate, the phytic acid is rapidly hydrolyzed by phytase, releasing zinc for plant growth and development. We speculate that the zinc-related transporter gene in the endosperm may promote the transportation of zinc from the endosperm to the embryo during zinc transport. As the zinc concentration in the seeds and embryos in the o213 mutant is significantly reduced, an insufficient zinc supply during seed germination may affect the plants’ growth and development.
References
Albenne C, Canut H, Boudart G, Zhang Y, San Clemente H, Pont-Lezica R, Jamet E (2009) Plant cell wall proteomics: mass spectrometry data, a trove for research on protein structure/function relationships. Mol Plant 2:977–989. https://doi.org/10.1093/mp/ssp059
Bhatnagar S, Betrán F, Rooney L (2004) Combining abilities of quality protein maize inbreds. Crop Science - CROP SCI 44(6):1997–2005. https://doi.org/10.2135/cropsci2004.1997
Bogs J, Bourbouloux A, Cagnac O, Wachter A, Rausch T, Delrot S (2003) Functional characterization and expression analysis of a glutathione transporter, BjGT1, from Brassica juncea: evidence for regulation by heavy metal exposure. Plant, Cell Environ 26:1703–1711. https://doi.org/10.1046/j.1365-3040.2003.01088.x
Boi M, Porceddu M, Cappai G, Giudici G, Bacchetta G (2020) Effects of zinc and lead on seed germination of Helichrysum microphyllum subsp. tyrrhenicum, a metal-tolerant plant. Int J Environ Sci Technol 17:1917–1928. https://doi.org/10.1007/s13762-019-02589-9
Chen F, Zhu SW, Xiang Y, Jiang Y, Cheng BJ (2010) Molecular marker-assisted selection of the ae alleles in maize. Genetics and Molecular Research : GMR 9:1074–1084. https://doi.org/10.4238/vol9-2gmr799
Curie C, Panaviene Z, Loulergue C, Dellaporta S, Briat J-F, Walker E (2001) Maize Yellow Stripe 1 (YS 1) encodes a membrane protein directly involved in Fe(III) uptake. Nature 409:346–349. https://doi.org/10.1038/35053080
Dangol S, Singh R, Chen Y, Jwa NS (2017) Visualization of multicolored in vivo organelle markers for co-localization studies in Oryza sativa. Mol. Cells 40(11):828–836. https://doi.org/10.14348/molcells.2017.0045
Delrot S, Atanassova R, Maurousset L (2000) Regulation of sugar, amino acid and peptide plant membrane transporters. Biochem Biophys Acta 1465:281–306. https://doi.org/10.1016/S0005-2736(00)00145-0
Deng Q, Shi-Quan W, Al-Ping Z, Hong-Yu Z, Ping L (2007) Breeding rice restorer lines with high resistance to bacterial blight by using molecular marker-assisted selection. Int Rice Res Notes 32
Ester R, Ang J (2021) Embryo and endosperm function and failure in Solanum species and hybrids.
Falchuk KH, Hardy C, Ulpino L, Vallee BL (1978) RNA metabolism, manganese, and RNA polymerases of zinc-sufficient and zinc-deficient Euglena gracilis. Proc Natl Acad Sci 75(9):4175. https://doi.org/10.1073/pnas.75.9.4175
Feng F, Qi W, Lv Y, Yan S, Xu L, Yang W, Yuan Y, Chen Y, Zhao H, Song R (2018) OPAQUE11 is a central hub of the regulatory network for maize endosperm development and nutrient metabolism. Plant Cell 30(2):375–396. https://doi.org/10.1105/tpc.17.00616
Gupta H, Pk A, Mahajan V, Gs B, A K, P V, A S, Saha S, Babu R, Mc P, Vp M, (2009) Quality protein maize for nutritional security: rapid development of short duration hybrids through molecular marker assisted breeding. Curr Sci 96:230–237
He Y, Yang Q, Yang J, Wang Y-F, Sun X, Wang S, Qi W, Ma Z, Song R (2021) shrunken4 is a mutant allele of ZmYSL2 that affects aleurone development and starch synthesis in maize. Genetics. https://doi.org/10.1093/genetics/iyab070
Janick J (2021) New crops and the search for new food resources.
