Abstract
Recent suggestions for mandated lowering of nicotine content in cigarettes have prompted tobacco breeders to search for N. tabacum germplasm with allelic variability contributing to low alkaloid accumulation. In this research, we phenotyped a series of 81 selected diverse tobacco introductions (TIs) to identify a sub-group with authentic low alkaloid phenotypes. We also genotyped these materials for sequences associated with the Nic1 and Nic2 loci previously reported to influence tobacco alkaloid biosynthesis. Only five low alkaloid TIs possessed previously described deletions of Ethylene Response Factor (ERF) genes at the Nic2 locus that contribute to lower alkaloid accumulation. Eleven TIs possessed an apparent deletion of ERF199, a gene recently reported to underlie the effect at the Nic1 locus. Quantitative trait locus (QTL) mapping was performed using populations derived from three selected low alkaloid TIs to possibly identify new genomic regions affecting alkaloid accumulation. A major QTL was identified on linkage group 7 in all three populations that aligned with the Nic1 locus. A newly discovered 5 bp deletion in the gene MYC2a on linkage group 5 was found to likely partially underlie the ultra-low alkaloid phenotype of TI 313. This new information is useful for tobacco breeders attempting to assemble novel genetic combinations with the potential for meeting future levels of tolerance for nicotine concentration in cigarette tobacco.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tobacco, Nicotiana tabacum L., produces a diversity of plant natural products that affect how members of the species interact with their environment. Such chemistry also affects sensory characteristics for users of derived tobacco products. Perhaps the most studied tobacco natural product is nicotine, which serves as a deterrent to insect herbivory and as a stimulant to consumers of tobacco products. Nicotine is usually the most abundant pyridine alkaloid found in tobacco leaves, with nornicotine, anatabine, and anabasine typically being found in much lower amounts. However, nornicotine can predominate in plants highly expressing the nicotine demethylase gene, CYP82E4 (Lewis et al. 2010).
The concentration of nicotine in cured leaves of commercial tobacco cultivars can range from ~ 10 to 60 mg/g (1.0 to 6.0% on a dry weight basis), depending on market type, growing conditions, management approaches, and stalk position (Lewis 2018; Henry et al. 2019; Lewis et al. 2020). As part of an overall strategy to reduce human addiction to combustible cigarettes and exposure to tobacco-related smoke toxicants, the World Health Organization (WHO) has recommended that nicotine levels in cigarette filler be reduced to below 0.4 mg/g (WHO 2015). However, there are currently no commercially available tobacco cultivars that routinely produce this ultra-low level of nicotine, when averaged over all stalk positions.
Various types of genetic variation have been evaluated for their utility in reducing nicotine levels in cured tobacco leaves (Lewis 2018). First, substantial naturally occurring genetic variation for alkaloid accumulation has been observed among diverse tobacco materials (Sisson and Saunders 1982). Tobacco alkaloid accumulation is considered to be under complex genetic control, but two loci with large effects, Nic1 and Nic2 (sometimes also designated as the A and B loci), have been described as having a large influence on the accumulation of all four major pyridine alkaloids (Legg and Collins 1971). The Nic2 locus has been found to be comprised of a series of genes encoding for Ethylene Response Factor (ERF) transcription factors on linkage group 19 that globally influence the expression of genes in the alkaloid biosynthesis pathway (Shoji et al. 2010). This cluster of genes has been found to be deleted in ‘LA Burley 21’ (a backcross-derived, nic1/nic1 nic2/nic2 version of Burley 21). Increased insight has also recently been gained on the nature of genetic variability underlying the effect at the Nic1 locus located on linkage group 7 (partially homologous to linkage group 19), where a series of similar genes coding for ERF transcription factors also reside (Adams et al. 2016, 2019; Humphry et al. 2018; Pramod et al. 2019a, b; Sui et al. 2020b; Qin et al. 2021). Qin et al. (2021) suggest that an epigenetically silenced allele of ERF199 explains the effect of the Nic1 locus in LA Burley 21, an outcome that could potentially be influenced by a large neighboring deletion (Sui et al. 2020b). Naturally occurring variability affecting transcription of CYP82E4 can also affect nicotine levels by virtue of increasing/decreasing its demethylation to form nornicotine (Lewis et al. 2010). Conversion of nicotine to nornicotine is not an overly attractive method to reduce nicotine levels in the tobacco plant, however, because of increased tendency for nornicotine to form its corresponding carcinogenic tobacco-specific nitrosamine (TSNA), N’-nitrosonornicotine (NNN) (Lewis et al. 2008).
