Abstract
Mature pod color (PC) and pod size (PS) served as important characteristics are used in the soybean breeding programs. However, manual phenotyping of such complex traits is time-consuming, laborious, and expensive for breeders. Here, we collected pod images from two different populations, namely, a soybean association panel (SAP) consisting of 187 accessions and an inter-specific recombinant inbred line (RIL) population containing 284 RILs. An image-based phenotyping method was developed and used to extract the pod color- and size-related parameters from images. Genome-wide association study (GWAS) and linkage mapping were performed to decipher the genetic control of pod color- and size-related traits across 2 successive years. Both populations exhibited wide phenotypic variations and continuous distribution in pod color- and size-related traits, indicating quantitative polygenic inheritance of these traits. GWAS and linkage mapping approaches identified the two major quantitative trait loci (QTL) underlying the pod color parameters, i.e., qPC3 and qPC19, located to chromosomes 3 and 19, respectively, and 12 stable QTLs for pod size-related traits across nine chromosomes. Several genes residing within the genomic region of stable QTL were identified as potential candidates underlying these pod-related traits based on the gene annotation and expression profiling data. Our results provide the useful information for fine-mapping/map-based cloning of QTL and marker-assisted selection of elite varieties with desirable pod traits.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soybean (Glycine max L. Merr.) is a crucial crop in terms of its agronomic and economic importance due to its nutritional qualities for human consumption, animal feed, and industrial utilization. The soybean pod is regarded as a key fruit tissue determining the seed yield via its photosynthetic ability (Yang et al. 2008). Pod-related traits such as mature pod color (PC) and pod size (PS) are also served as the critical characteristics that are usually used to identify and describe a new variety in breeding programs (Bernard 1967). It is extensively believed that modern cultivated soybean was domesticated from wild soybean in China 5000 years ago (Wang et al. 2016a). The allelic diversity in wild soybeans is higher than cultivated soybeans. According to the USDA-Germplasm Resources Information Network (GRIN, https://www.ars-grin.gov/), all wild soybeans have black and small pods, while most improved cultivars have light-colored (tan or brown) and relatively large pods, and the landraces are between them, with a certain proportion of black pods. Thus, such pod-related characteristics could also be considered as domestication-related traits and that evolved from wild soybean to modern varieties.
Phenotyping of large population is essential for most plant breeding programs or genetic studies (Ghanem et al. 2015; Zhou et al. 2019). Traditional manual phenotyping approaches are mostly time-consuming, laborious, and costly and prone to subjective errors (Chen et al. 2014; Uzal et al. 2018). According to the descriptors and data standard for soybean proposed by Qiu and Chang (Qiu and Chang 2006), based on visual evaluation for PC, it can be classified broadly into five categories, i.e., grayish brown, yellowish-brown, brown, dark brown, and black. However, from a practical perspective, only three main classes, i.e., tan (Tn), brown (Br), and black (Bl), could readily be distinguished. As for pod size-related parameters such as pod length (PL), pod width (PW), pod area (PA), and pod shape (straight, sickle, or bow-shaped), these attributes still are evaluated by using the caliper and visual evaluation at harvest. Thus, pod phenotyping based on manual phenotyping approaches are time-consuming and have greatly hampered the progress of genetic studies and molecular breeding for target traits.
The fruit-related traits are usually complex and controlled by large numbers of quantitative trait loci (QTL) with both major and minor effects. Genotyping and phenotyping are two major determinants affecting the effectiveness and efficiency of genome-wide association studies (GWAS) and linkage mapping (Ibrahim et al. 2020). Recently, the cost of genotyping has become relatively lower compared to the past decades (Barabaschi et al. 2016). Given that thousands or millions of molecular markers can now be easily developed, phenotyping has increasingly become the bottleneck for large-scale genetic analyses in crops (Zhou et al. 2019; Furbank and Tester 2011). Although lots of commercial platforms and equipment have been developed in the last decade for plant phenotyping, the high cost is one of major restricting factors for their widespread utilization (Vasseur et al. 2018). In recent years, image-based phenotyping as a popular and accessible technique has opened new opportunity to evaluate various phenotypes that had previously to be manually quantified or could not be easily quantified (Gage et al. 2018; Diaz-Garcia et al. 2018a). This approach overcomes the drawbacks of the manual phenotyping approach with relatively low cost, wide applicability, and high efficiency (Yang et al. 2020a). So far, a variety of image-based analysis approaches and software have been developed to carry on the image-based plant phenotyping in many species (Fahlgren et al. 2015; Guo et al. 2018; Fernandez-Gallego et al. 2018; Ali et al. 2020; Neumann et al. 2017; Diaz-Garcia et al. 2018b; Turner et al. 2018; Baek et al. 2020).
GWAS and linkage mapping have been widely used to dissect the genetic architecture of quantitatively inherited traits in multiple crops (Korte and Farlow 2013; Tian et al. 2020; Yano and Tuberosa 2009; Würschum 2012). In general, GWAS takes full advantages of all recombination events occurred in a natural population, while linkage mapping is suitable for bi-parental population. Combining these two mapping strategies in one study can complement each other in overcoming the limitations associated with either of them for dissecting the genetic basis of a given trait (Pascual et al. 2016). Several pod-related traits as complex quantitative traits have been investigated by previous researches either by GWAS or linkage mapping, such as pod wall thickness, pod wall weight and ratio of pod wall to pod (Guo et al. 2011), pod number and pod maturity date (Zhang et al. 2010; Yang et al. 2013; Fang et al. 2017), pod shattering/dehiscence (Hu et al. 2019) and pod color (Bandillo et al. 2017; He et al. 2015; Song et al. 2004). Although numerous QTLs underlying these traits have been identified, no promising progress has been achieved in the past decade except for a few studies identified the gene responsible for the QTL detected (Dong et al. 2014). Hence, we employed both GWAS and linkage mapping to identify the genomic regions underlying these pod-related traits. Then, the available access to bioinformatics platforms/tools facilitated in predicting the putative candidate genes for them (Abou-Elwafa and Shehzad 2018).
Therefore, this study was aimed to (1) employ a high-resolution imaging pipeline to collect pod images from a soybean association panel (SAP) consisting of 187 accessions as well as an inter-specific recombinant inbred line (RIL) population designated NJRINP with 284 lines, (2) develop a new phenotyping approach based on the imaging analysis to rapidly generate phenotypic data for basic soybean pod color- and size-related parameters for breeding programs, (3) implement the phenotypic data obtained from these two populations to conduct GWAS and linkage mapping to identify the genomic regions associated with target traits, and (4) predict the putative candidate genes for relatively stable QTLs detected in the present study. These findings will provide valuable information to unravel the genetic basis of these traits meanwhile developing marker-assisted selection (MAS) to breed the soybean varieties with desirable pod-related traits and providing the support for applying the similar procedures in exploiting the genetic basis of fruit-related traits in other crops.
Materials and methods
Plant materials and experimental design
The SAP consists of 187 cultivated soybeans, and an inter-specific RIL population was designated as NJRINP. The NJRINP population was generated by a cross between the elite cultivar NN86-4 and the wild soybean accession PI 342618B contains of 284 RILs. The NN86-4 cultivar is characterized by tan pod color and large pod size and 100-seed weight (~17.9g), while the wild accession PI342618B exhibits black pod color and small pod size and 100-seed weight (~1.1g). The detailed constructive procedures of the SAP and NJRINP populations were described by Li et al. (Li et al. 2016a) and Wang et al. (Wang et al. 2016b), respectively. All materials were obtained from the National Center for Soybean Improvement, Nanjing Agricultural University, Nanjing, China. Field trials of the SAP and NJRINP were conducted at the Jiangpu Experimental Station of Nanjing Agricultural University (31 °02′ N and 118° 04′ E) in the summer of 2017 and 2018 using a completely randomized experimental design with three replications. The SAP was sown in 1-m-long single-row plots with 0.5 m spacing between rows, while NJRINP was planted in 2-m-long row with five hills per row. Field management was performed under normal conditions.