Jin X, Fu Z, Ding D, Li W, Liu Z, Tang J (2013) Proteomic identification of genes associated with maize grain-filling rate. PLoS ONE 8:e59353. https://doi.org/10.1371/journal.pone.0059353
Kiesselbach TA (1980) The structure and reproduction of corn. Research Bulletin 161:1–96
Letchworth M, Lambert R (1998) Pollen parent effects on oil, protein, and starch concentration in maize kernels. Crop Science - CROP SCI 38(2):363–367. https://doi.org/10.2135/cropsci1998.0011183X003800020015x
Lubkowitz M (2006) The OPT family functions in long-distance peptide and metal transport in plants. Genet Eng 27:35–55. https://doi.org/10.1007/0-387-25856-6_3
Lubkowitz M, Hauser L, Breslav MF, Naider F, Becker J (1997) An oligo transport gene from Candida albicans. Microbiology (reading, England) 143(Pt 2):387–396. https://doi.org/10.1099/00221287-143-2-387
Massey L (2003) Dietary animal and plant protein and human bone health: a whole foods approach. J Nutr 133:862S-865S. https://doi.org/10.1093/jn/133.3.862S
Mertz E, Bates LSET, Nelson O (1964) Mutant gene that changes protein composition and increases lysine content of maize endosperm. Science (new York, NY) 145:279–280. https://doi.org/10.1126/science.145.3629.279
Myers AM, James MG, Lin Q, Yi G, Stinard PS, Hennen-Bierwagen TA, Becraft PW (2011) Maize opaque5 encodes monogalactosyldiacylglycerol synthase and specifically affects galactolipids necessary for amyloplast and chloroplast function. Plant Cell 23(6):2331–2347. https://doi.org/10.1105/tpc.111.087205
Olsen O-A (2020) The modular control of cereal endosperm development. Trends Plant Sci 25(3):279–290. https://doi.org/10.1016/j.tplants.2019.12.003
Ozturk L, Yazici A, Yucel C, Alkantorun A, Çekiç C, Bağcı S, Ozkan H, Braun HJ, Sayers Z, Cakmak I (2006) Concentration and localization of zinc during seed development and germination in wheat. Physiol Plant 128:144–152. https://doi.org/10.1111/j.1399-3054.2006.00737.x
Sabelli PA, Larkins BA (2009) The development of endosperm in grasses. Plant Physiol 149(1):14–26. https://doi.org/10.1104/pp.108.129437
Schaaf G, Ludewig U, Erenoglu BE, Mori S, Kitahara T, von Wiren N (2004) ZmYS1 functions as a proton-coupled symporter for phytosiderophore- and nicotianamine-chelated metals. J Biol Chem 279(10):9091–9096. https://doi.org/10.1074/jbc.M311799200
Schmidt R, Burr F, Burr B (1987) Transposon tagging and molecular analysis of the maize regulatory locus opaque-2. Science (new York, NY) 238:960–963. https://doi.org/10.1126/science.2823388
Segal G, Song R, Messing J (2003) A new opaque variant of maize by a single dominant RNAi-inducing transgene. Genetics 165:387–397. https://doi.org/10.1093/genetics/165.1.387
Sene M, Thevenot C, Prioul JL (1997) Simultaneous spectrophotometric determination of amylose and amylopectin in starch from maize kernel by multi-wavelength analysis. J Cereal Sci 26(2):211–221. https://doi.org/10.1006/jcrs.1997.0124
Snehi Dwivedi R, Takkar PN (1974) Ribonuclease activity as an index of hidden hunger of zinc in crops. Plant Soil 40(1):173–181. https://doi.org/10.1007/BF00011420
Stacey MG, Koh S, Becker J, Stacey G (2002) AtOPT3, a member of the oligopeptide transporter family, is essential for embryo development in Arabidopsis. Plant Cell 14(11):2799–2811. https://doi.org/10.1105/tpc.005629
Stacey MG, Osawa H, Patel A, Gassmann W, Stacey G (2006) Expression analyses of Arabidopsis oligopeptide transporters during seed germination, vegetative growth and reproduction. Planta 223(2):291–305. https://doi.org/10.1007/s00425-005-0087-x
Tunes L, Muniz M, Torales-Salinas J, Suarez-Castellanos C, Lemes E (2015) Response of wheat (Triticum aestivum L.) seedlings to seed zinc application. Agrociencia 49:623–636
Ullah A, Farooq M, Nadeem F, Rehman A, Nawaz A, Naveed M, Wakeel A, Hussain M (2020) Zinc seed treatments improve productivity, quality and grain biofortification of desi and kabuli chickpea (Cicer arietinum). Crop Pasture Sci 71:725–738. https://doi.org/10.1071/CP19266
Vasal SK (2000) High quality protein corn. In. pp 85–129
von Wirén N, Klair S, Bansal S, Briat JF, Khodr H, Shioiri T, Leigh RA, Hider RC (1999) Nicotianamine chelates both Fe<sup>III</sup> and Fe<sup>II</sup> Implications for metal transport in plants. Plant Physiology 119(3):1107. https://doi.org/10.1104/pp.119.3.1107
von Wiren N, Marschner H, Romheld V (1996) Roots of iron-efficient maize also absorb phytosiderophore-chelated zinc. Plant Physiol 111(4):1119. https://doi.org/10.1104/pp.111.4.1119
von Wiren N, Mori S, Marschner H, Romheld V (1994) Iron inefficiency in maize mutant ys1 (Zea mays L. cv Yellow-Stripe) is caused by a defect in uptake of iron phytosiderophores. Plant Physiology 106 (1):71. https://doi.org/10.1104/pp.106.1.71