De novo genetic variation generated via induced mutation, gene editing, or RNA interference has also been evaluated for its potential to affect alkaloid levels and relative composition (Chintapakorn and Hamill 2003; Xie et al. 2004; Kudithipudi et al. 2017; Lewis 2018; Lewis et al. 2015, 2020). Although these publications describe the development of tobacco genotypes with dramatically lower potential for nicotine accumulation, such tobaccos do not routinely accumulate nicotine at levels below that suggested by the WHO under conventional methods of cultivation. Commercial growing of tobacco cultivars derived via genetic engineering or gene editing is also complicated by a diversity of regulatory requirements in different tobacco-producing countries.
Due to a general lack of intellectual property or regulatory concerns, there is continued interest in identifying naturally occurring genetic variability that could be used to assist tobacco breeders attempting to lower cured leaf nicotine to levels being recommended by the WHO. Substantial phenotypic variability has been reported for alkaloid accumulation among 1,091 diverse tobacco introductions (TIs) maintained by the US Nicotiana Germplasm Collection (Sisson and Saunders 1982). Some materials have been reported to accumulate nicotine at levels below that for ‘LAFC53’ (https://npgsweb.ars-grin.gov/gringlobal/site.aspx?id=25), a nic1/nic1 nic2/nic2 nearly isogenic version of conventional flue-cured tobacco cultivar ‘NC 95’ (Chaplin 1975). All of these materials were not previously compared in a common environment, however, and some TIs are not believed to be authentic low alkaloid accumulators. A re-evaluation of materials previously reported to accumulate nicotine at low levels seems justified. It seems likely that allelic variability with large effects on nicotine accumulation, besides that present at the Nic1 and Nic2 loci, exists within the US Nicotiana Germplasm Collection.
The first objective of the research described here was to verify the potential for alkaloid accumulation in a subset of 81 putative low alkaloid TIs in a common environment and using common experimental methodology. A second objective was to genotype these materials to gain insight into their genetic condition at the Nic1 and Nic2 loci. A third objective was to gain insight on the genetic control of the low alkaloid accumulation trait in three verified low alkaloid TIs through quantitative trait locus (QTL) mapping, with the goal of potentially identifying large-effect QTLs affecting alkaloid accumulation alternative to the Nic1 and Nic2 loci. The final objective was to validate newly identified, gene-specific nucleotide variability for its association with the low alkaloid trait. The findings have implications for tobacco researchers with the goal of developing new tobacco cultivars with ultra-low nicotine levels.
Materials and methods
Phenotypic and genotypic evaluations of tobacco introductions
Eighty-one TIs (Table 1) from the US Nicotiana Germplasm Collection (Lewis and Nicholson 2007) previously reported to accumulate nicotine at relatively low levels (Sisson and Saunders 1982; https://npgsweb.ars-grin.gov/gringlobal/site.aspx?id=25) were selected for a 2017 field experiment near Clayton, NC, to authenticate low alkaloid phenotypes in a common environment. Accessions with high levels (greater than ~ 20%) of nicotine conversion, calculated as [((nornicotine content)/(nicotine content + nornicotine content)) × 100], were selected against. Standard flue-cured tobacco cultivar ‘K326’ and LAFC 53 were also included as control genotypes. Ten plants of each genotype were grown in single rows with 56 cm plant and 114 cm row spacing. Individual plots were topped (removal of the apical inflorescence) when ~ 80% of the plants within the plot were flowering. Sucker (lateral meristem) control was achieved via downstalk application of Prime + EC (flumetralin) according to manufacturer’s recommendations (Syngenta Crop Protection, Greensboro, NC). Fourteen days after topping, the top two leaves of each plant were collected, treated with ethephon (a chemical ripening agent used to stimulate expression of nicotine demethylase genes, if active) according to manufacturer’s recommendations (Arysta LifeScience, Cary, NC), air-cured, stripped of their midveins, oven-dried, ground, and analyzed for alkaloid composition using previously described gas chromatographic methodology (Lewis et al. 2015). Total alkaloid content (mg/g) was calculated as the sum of the four major tobacco alkaloids (nicotine, nornicotine, anatabine, and anabasine).
Each of the 81 TIs, K326, and LAFC53 were also genotyped for the presence/absence of three genes (ERF115, ERF179, and ERF189) reported to be deleted at the Nic2 locus in LA Burley 21 using previously reported primers and PCR reaction conditions (Shoji et al. 2010). These materials were also genotyped for the presence/absence of seven sequences (Table 1) reported to be deleted at, or near, the Nic1 locus in LA Burley 21 (Adams et al. 2016); and also for the presence/absence of ERF199 recently suggested to be the causal gene underlying the Nic1 effect on alkaloid accumulation (Qin et al. 2021). Primers used for PCR-based presence/absence assays are provided in Supplementary Table 1.