Imaging-based phenotyping
Image acquisition
Soybean pod is similar to that of some legumes, which usually contains 1~3 seeds and rarely 4 seeds. Given the practical operability, the 3-seeded pod was regarded as the object in the current research. The phenotypic evaluation of soybean pod-related traits was performed based on the generated pod images. Five soybean pods harvested from each genotype at maturity stage (around the October 15 for SAP and September 26 for NJRINP) were photographed using an industrial camera (MERCURY, MER-310-12UC, Daheng Group, Inc., China) with a lens (M5018-MP2, 50mm, F1.8-F16C, computer, Inc., Japan) on a white background with the controllable lighting source (OPT-LI38037-w/AP1024-2, OPT, Inc. China), and a 7-cm ruler was used as scale bar information to calculate the data corresponding to the actual size. To ensure a consistent height relative to the sample, the camera was mounted to a fixed camera stand (DH-FP380/500, Daheng Group, Inc., China). A backlight and light-emitting diode (LED) lighting was used to remove the shadows of soybean pods and hence reduced image errors caused by shadows during image generation. Pod samples were always laid out horizontally along the longest axis of the pod and upward at both ends in the center of the field below the camera. The software Daheng MER-Series Viewer was employed to capture images via the attached laptop computer. To facilitate separation of pods and the background, the initial values of the red, green, and blue (RGB) channel were set as 255, 255, and 255, respectively, for the white balance correction. The size of finally generated image was 2048 × 1536 pixels.
Image processing and analysis
The user-friendly Soybean Pod and Seed Phenotyping software SPSP version 1.0 was implemented to process and analyze the images and extract the pod color- and size-related parameters from each image. This software was developed based on the Windows 10 operating system, using Python and PyCharm as the programming language and platform, respectively, and a series of toolkits including PyQt toolkit, Qt Designer, and Model-View-Controller (MVC) were adopted to design and create GUI, which took the soybean pod and seed as the measured object, owning the main functions in pod color recognition and pod size-related parameter determinations including pod length (PL), pod width (PW), and pod area (PA), as well as seed counting. Image segmentation as a crucial step in image processing was applied to separate the pod from the background. Briefly, SPSP V1.0 software estimated PL (cm) and PW (cm) as the length and width of the minimum enclosing rectangle (MER) of the pod in the image, respectively, and PA (cm2) as the area of pod body. The RGB channel values of each pixel for a given image were recorded to measure the pod color using the SPSP V1.0. To accurately measure the pod color, the color space was then converted from RGB to hue (H), saturation(S), and value (V), and the HSV values were also recorded for the same pixel. Scanning all pixels and counting the saturation of each pixel for the given image were performed, and the color of the pixel with the highest saturation was recorded and regarded as the dominant color of the image. Because of the white background color of the image (RGB values of 255, 255, and 255), image showing RGB pixel values greater than 200 was eliminated from further processing and analysis. The RGB and HSV values of the corresponding dominant color of each image for pod were generated. The extracted value of the pod color- and size-related traits for each genotype was exported and saved as a .txt file. Details of these traits and their definitions were listed in Table S1. The workflow for the image-based phenotyping was simplified and shown in Fig. S1. The data from a few images was manually refined or corrected where faulty.
Manual measurements
Although image-based phenotyping method was mainly used to measure the pod color and size for SAP and NJRINP, the manual measurements were also needed for the test samples for verification purpose. Visual assessment for the PC was performed in the NJRINP population in 2017. Pod size-related parameters including PL and PW were measured manually by using a ruler for the subsets of both SAP and NJRINP in 2017 and 2018. Subsequently, the ratio of PL to PW (PRLW) was calculated.
Genotypic data
The SAP and NJRINP populations were genotyped by a restriction-site associated DNA sequencing (RAD-Seq) technique. The detailed information on the sequencing of SAP and NJRINP were same as the earlier studies reported by Li et al. (Li et al. 2016a) and Wang et al. (Wang et al. 2016b), respectively. Briefly, a total of 61, 541 single nucleotide polymorphisms (SNPs) with a minor allele frequency (MAF)≥0.05 for 187 genotypes of SAP and a high-density genetic map composed of 5728 bin markers for 284 RILs of NJRINP were used in this study.
Statistical analysis
Descriptive statistics, analysis of variance (ANOVA), and correlation analysis were performed for the phenotypic data of the SAP as well as NJRINP using the PROC MEANS, GLM, and CORR of SAS Version 9.4, respectively. Descriptions of all traits were determined by the mean value of each trait over five replicates. The broad-sense heritability (h2) for combined environments was calculated according to the method proposed by Nyquist and Baker (Nyquist and Baker 1991) and shown as the following:
Meanwhile the h2 for the individual environment was calculated as the following:
where \( {\sigma}_{\mathrm{g}}^2 \) is the genotypic variance, \( {\sigma}_{\mathrm{g}e}^2 \) is the genotype by environment interaction variance, \( {\sigma}_e^2 \) is the error variance, n is the number of environments, and r is the number of replications. The best linear unbiased predictors (BLUPs) for each genotype for all traits were calculated using the R package lme4 (Bates et al. 2015) taking the effect of environment, replicate within environment, genotype, interaction between genotype and environment, and error as random effects.
Genome-wide association study
The mixed linear model (MLM) algorithm was implemented with GAPIT package (Version: 2.0) (Tang et al. 2016) in R to perform GWAS with high-throughput SNP markers. After filtering the SNPs with a minor allele frequency (MAF) < 0.05, a total of 61541 SNPs was used for GWAS in the SAP population. The significant cutoff of P = 8.12×10−7 [-log (0.05/61541)] based on the Bonferroni correction method and a suggestive threshold of P-value of 1×10−4 were also adopted for SNP-trait association. Linkage disequilibrium (LD) was measured by the squared correlation coefficient (r2) of the pairwise SNPs across the whole genome using the RTM-GWASV1.1 software (He et al. 2017). LD decay rate was estimated by the previously proposed method (Huang et al. 2010). The LD block was identified using the Haploview V4.2 software with default algorithm and plotted by R package LD heatmap (Barrett et al. 2005; Shin et al. 2006). The association results were displayed using the Manhattan plots with an R script and package CMplot (Yin et al. 2020). The peak SNPs were chosen to represent the loci for the final GWAS result.
QTL mapping
Linkage mapping was performed by composite interval mapping (CIM) model in Windows QTL Cartographer V2.5 software (Wang et al. 2012) with a high-density bin published genetic map (Wang et al. 2016b). Briefly, the algorithm with a 10-centimorgan (cM) window and a 1-cM walking step were used in this model. The logarithm of odds (LOD) cutoff was determined with 1000 times permutation at P = 0.05. The QTL boundary was determined with a 1-drop of the LOD score from the peak score. For individual environment, the average value of the five biological replicates for 284 RILs was used for QTL mapping, whereas the BLUP values were used for the combined environment. QTL detected in different environments and exhibited full or partial overlapping confidence intervals were regarded as the same QTL. The QTL identified in at least two environments was treated as a stable one. Nomenclature of QTL was designated following McCouch’s description with minor modifications (McCouch et al. 1997).
Identification of putative candidate genes
A genomic region flanking the physical position of the identified major QTLs in the GWAS panel (500-kb upstream and downstream of the peak SNP) was used to identify candidate genes. The annotated genes within the physical interval of major QTLs were used for identification of candidate genes in the NJRINP population. The loci identified by GWAS were aligned to the reported QTL using the flanked markers information. The physical interval of a QTL was determined based on the physical position of the flanking markers located in the confidence interval of that QTL. Genes annotated in G. max Williams 82 reference gene model 1.0 were the source of candidate genes. Gene annotation and previously published QTLs/genes for relevant traits in soybean and other crop species as well as the orthologs in Arabidopsis with known functions of regulating the fruit color, size, and shape were retrieved from the SoyBase (http://www.soybase.org) and available literature. The expression profiling data of “ZH13,” a Chinese soybean reference genome, was also used to further investigate the expression level of candidate genes at different stages of pod development (Shen et al. 2019). In addition, the known information on the potential biosynthetic pathway responsible for target traits was used as a reference.
Results
Phenotypic variation and correlations among trait
Overall, abundant variations and continuous distribution in the three pod color components (H, S, and V) and pod size-related traits (PL, PW, PRLW, and PA) were observed in the SAP and NJRINP (Fig. S2 and S3), indicating polygenic inheritance of these traits. Moreover, the significant differences of pod size-related traits were also observed between the two populations (Fig. S4). In the SAP population, H, S, and V varied from 29.66 to 52.60, 0.12 to 0.86, and 0.25 to 1.00, with average values of 40.40, 0.67, and 0.81, respectively. PL, PW, PRLW, and PA varied from 3.83 to 7.07 cm, 1.10 to 2.73 cm, 2.33 to 4.55, and 2.84 to 8.24 cm2, with average values of 5.36 cm, 1.67 cm, 3.27, and 4.65 cm, respectively, over 2 years (Table S2). For the NJRINP population, H, S, and V ranged from 22.73 to 82.25, 0.19 to 0.87, and 0.26 to 1.00, with average values of 50.00, 0.50, and 0.60, respectively, over 2 years. PL, PW, PRLW, and PA ranged from 2.61 to 5.52 cm, 0.67 to1.55 cm, 2.45 to 4.65, and 1.14 to 3.69 cm2, with average values of 3.48 cm, 1.06 cm, 3.35, and 2.01 cm2, respectively, over 2 years (Table S3). The absolute values of skewness and kurtosis were less than 1 for these traits except for the S values in the SAP population (skewness varied from 2.56 to 3.05 over 2 years) and PL in the NJRINP population (kurtosis=2.67 in 2017), suggesting that these phenotypes conformed to a normal or skew-normal distribution in the two populations. ANOVA indicated that the effects of genotypes, environments, and genotype-by-environment interaction exhibited significant differences on all measured traits in the two populations (Tables S1 and S2). The h2 of the three-color components and four-size parameters ranged from 52.42 to 91.64% in the SAP population. While in the NJRINP population, a relatively high h2 (>70 %) was estimated for all traits except H component for the PC. Due to the large difference in the H component between 2 years, the h2 was separately estimated for on a per-year basis as 42.71% in 2017 and 78.45% in 2018. The range of h2 suggested that the genetic effects mainly dominated the performance of these traits in both populations.