Wang C, Kovacs M (2002) Swelling index of glutenin test. II. Application in prediction of dough properties and end-use quality 1. Cereal Chemistry - CEREAL CHEM 79:190–196. https://doi.org/10.1094/CCHEM.2002.79.2.190
Wang G, Sun X, Wang G, Wang F, Gao Q, Sun X, Tang Y, Chang C, Lai J, Zhu L, Xu Z, Song R (2011) Opaque7 encodes an acyl-activating enzyme-like protein that affects storage protein synthesis in maize endosperm. Genetics 189(4):1281–1295. https://doi.org/10.1534/genetics.111.133967
Wang G, Wang F, Wang G, Wang F, Zhang X, Zhong M, Zhang J, Lin D, Tang Y, Xu Z, Song R (2012) Opaque1 encodes a myosin XI motor protein that is required for endoplasmic reticulum motility and protein body formation in maize endosperm. Plant Cell 24(8):3447–3462. https://doi.org/10.1105/tpc.112.101360
Williams L, Miller A (2001) Transporters responsible for the uptake and partitioning of nitrogenous solutes. Annu Rev Plant Physiol Plant Mol Biol 52:659–688. https://doi.org/10.1146/annurev.arplant.52.1.659
Wu Y, Messing J (2014) Proteome balancing of the maize seed for higher nutritional value. Front Plant Sci 5:240. https://doi.org/10.3389/fpls.2014.00240
Yao D, Qi W, Li X, Yang Q, Yan S, Ling H, Wang G, Wang G, Song R (2016) Maize opaque10 encodes a cereal-specific protein that is essential for the proper distribution of zeins in endosperm protein bodies. PLOS Genet 12(8):e1006270. https://doi.org/10.1371/journal.pgen.1006270
Yen M-R, Tseng YH, Saier MH (2001) Maize Yellow Stripe 1, an iron-phytosiderophore uptake transporter, is a member of the oligopeptide transporter (OPT) family. Microbiology (reading, England) 147:2881–2883. https://doi.org/10.1099/00221287-147-11-2881
Zang J, Huo Y, Liu J, Zhang H, Liu J, Chen H (2020) Maize YSL2 is required for iron distribution and development in kernels. J Exp Bot 71(19):5896–5910. https://doi.org/10.1093/jxb/eraa332
Zhang MY, Bourbouloux A, Cagnac O, Srikanth CV, Rentsch D, Bachhawat AK, Delrot S (2004) A novel family of transporters mediating the transport of glutathione derivatives in plants. Plant Physiol 134(1):482–491. https://doi.org/10.1104/pp.103.030940
Zhang S, Ni D, Yi C, Li L, Wang X, Wang Z, Yang J (2005) Reducing amylose content of indica rice variety 057 by molecular marker-assisted selection. Zhongguo Shuidao Kexue 19(5):467–470
Zhang X, Zhang H, Li L, Lan H, Ren Z, Liu D, Wu L, Liu H, Jaqueth J, Li B, Pan G, Gao S (2016) Characterizing the population structure and genetic diversity of maize breeding germplasm in Southwest China using genome-wide SNP markers. BMC Genomics 17(1):697. https://doi.org/10.1186/s12864-016-3041-3
Funding
This research was funded and supported by China Agriculture Research System of MOF and MARA, and Sichuan Science and Technology Support Project (2021YFYZ0020, 2021YFYZ0027, 2021YFFZ0017).
Author information
Authors and Affiliations
Contributions
Y.W. designed the research and performed the experiment, analyzed data, and wrote the manuscript. H.H., H.Z., H.Z., Z.Z., and J.G. investigate the agronomic traits. D.L. and L.W. provide reagents and consumables. S.G. and D.G. contributed to plant materials management. X.Z. and B.L. revised the manuscript. S.G. contributed to critically reading this manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.

11032_2022_1278_MOESM1_ESM.jpg
Figure S1. Comparison of the difference of metal ion concentration between WT and o213 kernels a Copper concentration of WT and o213 mutant in embryo, endosperm and seed of 18DAP and 27DAP. b Manganese concentration of WT and o213 mutant in embryo, endosperm and seed of 18DAP and 27DAP. c Zinc concentration of WT and o213 mutant in embryo, endosperm and seed of 18DAP and 27DAP. Student's t test, (*) P<0.05, (***) P<0.001, and n, not significant. (JPG 227 KB)




11032_2022_1278_MOESM5_ESM.jpg
Figure S5. Analysis of the transmembrane domain of the four genotypes, ZmYSL2 was used as a control. The red box is the deleted amino acid (JPG 862 KB)

11032_2022_1278_MOESM7_ESM.pdf
Table S1 Information on specific primers for gene mapping, cloning, qRT–PCR, subcellular localization and evolution analysis (PDF 110 KB)
11032_2022_1278_MOESM9_ESM.pdf
Table S3 Related agronomic traits and quality content data of the four genotypes, t-test among the traits of the four genotypes (PDF 23 KB)
Rights and permissions
About this article
Cite this article
Wang, Y., Zhang, X., Luo, B. et al. Identification of a new mutant allele of ZmYSL2 that regulates kernel development and nutritional quality in maize. Mol Breeding 42, 7 (2022). https://doi.org/10.1007/s11032-022-01278-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-022-01278-9