QTL mapping in three K326 × TI populations
Based upon their low-nicotine phenotypes and lack of an obvious deletion at the Nic2 locus, we selected TI 313, TI 508, and TI 1211 to investigate the potential that they may carry large-effect allelic variability affecting nicotine accumulation at loci other than that conferred by the Nic1 and Nic2 loci. At the time of making these selections, the aforementioned information pertaining to the Nic1 locus was not available to the primary authors.
Each of the three selected TIs was hybridized with standard flue-cured tobacco cultivar K326 during the 2017 growing season. Resulting F1 plants were self-pollinated during 2018 to produce respective F2 populations that were grown in field experiments near Clayton, NC, during the 2019 growing season. Approximately 200 plants per population were grown using previously described plant and row spacings. Each population was surrounded by border plants.
Plants of each population were topped when approximately 80% of plants were in flower. Individual plants were topped above the highest leaf that exceeded ~ 10 cm. Suckers were removed from leaf axils by hand approximately every other day until leaf collection (22 days for the K326 × TI 313 F2 population; 27 days for the K326 × TI 508 and K326 × TI 1211 F2 populations). After this period of time, the top two leaves from every plant were harvested, treated with ethephon according to manufacturer’s recommendations, and air cured for approximately 12 days. Cured leaves were stripped of their midveins, oven-dried, ground, and analyzed for alkaloid profiles using previously described methodology (Lewis et al. 2015).
DNA was isolated from individuals of each of the F2 populations and parental lines using a customized kit designed for LGC oKtopure automated DNA extraction instrumentation (LGC Genomics, Middlesex, UK). Individuals were genotyped using an Affymetrix Axiom custom tobacco array containing 178,000 SNPs by Eurofins BioDiagnostics (River Falls, WI). Raw data sets were processed to remove duplicate loci, monomorphic loci, loci with large fractions of missing data, or that exhibited significant segregation distortion using a Bonferroni correction (P = 0.05 significance threshold). Remaining markers, approximately 6,000 per population, were used for subsequent linkage mapping and QTL analyses.
The R package ‘ASMap’ (Taylor & Butler 2017) with a threshold P value of 1 × 10−9 was used to generate linkage groups corresponding to each of the 24 chromosomes of N. tabacum. The few markers that did not cluster with any of the 24 major linkage groups were removed from the data set. Linkage groups were numbered to reflect their association with the reference genome pseudomolecules of Edwards et al. (2017).
QTL analyses were performed via interval mapping and composite interval mapping for nicotine and total alkaloid accumulation (mg/g) for each of the three populations using the R package R/QTL (Broman and Sen 2009). Both ‘suggestive’ and ‘significant’ logarithm of the odds (LOD) threshold levels (P = 0.05 chromosome-wide and genome-wide significance levels, respectively) were used for QTL identification (Lander and Kruglyak 1995; van Ooijen 1999) and were determined using the 1000 permutation method (Broman et al. 2003). Ninety-five percent Bayesian confidence intervals (CIs) for approximate genomic location of genes underlying major QTLs were also determined.
Identified QTLs were fit to defined QTL models for nicotine and total alkaloid accumulation using the ‘fitqtl’ function in R/QTL, specifying the use of the multiple imputation approach. QTLs were fit to two different models: an additive only model and a model including interactions between QTLs. Model selection was determined by calculating and comparing Akaike information criterion (AIC) scores (Akaike 1974) for the full models. Best fitting models were those with lower AIC scores. Estimated QTL effects were derived for selected models.
Validation of association between newly identified gene-specific nucleotide variation and nicotine accumulation
During the progression of this research, genomic regions of interest were identified that exhibited significant associations with nicotine accumulation. Based upon existing publicly available genome sequence information (Edwards et al. 2017), genes within confidence intervals for discovered QTL were identified. Gene lists were screened for candidate genes of interest (known alkaloid biosynthetic genes; or those encoding for ERF or MYC transcription factors, or defense related proteins). Existing genomic databases and sequencing were used to assess the presence of nucleotide variation in parental materials that could cause altered function of candidate genes.