Pairwise correlations among the three pod color-related components and four pod size-related parameters in the two populations exhibited significant correlations either within or between years except for the correlations between H and S as well as PRLW and PA in 2018 (Fig. 1). Specifically, for pod color-related components in the SAP population, V showed a positive correlation with S either within or between years. H exhibited a low correlation with both S and V between years. However, the correlation between H and S was inconsistent between years (Fig. 1a). For pod size-related parameter, PL was highly correlated with PA within years (r=0.92–0.93, P<0.05) and showed moderate to high correlation with that between years (r=0.79–0.82, P<0.05). PW was moderately correlated with both PL and PRLW within years and exhibited low to moderate and low correlations with PL and PRLW, respectively, between years. Additionally, low and moderate correlations were observed between PL and PRLW, PW, and PA, respectively, either within or between years (Fig. 1b). In the NJRINP population, except for the correlations between H as well as PL and PRLW either within or between years, correlations among all traits either within or between years were significant. In particular, the high and moderate positive correlations were observed between V and S within years (r=0.92–0.96, P<0.05) and between years (r=0.57–0.68, P<0.05), respectively. However, H showed positive and negative correlation with both S and V in 2017 and 2018, respectively (Fig. 1c). Among pod size-related parameters, PL was highly correlated with PA within years (r=0.87–0.92, P<0.05), and moderately correlated with that between years (r=0.51–0.62, P<0.05). PW showed moderate to high positive correlation with PA within years (r=0.77–0.82, P<0.05) and low to moderate correlation with that between years (r=0.45–0.51, P<0.05). PW was moderately correlated with both PL and PRLW within years and exhibited low positive and negative correlations with PL and PRLW between years, respectively. In addition, low negative correlations (lower than 0.5) were observed between PA and PRLW both within and between years except for the correlation between PA in 2018 and PRLW in 2017 (Fig. 1d).
Pearson correlation among traits in two populations. Pod color-related traits including hue (H), saturation (S), and value (V) in SAP and NJRINP are shown in a and c, respectively. Pod size-related traits including pod length (PL), pod width (PW), ratio of PL to PW (PRLW), and pod area (PA) in SAP and NJRINP are shown in b and d, respectively. Significant at P<0.05; both circle color and size illustrate the correlation between pairs of traits; non-significant correlations are indicated by a blank
To validate the results of the image-based phenotyping for pod size-related traits, we randomly selected a sample from each of the populations across 2 years for hand measurement of these traits. As expected, high correlations were observed between size-related traits generated from image-based analysis and that measured manually by hand either in the SAP (r=0.81–0.98, P<0.05) or NJRINP population (r=0.88–0.98, P<0.05) over 2 years (Fig. S5), which demonstrated that the image-based phenotyping approach could be efficiently employed to measure these traits without the laboriously manual work.
Genetic dissection of pod-related traits via GWAS and linkage mapping
GWAS for pod-related traits in the SAP
To identify genomic regions governing pod-related traits, GWAS was performed in the SAP for the three pod color-related components and four pod size-related parameters using MLM approach with both the mean value of each trait in each year and BLUPs of individual performance over 2 years. Because the multiple significant trait-associated SNPs located in close physical proximity, we used the most peak SNP (the strongest trait-associated SNP) to represent the trait-associated loci. A total of one locus for PC and 38 loci for PS were identified across 14 of 20 soybean chromosomes at the suggestive significant P-value level of 1×10−4 (Table 1; Figs. 2 and 3). Among these 38 loci, 14 for PL, 6 for PW, 8 for PRLW, and 10 for PA were detected (Table 1). Out of these, seven, 2, 2, and 6 loci were repetitively detected across the two environments (i.e., 2017 and 2018) for PL, PW, PRLW, and PA, respectively. Some loci were found to be significantly associated with more than one pod size-related parameter. For instance, five loci significantly associated with PL (i.e., PL2-2, PL7, PL14, PL15, and PL19-1) were also significantly associated with PA (Table 1). The locus at the 40.4 Mb position on chromosome 15 was associated with three PS parameters (PL, PW, and PA) with similar effects. Due to the existence of linkage disequilibrium (LD) (Fig. S6), the physical region of PC3 was finally identified at the 0.2~1.2 Mb on chromosome 3 for further identification of putative candidate genes. There were thirty-six SNPs within this 1Mb region. Among them, six SNPs were significantly associated with target trait. In addition, the haplotype block analysis revealed that seven blocks located in this region, and the high LD level (r2=0.87) was observed between the peak SNP and its surrounding SNPs (Fig. 4). Except the PC3 for H component, no loci associated with other pod color components were detected in the SAP population over two years.
Manhattan plots of the association analysis for V component in SAP. Manhattan plot of V on 20 chromosomes across different environments. The dotted red line indicates the significance threshold of P-value 1×10−4. The significant SNPs are labeled with red dots. The outermost ring strip represents the distribution of SNP markers on 20 chromosomes of soybean. The color represents the density of the SNP markers; the inner three circles from the inside to the outside represent three environments, 2017, 2018, and BLUP, respectively
Manhattan plots of the association analysis for pod size-related traits in SAP. Manhattan plots of pod size-related traits on 20 chromosomes across different environments. The dotted red line indicates the significance threshold of P-value 1×10−4. The significant SNPs are labeled with red dots. The outermost ring strip represents the distribution of SNP markers on 20 chromosomes of soybean. The color represents the density of the SNP markers; the inner three circles from the inside to outside represent three environments, 2017, 2018, and BLUP, respectively. a PL. b PW. c PRLW. d PA
Local Manhattan plot and LD heatmap of PC3. The top panel shows the local Manhattan plot for V component using the BLUP value, with a 0.5-Mb region on each side of the peak SNP (SNP with the lowest P value). The red line indicates the significance threshold of P-value (8.12×10−7, 0.05/61541), and the blue one indicates the suggestive significance threshold of P-value of 1×10−4. The two of six significant SNP are highlighted with red dots. The bottom panel depicts the extent LD and LD block in this candidate region. The LD blocks within this region are indicated with the black border. The blue asterisks indicate the position of the above significant SNPs
Linkage mapping of pod-related traits in the NJRINP
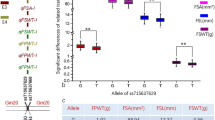
A total of two major QTLs, qPC3 and qPC19, associated with both V component and PC were identified on chromosomes 3 and 19, respectively, in the NJRINP across all environments (Table 2; Fig. 5). The qPC3 was mapped to the interval of 3.3–7.1 cM on chromosome 3, with LOD values ranged from 5.74 to 11.11 and accounted for 6.89 to 9.73% phenotypic variation (R2). The qPC19 was detected in the interval of 51.2–52.2 cM on chromosome 19, with LOD scores and R2 values in the range of 13.21–36.37 and 17.06–40.41 %, respectively. To verify the accuracy of the mapped QTLs for pod color based on the quantitative phenotypic data generated from image analysis, we employed the qualitative phenotypic data obtained from visual inspection to perform QTL mapping. The results obtained from the qualitative data were similar to those from the quantitative data based on image analysis. Interestingly, the relatively higher LOD values were observed for the QTLs detected by the quantitative data than those detected by the qualitative data in most environments (Fig. 5), indicating the more powerful of employing the quantitative phenotypic data for mining QTL. However, surprisingly, these two QTLs revealed the negative additive effect when using the qualitative phenotypic data, while the positive effect was detected when the quantitative phenotypic data were used, implying that the additive alleles were inherited from the opposite parents when using these two different types of phenotypic data.
In total, twenty-three QTLs for pod size-related parameters were identified in the NJRINP across different environments (Table 2; Fig. S7). Out of these QTLs, 8 for PL, 5 for PW, 3 for PRLW, and 7 for PA were detected (Table 2). Eleven QTLs were repetitively detected for pod size-related parameters in at least two environments (either 1 year with BLUP or both years with or without BLUP). Of these, four QTLs (qPL10-1, qPL17-1, qPW17-1, qPA17) were stably detected in 2 years and BLUP. Besides, more than half of these QTLs were only detected in an individual environment. The R2 of these detected QTLs ranged from 3.66 (qPL2) to 14.17 % (qPA17). Notably, one QTL was identified for both PL (qPL17-1) and PA (qPA17) and located in the interval of 41.8–45.4 cM on chromosome 17 with LOD scores and R2 values ranging from 6.13 to 11.03 and 7.17 to 14.17%, respectively. Most identified QTLs such as qPL17-1 and qPA17 overlapped with previously mapped QTLs for soybean seed size-related traits (Table 2), suggesting a potential genetic relationship between pod size and seed size in soybean. These QTLs exhibited the positive additive effects, indicating that female parent NN86-4 contributed the beneficial alleles with exception of qPRLW9 that had its beneficial alleles from the male parent PI 342618B.
Identification of candidate genes
Identification of candidate genes for major QTLs underlying pod color
To further dissect the molecular basis of pod color- and size-related traits, we identified the candidate genes for two major QTLs detected either through GWAS and linkage mapping. Annotation of the physical interval of 491,890–847,720 and 37,463,301–37,938,066 of qPC3 (PC3) and qPC19 to the SoyBase database identified 35 and 48 candidate genes, respectively. Among the model genes within the physical regions of the qPC3, the MATE (multidrug and toxic compound extrusion) gene cluster that contained four MATE genes (Glyma03g00770, Glyma03g00780, Glyma03g00790, and Glyma03g00830) and two MYB (myeloblastosis) genes (Glyma03g00890 and Glyma03g00980) were identified. Similarly, another four MATE genes (Glyma19g29860, Glyma19g29870, Glyma19g29940, and Glyma19g29970) and two MYB genes (Glyma19g29750 and Glyma19g30220) were identified within the physical region of qPC19 (Table S4). The expression profiling of the corresponding gene models in the Chinese soybean (ZH13) genome was utilized to investigate the pod-related tissues expression level of the candidate genes. A BLAST search against the ZH13 reference genome revealed that among the identified MATE genes, two genes, i.e., Glyma03g00830 and Glyma19g29970, whose corresponding genes in ZH13 genome have higher expression level in pod tissue at four different developmental stages (2-week-old pods with seed, 3-, 4-, and 5-week-old pods only). Furthermore, another gene within the qPC19 genomic region, Glyma19g29880, and the corresponding gene in ZH13 genome, SoyZH13_19G106100, have the highest expression level in pod tissue at the same four different developmental stages (Table S5).
Identification of candidate genes for major QTLs associated with pod size-related traits
As to pod size-related traits, among the 38 loci identified in the SAP, eight loci including 2 for PL (PL2-2 and PL7), 2 for PW (PW1 and PW5), 1 for PRLW (PRLW20-1), and 3 for PA (PA2, PA16 and PA19), marked in bold in Table 1, were simultaneously identified in all environments, indicating that they were more stable. These loci were used to retrieve model genes from the SoyBase database. Considering the LD decay distance of the SAP used in this study, the regions within 500 kb on either side of the eight loci were used for identification of candidate genes. As a result, the corresponding 228, 213, 99, and 389 annotated genes were found in these regions, of which, 34 candidate genes were further identified within these QTLs based on gene ontology (GO) annotation, implicated mainly on the GO biological processes such as cell proliferation, cell division, elongation, multiplication, cell wall biosynthesis, and fruit growth and development, as well as response to phytohormone signaling and biosynthesis pathways (Table S6).
A total of 483 candidate genes were identified within the physical regions of four major pod size-related QTLs (159 for qPL10-1, 78 for qPL17-1(qPA17), and 246 for qPW17-1) detected in the NJRINP. Among these identified candidate genes, thirty-five genes were further identified underlying these QTLs based on the same terms of biological process (Table S6). On the other hand, the expression profiles of those candidate genes in pod tissue were further examined. According to the SoyBase RNA-seq Atlas, among the selected candidate genes, five genes, i.e., Glyma01g05410, Glyma05g37190, Glyma05g37470, Glyma17g12880, and Glyma17g13470, have high expression levels at three stages of pod development (1-cm-long pod, pod shell 10 days after flowering (DAF), and pod shell 14 (DAF)). Moreover, the expression level of these 5 genes is twice higher than the average expression level of the genome-wide genes over these three stages of pod development (the normalized FPKM value is 7.74).
Furthermore, several promising candidate genes were mined based on the function of their homologs in other plant species. For instance, two protein families, i.e., ovate protein family (OPF) and IQ67-domain (IQD) protein which coordinate organ development, have been reported to regulate organ size and shape in multiple plant species (Snouffer et al. 2020). A candidate gene in the qPA19, Glyma19g38480, was annotated as an OFP homolog and homologous to Arabidopsis AT2G36026 (OFP13) (Table S7). Three candidate genes, Glyma01g05100, Glyma20g31800, and Glyma17g10660, encode IQD proteins two of which were associated with PW1 and PR20-1 in the SAP, respectively, and the other one associated with the qPW17-1 in the NJRINP are homologs to Arabidopsis AT1G14380, AT1G74690, and AT5G03040, respectively (Table S7). Moreover, the candidate gene Glyma17g10660 has high expression level at the three pod developmental stages mentioned above, with the normalized FPKM values ranging from 9 to 20 (Table S5).
Discussion
Image-based phenotyping accelerates the genetic dissection for pod-related traits
Pod-related traits such as PC and PS are essential morphological characteristics which are closely related to the variety description and seed yield in soybean breeding programs (Yang et al. 2008; Bernard 1967). However, because of being time-consuming, labor-intensive, and often influenced by the changing environmental factors and the observer subjectivity, the conventional phenotyping approaches are becoming one of the limiting factors for soybean genetic analysis and molecular breeding in the past few decades (Furbank and Tester 2011). With the rapid development of crop phenomics, several phenotyping platforms and approaches have been developed (Vasseur et al. 2018). Image-based phenotyping has been widely applied in plant phenotyping across multiple scales from cellular to organ level and from single plants to population level. Compared to the conventional phenotyping approach, it provides advantages in terms of cost, efficiency, and level of throughput (Zhao et al. 2019). In the current study, image-based phenotyping using high-resolution images of the soybean three-seeded pods was employed to extract the basic parameters of pod color and size in two different types of population. To validate the reliability of the generated phenotypic data on pod size-related parameters, we manually measured in a subset containing random samples from each of the two populations. A highly positive correlation between the phenotypic data obtain from either phenotyping approaches was observed, suggesting that the image-based phenotyping could be efficiently implemented to generate the accurate and reliable quantitative and high-throughput phenotypic data in a shorter time with less effort and costs.
GWAS and linkage mapping have been extensively used to analyze the genetic basis of quantitative traits. GWAS is a powerful tool for identification of the chromosomal regions associated with target traits by taking full advantage of historical recombination in natural populations (Che et al. 2020). Conventional QTL mapping usually relies on linkage analysis in biparental populations. A combination of the two approaches complemented with a high-throughput phenotyping approach might comprehensively elucidate the genetic architecture of complex traits.
Dissecting the genetic basis of pod-related traits
Although previous studies have dissected the genetic basis of several pod-related traits including pod wall thickness, pod number and maturity date, and pod color in soybean (Guo et al. 2011; Zhang et al. 2010; Yang et al. 2013; Fang et al. 2017; Bandillo et al. 2017; Song et al. 2004), uncovering the genetic control for pod size-related traits is still very limited. Since the PS is the important component of soybean seed yield, the identification of major QTL controlling them across different environments is of great importance for fine mapping and MAS breeding of new varieties with the desirable pod traits. In the present study, two diverse populations, including a natural population with 187 breeding lines and an inter-specific bi-parental population derived from a cross between two genotypes possessing the distinct pod characteristics with 284 lines, were evaluated for pod color- and size-related parameters. Two QTL mapping approaches were implemented to investigate the power and reliability of the data generated by image-based phenotyping approach and to mine the potential genomic regions for pod size-related traits as much as possible. Both populations exhibited wide phenotypic variations and continuous distribution in pod color- and size-related traits, indicating quantitative polygenic inheritance of these traits. The high heritability (0.80 and 0.79) were observed for the V component in the SAP and NJRINP populations, respectively, indicating that the V component could be implemented as a stable phenotypic indicator facilitated GWAS and QTL mapping for pod color. Meanwhile, pod size-related parameters revealed the high heritability higher than 0.80 and 0.70 in the SAP and NJRINP populations, respectively. The estimated heritability in both populations indicated that the genetic effects mainly dominated the performance of the traits.