The association between one gene variant of particular interest (a 5 bp deletion in MYC2a in TI 313) and alkaloid accumulation was studied. First, a Cleaved Amplified Polymorphic Sequence (CAPS) marker was generated to permit identification of all three genotypic classes for this polymorphism. This marker was genotyped using PCR to screen for the presence/absence of the 5 bp deletion (5′-TTCTG-3′) in the MYC2a gene. PCR reactions were carried out in a total reaction volume of 20 μl containing 150 ng DNA, 1× Thermopol buffer, 2 mM MgSO4, 0.4 mM each dNTP, 4 μM each primer (Forward 5′-GTTTTGGCCCGGAACAACTA-3′ and Reverse 5′-CTGAATAGCACATGAGCCCGA-3′), and 1 unit Taq polymerase (New England Biolabs, Beverly, MA). Amplification conditions consisted of an initial denaturation step at 94°C for 2:30 min, followed by 30 cycles of 94°C for 30 s, 63°C for 30 s, and 72°C for 1 min, with a final extension at 72°C for 5 min. Restriction enzyme digests were performed in the same tube as the PCR reaction. The digestion contained the 20 μl amplified product, 1X Cutsmart buffer and 5 units Hpy188i (New England Biolabs) in a total volume of 30 μl. The reaction was incubated at 37°C for 3 h. Products were separated on an ethidium bromide–stained 2% agarose gel.
Association between CAPS marker genotypes and the low nicotine trait was verified using a separate F2 population of 173 individuals derived from the K326 × TI313 cross. This population was evaluated during the 2020 growing season using previously described plant and row spacing. Plants were topped when ~ 80% of plants were in flower, and suckers were controlled via downstalk applications of Prime + . The top leaf of each plant was collected 21 days after topping and analyzed for alkaloid content as previously described. DNA was also collected from each plant, and plants were genotyped using the MYC2a CAPS marker. The association between genotype and nicotine/total alkaloid content was analyzed using PROC TTEST in SAS 9.4. The presence/absence of this newly discovered variation was also assessed in the aforementioned set of 81 TIs.
Results
Preliminary phenotypic and genotypic evaluations
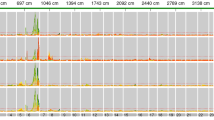
We evaluated 81 selected diverse TIs for alkaloid accumulation after topping for the purpose of potentially identifying those that accumulated lower levels of nicotine relative to the established low alkaloid standard genotype LAFC 53. First, a large number of the selected TIs exhibited nicotine levels that were in the vicinity of that produced by standard flue-cured cultivar ‘K326,’ suggesting that these materials are not authentic low alkaloid genotypes. Several accessions (TI 785, TI 1573, and TI 313) exhibited nicotine levels that were lower than that for LAFC53, and several others were similar in alkaloid phenotype (TI 1562, TI 554, TI 1246, and TI 508, for example) (Fig. 1). These low nicotine phenotypes were not the result of high nicotine conversion, as only a single accession (TI 888) exhibited a high rate of nicotine conversion (Fig. 1).
The 81 selected diverse TIs were also genotyped for the presence/absence or three ERF genes (ERF115, ERF179, and ERF189) previously found to be deleted at the tobacco Nic2 locus in LA Burley 21 (Shoji et al. 2010). The majority of the genotypes did not possess obvious deletions of all three of these genes at the Nic2 locus (Table 1). In fact, only three TIs (TI 1573, TI 554, and TI 1246) were found to possess apparent deletions of all three of these genes. Two TIs (TI 1085 and TI 1228) were found to possess deletions of ERF179 and ERF189, but not ERF115. Based upon the phenotypic and genotypic results, three low-nicotine accessions (TI 313, TI 508, and TI 1211) were selected as not possessing obvious deletions of the ERF genes at the Nic2 locus for the purpose of creating mapping populations.
After the choice of parental materials for mapping populations was made, information became available regarding potential genetic variability at the Nic1 locus that may influence alkaloid accumulation. While LA Burley 21 and LI Burley 21 were found to carry a deletion reported to be associated or linked with the Nic1 locus in LA Burley 21 (Adams et al. 2016), only three low alkaloid TIs (TI 554, TI 1246, and TI 1573) also apparently possessed this deletion (Table 1). Both LA Burley 21 and LI Burley 21 possessed the ERF199 gene recently reported to be responsible for the effect at the Nic1 locus, but ten TIs did not (TI 95, TI 313, TI 508, TI 527, TI 516, TI 554, TI 785, TI 1014, TI 1211, and TI 1562) (Table 1).
QTL mapping
Average nicotine accumulations for the K326 × TI 313, K326 × TI 508, and K326 × TI 1211 F2 populations were 15.8, 27.6, and 23.3 mg/g, respectively (Table 2). Average total alkaloid content for the three populations was 16.3, 29.1, and 24.2 mg/g, respectively. Lastly, average percent nicotine conversion was very low, averaging 1.73, 1.79, and 1.45 for the three populations, respectively (Table 2).