Through performing GWAS with 61,541 SNP markers, we identified a major pod color locus and 38 pod size-related parameters loci in the SAP population (Table 1). Among the pod size-related parameter loci, eight loci (PL2-2, PL7, PW1, PW5, PRLW20-1, PA2, PA16, and PA19) were identified for pod size-related traits across all environments (2017, 2018, and BLUP). On the other hand, two and twenty-three QTLs were identified for pod color- and pod size-related traits, respectively, in the NJRINP population with 5728 bin markers and R2 values of 3.66–40.41%. Of these, two QTLs (qPC3 and qPC19) for PC and four QTLs (qPL10-1, qPL17-1, qPW17-1, and qPA17) for PS-related parameters were detected in all environments (Table 2). These QTLs identified by association and linkage mapping revealed that PC and PS are controlled by both major and minor QTL which partially agree with the report in other crop species where the minor QTLs also play important roles in the genetic architecture of fruit traits (Zhou et al. 2019).
The stable QTLs are requisite for their utilization in plant breeding, hence, the QTLs including PC3, PL2-2, PL7, PW1, PW5, PRLW20-1, PA2, PA16, and PA19 identified in the SAP and qPC3, qPC19, qPL10-1, qPL17-1, qPW17-1, and qPA17 detected in the NJRINP (Tables 1 and 2) might be targeted for future soybean breeding with the specific PC and PS traits. Besides, the two identified QTLs (qPC3 (PC3) and qPC19) for PC overlapped with the reported classical loci controlling pod color in soybean, L1 and L2, which located on the central genomic region of the chromosome 19 and the top of the chromosome 3, respectively (Bernard 1967; Bandillo et al. 2017; He et al. 2015; Song et al. 2004). Moreover, these two loci interact with each other in a dominant epistatic manner to produce a F2 phenotypic segregation ratio of 12 (Bl): 3 (Br): 1(Tn) (Bandillo et al. 2017). In addition, the qPC19 also overlapped with a previously reported QTL hotspot regulating the antioxidants, phenolics, and flavonoids contents in soybean seeds (Li et al. 2016b). These results give credence to the reliability of the current mapping for the image-based phenotypes. However, the detected QTLs under the specific environment could be explained by the significant influence of environment on these traits as revealed by ANOVA (Table S2 and S3). Similarly, the colocalization of genomic regions regulating more than one trait may be the genetic basis for the strong to weak correlation among some of the traits. This phenomenon may be due to the presence of tightly linked genes that control the independent traits or possible pleiotropic effects. For instance, PA2, PA7, PA14, PA15, and PA19 for PA colocalized with the genomic regions of PL2-2, PL7, PL14, PL15, and PL19-1 for PL on chromosomes 2, 7, 14, 15, and 19, respectively, in the GWAS study. Furthermore, the colocalization of qPA17 for PA and qPL17-1 for PL was identified on chromosome 17 in the conventional QTL analysis.
In comparison of the results from GWAS and linkage mapping for pod size-related traits, no genomic regions were overlapped between them possibly due to the genetic composition of the bi-parental population which exhibited wide variations in pod size, or the two populations used in this study have greatly phenotypic differences in pod size-related traits. The SAP population was composed of breeding lines generated by cultivated soybeans, while NJRINP population was derived from the inter-specific cross between the large-podded cultivar NN86-4 and the small-podded wild accession PI 342618B. Additionally, the results further showed that most QTLs identified in our study have been previously reported for seed-related traits, suggesting the presence of an inheritance relationship between the size of pod and seed in soybean. Besides, a previously reported improvement-selective sweep partially overlapped with the genomic region of qPC19 (37,463,301-37,938,066), and six selective sweep regions overlapped with the three loci identified for PL via GWAS, i.e., PL7, PL9, and PL14, and three QTLs detected for that by linkage mapping, i.e., qPL2, qPL10-1, and qPL15 (Table S8). Considering the pod-related traits such as PC and PS largely diverged from the wild to cultivated soybeans, thus some of these traits might be targeted by selection in breeding programs.
Identifying potential candidate genes within the stable QTLs
Candidate gene analyses are necessary for further gene cloning and functional verifications. Based on the two QTLs detected, thirteen candidate genes were identified for PC (Table S4). Among them, a gene cluster including four genes, Glyma03g00770, Glyma03g00780, Glyma03g00790, and Glyma03g00830, was annotated as multidrug and toxic compound extrusion (MATE) transporter, and the remaining two genes, Glyma03g00890 and Glyma03g00980, were annotated as MYB transcription factors (TFs). Interestingly, the six genes with the same annotations, including 4 MATE transporters of Glyma19g29860, Glyma19g29870, Glyma19g29940, and Glyma19g29970 and 2 MYB TFs of Glyma19g29750 and Glyma19g30220, were also located in the qPC19 region. In addition, based on the tissue expression level of the corresponding gene model in ZH13, we found that the corresponding genes of Glyma03g00830 and Glyma19g29970 had higher expression level in pod tissue at different developmental stages than the other MATE genes detected. We also found another gene, Glyma19g29880, whose corresponding gene in ZH13 had highest expression level in pod tissue at different developmental stages compared to all other genes, which was annotated as alpha-isopropylmalate synthase.
It is known that anthocyanins as one of major secondary metabolites responsible for the coloration of many fruits, flowers, leaves, and seeds (Lin-Wang et al. 2014). Previous studies have identified some key regulators of anthocyanin biosynthesis in various plants (Niu et al. 2010; Li et al. 2012; Espley et al. 2013; Zhang et al. 2014). Of them, a conserved MBW (MYB-Bhlh-WD40) complex, consisting of R2R3-MYB, basic-helix-loop-helix (bHLH), and WD-repeat (WDR) proteins, is widely believed to regulate the common pathway of anthocyanins and proanthocyanidins (PA) biosynthesis (Jaakola 2013; Albert et al. 2014). As a member of this complex, several MYB genes in Arabidopsis and fruits have been reported to influence the organ coloration by regulating anthocyanin biosynthetic pathway. For instance, in lettuce, AtMYB60 as an exogenous gene has been reported to participate in the regulation of leaf pigmentation by inhibiting anthocyanin biosynthesis (Park et al. 2008). In strawberry, FvMYB10 as a key activator has been reported to participate in the regulation of leaves, flowers, and fruits coloration (Lin-Wang et al. 2014). In pear and apple, PcMYB10 and MdMYB10, respectively, have also been shown to involve in the regulation of anthocyanin biosynthesis (Li et al. 2012; Espley et al. 2013). On the other hand, the fruit coloration may be related to not only the biosynthesis pathway, but also the transport and accumulation process. Previous studies have reported that MATE transporters were involved in the transport and accumulation of secondary metabolites such as flavonoid, anthocyanin, and PA (Takanashi et al. 2014). In Arabidopsis, AtTT12 was first MATE transporter found to transport flavonoids to alter seed coloration (Diener et al. 2001). In Medicago, MtMATE1 and MtMATE2 were involved in the transport of PA and flavonoid, which influence the pigmentation of seeds, flowers, and leaves (Zhao and Dixon 2009; Zhao et al. 2011). Fruits such as grape (VvAM1 and VvAM3) and in apple (MdMATE1 and MdMATE2) were involved in the transport and accumulation of anthocyanin and PA (Gomez et al. 2009; Gomez et al. 2011; Frank et al. 2011). Therefore, we speculate that one or several of the above genes might be the potential candidate genes involved in pod coloration in soybean based on the gene annotation and previous reports on the known genes involved in the biosynthesis pathway related to fruit color in other plants (Albert et al. 2014; Takanashi et al. 2014). More attention should be paid to the genes highly expressed in pod tissue in the future. Facing such results and considering the nature of soybean genome, we further speculated that the genes controlling PC might undergo the gene duplication and subsequent functional divergence in soybean evolutionary process, which eventually leads to the differentiation of pod color.
Based on the results of GWAS and linkage mapping in both populations, a total of 12 QTLs for pod size-related traits, including 4, 3, 1, and 4 for PL, PW, PRLW, and PA, respectively, were stably identified in all environments. Sixty-nine candidate genes were predicted based on the biological process of GO with the terms of “cell proliferation, cell division, elongation, multiplication, cell wall biosynthesis, and fruit growth and development as well as involved in phytohormones signaling and biosynthesis pathways” (Table S6). According to the RNA-Seq Atlas in SoyBase, five of these candidate genes, Glyma01g05410, Glyma05g37190, Glyma05g37470, Glyma17g12880, and Glyma17g13470 were highly expressed in pod tissue at three developmental stages, two of which have also been proven to be highly expressed in pod tissue at four stages of pod development by the previously published data (Shen et al. 2019) (Table S5). Among these five genes mentioned, the ZH13 corresponding gene to Glyma01g05410 has the highest expression level in pod tissue across the different developmental stages.
In addition, three genes, Glyma01g05100, Glyma17g10660, and Glyma20g31800, were found in the interval of PW1, qPW17-1, and PRLW20-1, respectively, which were annotated as IQ67 Domain (IQD) proteins. One gene, Glyma19g38480, was found in the interval of PA19, which was annotated as ovate family protein (OFP) (Table S7). The OFPs are the protein family containing a conserved OVATE domain that regulates organ size and shape in plants. The IQD family proteins are the calmodulin-binding proteins encoding a plant-specific domain of 67 conserved amino acid residues (designated as IQ67 domain), which often serves as the regulator to participate in the complexes of OFP to form organ shapes by regulating cytoskeleton activities (Snouffer et al. 2020; Abel et al. 2005). Several homologous genes of OFP and IQD in Arabidopsis and other crops have been reported to participate in the regulation of organ size and shape. For example, in Arabidopsis, AtOFP1 has been shown to suppress cell elongation (Wang et al. 2007). In rice, OsOFP19 has been reported to participate in the regulation of grain width by increasing cell length and cell number (Yang et al. 2018) and SlOFP20 in tomato (Wu et al. 2018). For IQD, GSE5, the causal gene of GW5 locus that is widely utilized by rice breeders to control grain size, which encodes a protein with IQ domain, has been demonstrated to participate in regulation of gain size predominantly by influencing cell proliferation in spikelet hulls (Duan et al. 2017; Liu et al. 2017). Another known gene in rice, OsIQD14, has been reported to control the shape and size of gain seeds (Yang et al. 2020b). In tomato, SUN encoding a member of IQ67-domain (IQD) family has been reported to govern the elongated fruit shape (Xiao et al. 2008). Thus, these four genes may also be potential candidate genes regulating pod size and shape in soybean. Although we cannot accurately determine which genes are directly related to pod color and size based on the data of this study, we provide a list of candidate gene models, which could be used as the useful information for future works. Further studies should be conducted via fine-mapping of stable QTLs and validating the molecular function of potential candidate genes discovered in this study.
Conclusion
A high-throughput image-based phenotyping approach has been successfully employed for dissecting the genetic control of pod color- and size-related traits using GWAS and linkage mapping strategies in two independent populations. Two QTLs, qPC3 (PC3) and qPC19, were identified to be associated with PC. Thirty-eight QTLs for pod size-related parameters, including 14 for PL, 6 for PW, 8 for PRLW, and 10 for PA, were identified by GWAS in the SAP population, and 23 QTLs containing 8 for PL, 5 for PW, 3 for PRLW, and 7 for PA were detected by linkage mapping in the NJRINP population. Twelve QTLs were stably identified in all environments for the two populations. Around these stable QTLs, 13 and 73 candidate genes were predicted for pod color-and size-related traits, respectively. These QTLs and corresponding candidate genes provide important information for fine-mapping/map-based cloning and MAS for breeding elite varieties with desirable pod traits.
Availability of data and materials
All data generated and analyzed during this study are included in this article.
References
Abel S, Savchenko T, Levy M (2005) Genome-wide comparative analysis of the IQD gene families in Arabidopsis thaliana and Oryza sativa. BMC Evol Biol 5:72–72. https://doi.org/10.1186/1471-2148-5-72
Abou-Elwafa SF, Shehzad T (2018) Genetic identification and expression profiling of drought responsive genes in sorghum. Environ Exp Bot 155:12–20. https://doi.org/10.1016/j.envexpbot.2018.06.019
Albert NW, Davies KM, Lewis DH, Zhang H, Montefiori M, Brendolise C, Boase MR, Ngo H, Jameson PE, Schwinn KE (2014) A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26:962–980. https://doi.org/10.1105/tpc.113.122069
Ali A, Ullah Z, Alam N, Naqvi SMS, Jamil M, Bux H, Sher H (2020) Genetic analysis of wheat grains using digital imaging and their relationship to enhance grain weight. Sci Agric 77. https://doi.org/10.1590/1678-992x-2019-0069
Baek J, Lee E, Kim N, Kim SL, Choi I, Ji H, Chung YS, Choi MS, Moon JK, Kim KH (2020) High throughput phenotyping for various traits on soybean seeds using image analysis. Sensors 20:248. https://doi.org/10.3390/s20010248
Bandillo NB, Lorenz AJ, Graef GL, Jarquin D, Hyten DL, Nelson RL, Specht JE (2017) Genome-wide association mapping of qualitatively inherited traits in a germplasm collection. Plant Genome 10:18. https://doi.org/10.3835/plantgenome2016.06.0054
Barabaschi D, Tondelli A, Desiderio F, Volante A, Vaccino P, Vale G, Cattivelli L (2016) Next generation breeding. Plant Sci 242:3–13. https://doi.org/10.1016/j.plantsci.2015.07.010
Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265. https://doi.org/10.1093/bioinformatics/bth457
Bates D, Mächler M, Bolker B, Walker S (2015) Fitting linear mixed-effects models using lme4. J Stat Softw 67:1–48. https://doi.org/10.18637/jss.v067.i01
Bernard RL (1967) The inheritance of pod color in soybeans. J Hered 58:165–168. https://doi.org/10.1093/oxfordjournals.jhered.a107575
Che Z, Yan H, Liu H, Yang H, Du H, Yang Y, Liu B, Yu D (2020) Genome-wide association study for soybean mosaic virus SC3 resistance in soybean. Mol Breed 40. https://doi.org/10.1007/s11032-020-01149-1
Chen D, Neumann K, Friedel S, Kilian B, Chen M, Altmann T, Klukas C (2014) Dissecting the phenotypic components of crop plant growth and drought responses based on high-throughput image analysis. Plant Cell 26:4636–4655. https://doi.org/10.1105/tpc.114.129601
Diaz-Garcia L, Covarrubias-Pazaran G, Schlautman B, Grygleski E, Zalapa J (2018a) Image-based phenotyping for identification of QTL determining fruit shape and size in American cranberry (Vaccinium macrocarpon L.). PeerJ 6:e5461. https://doi.org/10.7717/peerj.5461
Diaz-Garcia L, Schlautman B, Covarrubias-Pazaran G, Maule A, Johnson-Cicalese J, Grygleski E, Vorsa N, Zalapa J (2018b) Massive phenotyping of multiple cranberry populations reveals novel QTLs for fruit anthocyanin content and other important chemical traits. Mol Gen Genomics 293:1379–1392. https://doi.org/10.1007/s00438-018-1464-z
Diener AC, Gaxiola RA, Fink GR (2001) Arabidopsis ALF5, a multidrug efflux transporter gene family member, confers resistance to toxins. Plant Cell 13:1625–1638. https://doi.org/10.1105/tpc.010035
Dong Y, Yang X, Liu J, Wang BH, Liu BL, Wang YZ (2014) Pod shattering resistance associated with domestication is mediated by a NAC gene in soybean. Nat Commun 5:3352. https://doi.org/10.1038/ncomms4352
Duan P, Xu J, Zeng D, Zhang B, Geng M, Zhang G, Huang K, Huang L, Xu R, Ge S, Qian Q, Li Y (2017) Natural variation in the promoter of GSE5 contributes to grain size diversity in rice. Mol Plant 10:685–694. https://doi.org/10.1016/j.molp.2017.03.009
Espley RV, Bovy A, Bava C, Jaeger SR, Tomes S, Norling C, Crawford J, Rowan D, McGhie TK, Brendolise C, Putterill J, Schouten HJ, Hellens RP, Allan AC (2013) Analysis of genetically modified red-fleshed apples reveals effects on growth and consumer attributes. Plant Biotechnol J 11:408–419. https://doi.org/10.1111/pbi.12017
Fahlgren N, Gehan MA, Baxter I (2015) Lights, camera, action: high-throughput plant phenotyping is ready for a close-up. Curr Opin Plant Biol 24:93–99. https://doi.org/10.1016/j.pbi.2015.02.006