Linkage maps were established for the K326 × TI 313, K326 × TI 508, and K326 × TI 1211 F2 populations, whereby 128, 146, and 107 individuals were genotyped using 5,816, 6,103, and 5,781 polymorphic markers, respectively. Composite interval mapping analysis using a suggestive chromosome-wide significance threshold identified two genomic regions for each F2 population that were strongly associated with nicotine accumulation (Fig. 2; Table 3). Highly significant peaks were identified on linkage group 7 for all three populations. Additionally, significant peaks associated with nicotine accumulation were identified on linkage group 5 in the K326 × TI 313 and K326 × TI 508 populations. Lastly, a significant peak for nicotine content on linkage group 11 was unique to the K326 × TI 1211 F2 population (Fig. 2). In general, LOD score plots for nicotine accumulation roughly overlapped plots for total alkaloid accumulation. Interestingly, a peak exceeding the chromosome-wide threshold level of significance was identified on chromosome 19 for the K326 × TI 313 population for total alkaloid accumulation, whereas a significant peak was not observed in this region for nicotine content. Additionally, a peak significantly associated with nicotine content on linkage group 5 was not identified for total alkaloid accumulation for the K326 × TI 508 population. For all three populations, models considering only additive marker effects for nicotine and total alkaloid accumulation generated lower AIC scores as compared to models in which QTL interactions were included.
LOD score peaks generated from composite interval mapping (CIM) for nicotine (Nic) and total alkaloid (TA) accumulation in A K326 × TI 313, B K326 × TI 508, and C K326 × TI 1211 F2 populations. Horizontal dashed lines are the permutation-derived P = 0.05 chromosome-wide (CW) and genome-wide (GW) threshold levels of significance
The identified QTL on linkage group 7 exhibited the largest effect in each of the three populations and explained 35.3%, 19.7%, and 36.7% of the phenotypic variance for nicotine accumulation in the K326 × TI 313, K326 × TI 508, and K326 × TI 1211 F2 populations, respectively (Table 3). The QTL identified on linkage group 5 explained 15.9% and 5.9% of the phenotypic for nicotine content in the K326 × TI 313 and K326 × TI 508 populations, respectively. Lastly, the identified QTL on linkage group 11 explained 8.1% of the variation for nicotine accumulation for the TI 1211 × K326 population. R2 values for these QTLs for nicotine concentration were similar to those total alkaloid accumulation (Table 3). For the majority of identified QTLs, the allele contributed by the TI contributed to lower nicotine and/or total alkaloid accumulation.
Ninety-five percent Bayesian confidence intervals were determined for identified QTLs. Using markers spanning these intervals, scaffold sequences (Edwards et al. 2017) contained within the intervals were identified and scanned for genes that might reasonably be assumed to be involved in alkaloid biosynthesis or the control thereof. The QTL with the largest effect in each of the three studied populations was that on linkage group 7, previously documented to contain the Nic1 locus (Adams et al. 2016, 2019; Humphry et al. 2018; Pramod et al. 2019a, b; Sui et al. 2020b). Scaffolds within the Edwards et al. (2017) assembly that corresponded to this QTL’s confidence interval contained between 3 and 9 of the ERF genes reported to be associated with the Nic1 locus (Humphry et al. 2018; Pramod et al. 2019a, b; Sui et al. 2020b; Qin et al. 2021).
The Nic2 locus was previously described as being located on linkage group 19 (Shoji et al. 2010; Adams et al. 2016). A significant QTL peak for total alkaloid accumulation was found to be associated with this linkage group only for the K326 × TI 313 population. Sequence data available for markers within this QTL’s confidence interval mapped to a region near the end of linkage group 19 approximately 1 Mb downstream of a cluster of six ERF genes (ERF17L1, ERF179, ERF17, ERF115, ERF104, and ERF221) previously found to be deleted at the Nic2 locus in LA Burley 21 (Shoji et al. 2010; Kajikawa et al. 2017). However, we did not find evidence of the large deletion present in LA Burley 21 in TI 313 (Table 1).
A QTL associated with lower nicotine and total alkaloid accumulation was identified on linkage group 5 in the K326 × TI 313 population. Three sequences (Nitab4.5_0000876g0060, Nitab4.5_0000876g0070, and Nitab4.5_0000876g0080) with a high degree of similarity to JRE6, encoding a jasmonate responsive element present from tomato (Kajikawa et al. 2017), were identified within QTL confidence intervals for this linkage group. Fragments from these regions were PCR amplified and sequenced from K326, TI 313, TI 508, and TI 1211. No sequence variation was identified among these genotypes, however. Additionally, two ERF-like genes (Nitab4.5_0002180g0010.1 and Nitab4.5_0002683g0020.1) were identified within the QTL confidence interval for linkage group 5. A missense mutation was detected within the Nitab4.5_0002180g0010.1 fragment for TI 313, but the corresponding amino acid substitution was assumed to be inconsequential.
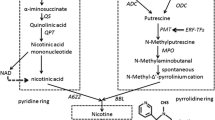
Only 64% of the sequence information of Edwards et al. (2017) is assigned to linkage groups. To increase the possibility of identifying causal DNA variation underlying discovered QTLs, proprietary genome sequences of TI 313 and K326 were compared for genes of interest within the linkage group 5 QTL confidence interval. Of particular interest was a 5 bp deletion identified within MYC2a (GenBank entry GQ859160.1 and gene model Nitab4.5_0002539g0040.1 of Edwards et al. 2017) for TI 313, a gene previously described as a main coordinator of jasmonate-mediated defense response in Arabidopsis (Kazan and Manners 2013) and activator of the expression of tobacco genes involved with alkaloid biosynthesis (Shoji and Hashimoto 2011; Sui et al. 2020a). The 5 bp deletion of this gene in TI 313 causes a frameshift mutation leading to premature termination of the gene product (Fig. 3).
Verification of association between MYC2a polymorphism and alkaloid accumulation
Sequencing was used to verify the authenticity of the 5 bp deletion in MYC2a in TI 313, and its absence in K326, TI 508, TI 1211, LAFC53, and LA Burley 21. The relationship between the deleterious mutation and alkaloid accumulation was verified in a separate K326 × TI 313 F2 population grown during the 2020 field season. Genotyping of 173 individuals with a derived CAPS marker indicated significant (P < 0.05) differences between the genotypic classes, and an additive effect on both nicotine and total alkaloid accumulation (Fig. 4). The presence/absence of the 5 bp deletion was also assessed in the remaining selected 78 TIs initially tested in 2017. The deletion was only found to be present in TI 311, TI 313, TI 384, and TI 407. All of these accessions were initially collected in Columbia in the 1930s (Chaplin et al. 1982).
Discussion
In the event of a mandated lowering of acceptable levels of nicotine content for cigarette tobacco filler to 0.4 mg/g, or below, generation of new cultivars with novel allelic combinations will be required to routinely produce such cured leaf. Due to regulatory and intellectual property issues associated with varietal outcomes of genetic engineering and gene editing, naturally occurring genetic variability is preferred by most breeding programs. To date, three types of large-effect, naturally occurring genetic variability affecting lower nicotine accumulation have been documented: (1) genetics-based demethylation of nicotine to form nornicotine, (2) that conferred by recessive genotypes at the Nic1 locus (Legg and Collins 1971), and (3) that conferred by recessive genotypes at the Nic2 locus (Shoji et al. 2010). Recessive alleles at the Nic1 and Nic2 alleles have already been combined into single breeding lines such as LA Burley 21 (Legg et al. 1970) and LAFC53 (Chaplin 1975), but such materials do not routinely exhibit nicotine levels below the 0.4 mg/g level, when averaged over all stalk positions (Lewis et al. 2020). Conversion of nicotine to nornicotine is not an overly attractive method to reduce nicotine levels in the tobacco plant because of a corresponding increased tendency for accumulation of the carcinogenic TSNA, NNN (Lewis et al. 2008).
In the research described here, we investigated alkaloid accumulation in 81 TIs previously reported to accumulate nicotine at relatively low levels. Many of these TIs were not found to be authentic low alkaloid accumulators. We studied the genetic control of the low-nicotine trait in three selected TIs (TI 313, TI 508, and TI 1211) via QTL mapping. Genetic variation within the vicinity of the Nic1 locus on linkage group 7 was found to have the largest effect on nicotine accumulation in all three of these accessions. None of these materials possessed obvious deletions of genes previously reported to be associated with the Nic1 locus in LA Burley 21 (Adams et al. 2016, 2019; Humphry et al. 2018; Pramod et al. 2019a, b; Sui et al. 2020b). Indeed, only three of the 81 evaluated TIs (TI 554, TI 1246, and TI 1573) apparently possessed this deletion. However, all three of the parental TIs did apparently possess deletions of ERF199 suggested by Qin et al. (2021) to be responsible for the genetic effect on alkaloid accumulation at the Nic1 locus and to be epigenetically silenced in LA Burley 21.