Fang C, Ma Y, Wu S, Liu Z, Wang Z, Yang R, Hu G, Zhou Z, Yu H, Zhang M, Pan Y, Zhou G, Ren H, Du W, Yan H, Wang Y, Han D, Shen Y, Liu S, Liu T, Zhang J, Qin H, Yuan J, Yuan X, Kong F, Liu B, Li J, Zhang Z, Wang G, Zhu B, Tian Z (2017) Genome-wide association studies dissect the genetic networks underlying agronomical traits in soybean. Genome Biol 18:161. https://doi.org/10.1186/s13059-017-1289-9
Fernandez-Gallego JA, Kefauver SC, Gutierrez NA, Nieto-Taladriz MT, Araus JL (2018) Wheat ear counting in-field conditions: high throughput and low-cost approach using RGB images. Plant Methods 14:22. https://doi.org/10.1186/s13007-018-0289-4
Frank S, Keck M, Sagasser M, Niehaus K, Weisshaar B, Stracke R (2011) Two differentially expressed MATE factor genes from apple complement the Arabidopsis transparent testa12 mutant. Plant Biol (Stuttg) 13:42–50. https://doi.org/10.1111/j.1438-8677.2010.00350.x
Furbank RT, Tester M (2011) Phenomics-technologies to relieve the phenotyping bottleneck. Trends Plant Sci 16:635–644. https://doi.org/10.1016/j.tplants.2011.09.005
Gage JL, de Leon N, Clayton MK (2018) Comparing genome-wide association study results from different measurements of an underlying phenotype. G3 (Bethesda) 8:3715–3722. https://doi.org/10.1534/g3.118.200700
Ghanem ME, Marrou H, Sinclair TR (2015) Physiological phenotyping of plants for crop improvement. Trends Plant Sci 20:139–144. https://doi.org/10.1016/j.tplants.2014.11.006
Gomez C, Terrier N, Torregrosa L, Vialet S, Fournier-Level A, Verries C, Souquet JM, Mazauric JP, Klein M, Cheynier V, Ageorges A (2009) Grapevine MATE-type proteins act as vacuolar H+-dependent acylated anthocyanin transporters. Plant Physiol 150:402–415. https://doi.org/10.1104/pp.109.135624
Gomez C, Conejero G, Torregrosa L, Cheynier V, Terrier N, Ageorges A (2011) In vivo grapevine anthocyanin transport involves vesicle-mediated trafficking and the contribution of anthoMATE transporters and GST. Plant J 67:960–970. https://doi.org/10.1111/j.1365-313X.2011.04648.x
Guo GY, Sun R, Hou M, Guo YX, Xin DW, Jiang HW, Liu CY, Hu GH, Chen QS (2011) Quantitative trait locus (QTL) analysis of pod related traits in different environments in soybean. Afr J Biotechnol 10:11848–11854
Guo Z, Yang W, Chang Y, Ma X, Tu H, Xiong F, Jiang N, Feng H, Huang C, Yang P, Zhao H, Chen G, Liu H, Luo L, Hu H, Liu Q, Xiong L (2018) Genome-wide association studies of image traits reveal genetic architecture of drought resistance in rice. Mol Plant 11:789–805. https://doi.org/10.1016/j.molp.2018.03.018
He Q, Yang H, Xiang S, Tian D, Wang W, Zhao T, Gai J, Singh R (2015) Fine mapping of the genetic locusL1conferring black pods using a chromosome segment substitution line population of soybean. Plant Breed 134:437–445. https://doi.org/10.1111/pbr.12272
He J, Meng S, Zhao T, Xing G, Yang S, Li Y, Guan R, Lu J, Wang Y, Xia Q, Yang B, Gai J (2017) An innovative procedure of genome-wide association analysis fits studies on germplasm population and plant breeding. Theor Appl Genet 130:2327–2343. https://doi.org/10.1007/s00122-017-2962-9
Hu D, Kan G, Hu W, Li Y, Hao D, Li X, Yang H, Yang Z, He X, Huang F, Yu D (2019) Identification of loci and candidate genes responsible for pod dehiscence in soybean via genome-wide association analysis across multiple environments. Front Plant Sci 10:811. https://doi.org/10.3389/fpls.2019.00811
Huang X, Wei X, Sang T, Zhao Q, Feng Q, Zhao Y, Li C, Zhu C, Lu T, Zhang Z, Li M, Fan D, Guo Y, Wang A, Wang L, Deng L, Li W, Lu Y, Weng Q, Liu K, Huang T, Zhou T, Jing Y, Li W, Lin Z, Buckler ES, Qian Q, Zhang QF, Li J, Han B (2010) Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet 42:961–967. https://doi.org/10.1038/ng.695
Ibrahim AK, Zhang L, Niyitanga S, Afzal MZ, Xu Y, Zhang L, Zhang L, Qi J (2020) Principles and approaches of association mapping in plant breeding. Trop Plant Biol 13:212–224. https://doi.org/10.1007/s12042-020-09261-4
Jaakola L (2013) New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci 18:477–483. https://doi.org/10.1016/j.tplants.2013.06.003
Korte A, Farlow A (2013) The advantages and limitations of trait analysis with GWAS: a review. Plant Methods 9:29. https://doi.org/10.1186/1746-4811-9-29
Li L, Ban ZJ, Li XH, Wu MY, Wang AL, Jiang YQ, Jiang YH (2012) Differential expression of anthocyanin biosynthetic genes and transcription factor PcMYB10 in pears (Pyrus communis L.). PLoS One 7:e46070. https://doi.org/10.1371/journal.pone.0046070
Li L, Guo N, Niu J, Wang Z, Cui X, Sun J, Zhao T, Xing H (2016a) Loci and candidate gene identification for resistance to Phytophthora sojae via association analysis in soybean [ Glycine max (L.) Merr.]. Mol Gen Genomics 291:1095–1103. https://doi.org/10.1007/s00438-015-1164-x
Li MW, Munoz NB, Wong CF, Wong FL, Wong KS, Wong WH, Qi X, Li KP, Ng MS, Lam HM (2016b) QTLs regulating the contents of antioxidants, phenolics, and flavonoids in soybean seeds share a common genomic region. Front Plant Sci 7:854. https://doi.org/10.3389/fpls.2016.00854
Lin-Wang K, McGhie TK, Wang M, Liu Y, Warren B, Storey R, Espley RV, Allan AC (2014) Engineering the anthocyanin regulatory complex of strawberry (Fragaria vesca). Front Plant Sci 5:651. https://doi.org/10.3389/fpls.2014.00651
Liu J, Chen J, Zheng X, Wu F, Lin Q, Heng Y, Tian P, Cheng Z, Yu X, Zhou K, Zhang X, Guo X, Wang J, Wang H, Wan J (2017) GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat Plants 3:17043. https://doi.org/10.1038/nplants.2017.43
McCouch SR, Cho YG, Yano M, Paul E, Blinstrub M, Morishima H, Kinoshita T (1997) Report on QTL nomenclature. Rice Genet Newsl 14:11–13. https://doi.org/10.1007/s10142-013-0328-1
Neumann K, Zhao Y, Chu J, Keilwagen J, Reif JC, Kilian B, Graner A (2017) Genetic architecture and temporal patterns of biomass accumulation in spring barley revealed by image analysis. BMC Plant Biol 17:137. https://doi.org/10.1186/s12870-017-1085-4
Niu SS, Xu CJ, Zhang WS, Zhang B, Li X, Lin-Wang K, Ferguson IB, Allan AC, Chen KS (2010) Coordinated regulation of anthocyanin biosynthesis in Chinese bayberry (Myrica rubra) fruit by a R2R3 MYB transcription factor. Planta 231:887–899. https://doi.org/10.1007/s00425-009-1095-z
Nyquist WE, Baker RJ (1991) Estimation of heritability and prediction of selection response in plant populations. Crit Rev Plant Sci 10:235–322. https://doi.org/10.1080/07352689109382313
Park JS, Kim JB, Cho KJ, Cheon CI, Sung MK, Choung MG, Roh KH (2008) Arabidopsis R2R3-MYB transcription factor AtMYB60 functions as a transcriptional repressor of anthocyanin biosynthesis in lettuce (Lactuca sativa). Plant Cell Rep 27:985–994. https://doi.org/10.1007/s00299-008-0521-1
Pascual L, Albert E, Sauvage C, Duangjit J, Bouchet JP, Bitton F, Desplat N, Brunel D, Le Paslier MC, Ranc N, Bruguier L, Chauchard B, Verschave P, Causse M (2016) Dissecting quantitative trait variation in the resequencing era: complementarity of bi-parental, multi-parental and association panels. Plant Sci 242:120–130. https://doi.org/10.1016/j.plantsci.2015.06.017
Qiu LJ, Chang RZ (2006) Descriptors and data standard for soybean (Glycine spp.). China Agriculture Press, Beijing
Shen Y, Du H, Liu Y, Ni L, Wang Z, Liang C, Tian Z (2019) Update soybean Zhonghuang 13 genome to a golden reference. Sci China Life Sci 62:1257–1260. https://doi.org/10.1007/s11427-019-9822-2
Shin J-H, Blay S, McNeney B, Graham J (2006) LDheatmap: An R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. J Stat Softw 16:1–9. https://doi.org/10.18637/jss.v016.c03