A second major QTL located on linkage group 5 was identified as being associated with the low-nicotine phenotype in accession TI 313. After examination of candidate gene sequences within the confidence interval for this QTL, a 5 bp deletion leading to a truncated gene product was discovered within a gene designated as MYC2a. This deletion was verified to be associated with the low-nicotine trait in a K326 × TI 313 F2 population and was also found to be present in three additional low-nicotine accessions. MYC2a belongs to the four-member MYC2 gene family that also includes MYC1a, MYC1b, and MYC2b. These genes have been shown to be involved in the regulation of jasmonate-inducible nicotine biosynthesis in tobacco and Myc2 transcription factors recognize G-box sequences found in the promoter regions of major nicotine biosynthetic genes (Shoji and Hashimoto 2011). Silencing of MYC2 genes via RNA interference in tobacco leads to a decrease in transcription of jasmonate-responsive structural genes (including alkaloid biosynthesis genes) in tobacco roots, as well as ERF genes associated with the Nic1 and Nic2 locus (Shoji and Hashimoto 2011; Sui et al. 2020a). Mutations introduced into MYC2a in tobacco via gene editing were also previously reported to lead to dramatic reductions (80%) in nicotine in tobacco leaves (Sui et al. 2020a).
The identification of new naturally occurring variability affecting the potential for lower levels of nicotine accumulation should allow for the generation of new allelic combinations with the potential to confer nicotine phenotypes lower than those that are currently observed for breeding lines such as LA Burley 21 and LAFC 53. The overall effect of combining the 5 bp MYC2a deletion identified in TI 313 with recessive alleles at the Nic1 and Nic2 loci is unknown at the current time, but it seems reasonable to predict that further reductions could be achieved over that exhibited by nic1/nic1 nic2/nic2 genotypes by themselves.
Data availability
Relevant raw data will be freely available to any researcher wishing to use them for non-commercial purposes, without breaching participant confidentiality.
References
Adams AC, Lusso MF, Pramod S, Xu D (2016) Compositions and methods for producing tobacco plants and products having altered alkaloid levels. United States Patent Application US2016/0374387. Published Dec. 2016
Adams AC, Lusso MF, Pramod S, Xu D (2019) Compositions and methods for producing tobacco plants and products having altered alkaloid levels. United States Patent Application US2019/0343166. Published Nov. 2019
Akaike H (1974) A new look at the statistical model identification. IEEE Trans Autom Control 19:716–723
Broman KW, Sen S (2009) A guide to QTL mapping with R/qtl, vol 46. Springer, New York. https://doi.org/10.1007/978-0-387-92125-9
Broman KW, Wu H, Sen S, Churchill GA (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19:889–890
Chaplin JF (1975) Registration of LAFC53 tobacco germplasm. Crop Sci 15:282
Chaplin JF, Stavely JR, Litton CC, Pittarelli GN, West WH (1982) Catalog of the tobacco introductions in the US Department of Agriculture's tobacco germplasm collection (Nicotiana tabacum). No. 583.790973 C357c. New Orleans, United States Department of Agriculture, Agricultural Research Service
Chintapakorn Y, Hamill JD (2003) Antisense-mediated down-regulation of putrescine N-methyltransferase activity in transgenic Nicotiana tabacum L. can lead to elevated levels of anatabine at the expense of nicotine. Plant Mol Biol 53:87–105
Edwards KD, Fernandez-Pozo N, Drake-Stowe K, Humphry M, Evans AD, Bombarely A, Allen F, Hurst R, White B, Kernodle SP, Bromley JR, Sanchez-Tamburrino JP, Lewis RS, Mueller LA (2017) A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genomics 18:448
Henry JB, Vann MC, Lewis RS (2019) Agronomic practices affecting nicotine concentration in flue-cured tobacco: a review. Agron J 111:1–9
Humphry ME, Yang S, Qin Q (2018) International Patent Publication WO 2018/237107
Kajikawa M, Sierro N, Kawaguchi H, Bakaher N, Ivanov NV, Hashimoto T, Shoji T (2017) Genomic insights into the evolution of the nicotine biosynthesis pathway in tobacco. Plant Physiol 174:999–1011
Kazan K, Manners JM (2013) MYC2: the master in action. Mol Plant 6:686–703
Kudithipudi C, Qi D, Shen Y, Payyavula R, Xu D, Warek U, Strickland JA (2017) Genetic and biochemical analysis of low alkaloid lines with improved leaf quality. In: CORESTA 2017 Meeting; 22–26 October, 2017; Santa Cruz Do Sul, Brazil (abstract AP12)
Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nature Genet 11:241–247