Snouffer A, Kraus C, van der Knaap E (2020) The shape of things to come: ovate family proteins regulate plant organ shape. Curr Opin Plant Biol 53:98–105. https://doi.org/10.1016/j.pbi.2019.10.005
Song QJ, Marek LF, Shoemaker RC, Lark KG, Concibido VC, Delannay X, Specht JE, Cregan PB (2004) A new integrated genetic linkage map of the soybean. Theor Appl Genet 109:122–128. https://doi.org/10.1007/s00122-004-1602-3
Takanashi K, Shitan N, Yazaki K (2014) The multidrug and toxic compound extrusion (MATE) family in plants. Plant Biotechnol 31:417–430. https://doi.org/10.5511/plantbiotechnology.14.0904a
Tang Y, Liu X, Wang J, Li M, Wang Q, Tian F, Su Z, Pan Y, Liu D, Lipka AE, Buckler ES, Zhang Z (2016) GAPIT Version 2: an enhanced integrated tool for genomic association and prediction. Plant Genome 9:1–9. https://doi.org/10.3835/plantgenome2015.11.0120
Tian D, Wang P, Tang B, Teng X, Li C, Liu X, Zou D, Song S, Zhang Z (2020) GWAS Atlas: a curated resource of genome-wide variant-trait associations in plants and animals. Nucleic Acids Res 48:D927–D932. https://doi.org/10.1093/nar/gkz828
Turner SD, Ellison SL, Senalik DA, Simon PW, Spalding EP, Miller ND (2018) An automated image analysis pipeline enables genetic studies of shoot and root morphology in carrot (Daucus carota L.). Front Plant Sci 9:1703. https://doi.org/10.3389/fpls.2018.01703
Uzal LC, Grinblat GL, Namías R, Larese MG, Bianchi JS, Morandi EN, Granitto PM (2018) Seed-per-pod estimation for plant breeding using deep learning. Comput Electron Agric 150:196–204. https://doi.org/10.1016/j.compag.2018.04.024
Vasseur F, Bresson J, Wang G, Schwab R, Weigel D (2018) Image-based methods for phenotyping growth dynamics and fitness components in Arabidopsis thaliana. Plant Methods 14:63. https://doi.org/10.1186/s13007-018-0331-6
Wang S, Chang Y, Guo J, Chen JG (2007) Arabidopsis Ovate Family Protein 1 is a transcriptional repressor that suppresses cell elongation. Plant J 50:858–872. https://doi.org/10.1111/j.1365-313X.2007.03096.x
Wang S, Basten CJ, Zeng ZB (2012) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh
Wang J, Chu S, Zhang H, Zhu Y, Cheng H, Yu D (2016a) Development and application of a novel genome-wide SNP array reveals domestication history in soybean. Sci Rep 6:20728. https://doi.org/10.1038/srep20728
Wang W, Liu M, Wang Y, Li X, Cheng S, Shu L, Yu Z, Kong J, Zhao T, Gai J (2016b) Characterizing two inter-specific bin maps for the exploration of the QTLs/genes that confer three soybean evolutionary traits. Front Plant Sci 7:1248. https://doi.org/10.3389/fpls.2016.01248
Wu S, Zhang B, Keyhaninejad N, Rodriguez GR, Kim HJ, Chakrabarti M, Illa-Berenguer E, Taitano NK, Gonzalo MJ, Diaz A, Pan Y, Leisner CP, Halterman D, Buell CR, Weng Y, Jansky SH, van Eck H, Willemsen J, Monforte AJ, Meulia T, van der Knaap E (2018) A common genetic mechanism underlies morphological diversity in fruits and other plant organs. Nat Commun 9:4734. https://doi.org/10.1038/s41467-018-07216-8
Würschum T (2012) Mapping QTL for agronomic traits in breeding populations. Theor Appl Genet 125:201–210. https://doi.org/10.1007/s00122-012-1887-6
Xiao H, Jiang N, Schaffner E, Stockinger EJ, van der Knaap E (2008) A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 319:1527–1530. https://doi.org/10.1126/science.1153040
Yang Y, Jing C, Wang X, Lin C, Wang X, Zhou ZS (2008) Photosynthetic characteristics of soybean pod and its contribution to yield. J Northeast Agric Univ 39:51–56 885
Yang Z, Xin D, Liu C, Jiang H, Han X, Sun Y, Qi Z, Hu G, Chen Q (2013) Identification of QTLs for seed and pod traits in soybean and analysis for additive effects and epistatic effects of QTLs among multiple environments. Mol Gen Genomics 288:651–667. https://doi.org/10.1007/s00438-013-0779-z
Yang C, Ma Y, He Y, Tian Z, Li J (2018) OsOFP19 modulates plant architecture by integrating the cell division pattern and brassinosteroid signaling. Plant J 93:489–501. https://doi.org/10.1111/tpj.13793
Yang W, Feng H, Zhang X, Zhang J, Doonan JH, Batchelor WD, Xiong L, Yan J (2020a) Crop phenomics and high-throughput phenotyping: past decades, current challenges, and future perspectives. Mol Plant 13:187–214. https://doi.org/10.1016/j.molp.2020.01.008
Yang B, Wendrich JR, De Rybel B, Weijers D, Xue HW (2020b) Rice microtubule-associated protein IQ67-DOMAIN14 regulates grain shape by modulating microtubule cytoskeleton dynamics. Plant Biotechnol J 18:1141–1152. https://doi.org/10.1111/pbi.13279
Yano M, Tuberosa R (2009) Genome studies and molecular genetics-from sequence to crops: genomics comes of age. Curr Opin Plant Biol 12:103–106. https://doi.org/10.1016/j.pbi.2009.01.001
Yin L, Zhang H, Tang Z, Xu J, Yin D, Zhang Z, Yuan X, Zhu M, Zhao S, Li X, Liu X (2020) rMVP: A memory-efficient, visualization-enhanced, and parallel-accelerated tool for genome-wide association study. bioRxiv. https://doi.org/10.1101/2020.08.20.258491
Zhang D, Cheng H, Wang H, Zhang H, Liu C, Yu D (2010) Identification of genomic regions determining flower and pod numbers development in soybean (Glycine max L.). J Genet Genomics 37:545–556. https://doi.org/10.1016/s1673-8527(09)60074-6
Zhang Y, Butelli E, Martin C (2014) Engineering anthocyanin biosynthesis in plants. Curr Opin Plant Biol 19:81–90. https://doi.org/10.1016/j.pbi.2014.05.011
Zhao J, Dixon RA (2009) MATE transporters facilitate vacuolar uptake of epicatechin 3'-O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and Arabidopsis. Plant Cell 21:2323–2340. https://doi.org/10.1105/tpc.109.067819
Zhao J, Huhman D, Shadle G, He XZ, Sumner LW, Tang Y, Dixon RA (2011) MATE2 mediates vacuolar sequestration of flavonoid glycosides and glycoside malonates in Medicago truncatula. Plant Cell 23:1536–1555. https://doi.org/10.1105/tpc.110.080804
Zhao C, Zhang Y, Du J, Guo X, Wen W, Gu S, Wang J, Fan J (2019) Crop phenomics: current status and perspectives. Front Plant Sci 10:714. https://doi.org/10.3389/fpls.2019.00714
Zhou Y, Srinivasan S, Mirnezami SV, Kusmec A, Fu Q, Attigala L, Salas Fernandez MG, Ganapathysubramanian B, Schnable PS (2019) Semiautomated feature extraction from RGB images for sorghum panicle architecture GWAS. Plant Physiol 179:24–37. https://doi.org/10.1104/pp.18.00974
Acknowledgements
We appreciated Prof. Yufeng Wu and Dr. Wenkai Yan for their technical assistance. The work was supported in part by the Bioinformatics Center of Nanjing Agricultural University.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 31872847, 31871646), the National Key Research and Development Project (2018YFD0201006), the MOE Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT_17R55), the Fundamental Research Funds for the Central Universities (KYT201801), and the Jiangsu Collaborative Innovation Center for Modern Crop Production (JCIC-MCP) Program.
Author information
Authors and Affiliations
Contributions
TJZ and HYJ conceived and designed the research experiments. FGC, PYLv, LYZ, and PX implemented the field experiments. FGC, SC, WHLv, YTX, WLY, and LW performed phenotyping. FGC and SC analyzed data. TJZ and HYJ designed the overall project. FGC drafted the manuscript. TJZ, BK, and Abou-Elwafa revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chang, F., Lv, W., Lv, P. et al. Exploring genetic architecture for pod-related traits in soybean using image-based phenotyping. Mol Breeding 41, 28 (2021). https://doi.org/10.1007/s11032-021-01223-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-021-01223-2