Legg PD, Collins GB (1971) Inheritance of percent total alkaloids in Nicotiana tabacum L. II. Genetic effects of two loci in Burley 21 × LA Burley 21 populations. Can J Genet Cytol 13:287–291
Legg PD, Collins GB, Litton CC (1970) Registration of LA Burley 21 Tobacco Germplasm Registration No. (GP 8). Crop Sci 10:212–212
Lewis RS (2018) Potential mandated lowering of nicotine levels in cigarettes: a plant perspective. Nicotine Tob Res 2018:1–5
Lewis RS, Nicholson JS (2007) Aspects of the evolution of Nicotiana tabacum L. and the status of the United States Nicotiana Germplasm Collection. Genet Resour Crop Evol 54:727–740
Lewis RS, Jack AM, Morris JW, Robert VJM, Gavilano LB, Siminszky B, Bush LP, Hayes AJ, Dewey RE (2008) RNA interference (RNAi)-induced suppression of nicotine demethylase activity reduces levels of a key carcinogen in cured tobacco leaves. Plant Biotech J 6:346–354
Lewis RS, Bowen SW, Keogh MR, Dewey RE (2010) Three nicotine demethylase genes mediate nornicotine accumulation in tobacco: functional characterization of the CYP82E10 gene. Phytochem 71:1988–1998
Lewis RS, Lopez HO, Bowen SW, Andres KR, Steede WT, Dewey RE (2015) Transgenic and mutation-based suppression of a berberine bridge enzyme-like (BBL) gene family reduces alkaloid content in field-grown tobacco. PloS One 10(2):e0117273
Lewis RS, Drake-Stowe KE, Heim CB, Steede T, Smith W, Dewey RE (2020) A thorough analysis of tobacco genotypes exhibiting reduced nicotine accumulation due to induced mutations in Berberine Bridge Like (BBL) genes. Frontiers Plant Sci 11:368
Pramod S, Lusso MF, Frederick J, Adams AC, Xu D (2019a) Compositions and methods for producing tobacco plants and products having altered alkaloid levels. United States Patent Application US2019/0216037. Published July 2019
Pramod S, Lusso MF, Frederick J, Adams AC, Xu D (2019b) Compositions and methods for producing tobacco plants and products having altered alkaloid levels. United States Patent Application US2019/0246596. Published Aug. 2019
Qin Q, Humphry M, Gilles T, Fisher A, Patra B, Singh SK, Li D, Yang S (2021) NIC1 cloning and gene editing generates low nicotine tobacco plants. Plant Biotech J. https://doi.org/10.1111/pbi.13694
Shoji T, Hashimoto T (2011) Tobacco MYC2 regulates jasmonate-inducible nicotine biosynthesis genes directly and by way of the NIC2-Locus ERF genes. Plant Cell Physiol 52:1117–1130
Shoji T, Kajikawa M, Hashimoto T (2010) Clustered transcription factor genes regulate nicotine biosynthesis in tobacco. Plant Cell 22:3390–3409
Sisson VA, Saunders JA (1982) Alkaloid composition of the USDA tobacco (Nicotiana tabacum L.) introduction collection. Tob Sci 26:117–120
Sui X, He X, Song Z, Gao Y, Zhao L, Jiao F, Kong G, Li Y, Han S, Wang B (2020a) The gene NtMYC2a acts as a ‘master switch’ in the regulation of JA-induced nicotine accumulation in tobacco. Plant Biol. https://doi.org/10.1111/plb.13223
Sui X, Xie H, Tong Z, Zhang H, Song Z, Gao Y, Zhao L, Li W, Li M, Li Y, Li Y, Wang B (2020) Unravel the mystery of NIC1-locus on nicotine biosynthesis regulation in tobacco. bioRxiv. https://doi.org/10.1101/2020.07.04.187922
Taylor J, Butler D (2017) R package ASMap: efficient genetic linkage map construction and diagnosis. arXiv preprint arXiv:1705.06916
Van Ooijen JW (1999) LOD significance thresholds for QTL analysis in experimental populations of diploid species. Heredity 83:613–624
World Health Organization (2015) Advisory note: global nicotine reduction strategy. WHO study group on tobacco product regulation. WHO Press, Geneva
Xie JH, Song W, Maksymowicz W, Jin W, Cheah K, Chen WX, Carnes C, Ke J, Conkling MA (2004) Biotechnology: a tool for reduced risk tobacco products – the nicotine experience from test tube to cigarette pack. Rec Adv Tob Sci 30:17–37
Acknowledgements
This research was partially supported by Altria Client Services.
Author information
Authors and Affiliations
Contributions
NB, AM, SP, JF, TS, SPK, and RSL carried out the research and assisted in the analysis of data. RSL designed the research, helped analyze data, and drafted the manuscript. All co-authors reviewed the manuscript prior to submission.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
No animal or human research subjects were used in this research.
Consent for publication
All authors have given consent to publish results described in this manuscript.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Burner, N., McCauley, A., Pramod, S. et al. Analyses of diverse low alkaloid tobacco germplasm identify naturally occurring nucleotide variability contributing to reduced leaf nicotine accumulation. Mol Breeding 42, 4 (2022). https://doi.org/10.1007/s11032-021-01274-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-021-01274-5