Abstract
The soybean aphid (Aphis glycines Matsumura) is a pest of soybean (Glycine max (L.) Merr.) in many soybean-growing countries of the world, mainly in Asia and North America. While numerous aphid-resistance loci have been mapped, new and virulent aphid biotypes continue to be identified. PI 606390A confers resistance to all four characterized soybean aphid biotypes. The objectives of this study were to identify single nucleotide polymorphism (SNP) markers closely linked to the loci conferring resistance to soybean aphid biotypes 3 and 4. One hundred and eight F6:7 families from a cross between the susceptible experimental line HR09-018 and the resistant PI 606390A plus the two parental lines were screened with aphid biotypes 3 and 4 using greenhouse choice tests. Broad-sense heritability estimates ranged from 0.69 to 0.91. Correlation between biotype 3 and biotype 4 resistance was also moderate (45–55%). A total of 1847 SNP markers were mapped to 22 linkage groups, and loci for resistance to both aphid biotypes were mapped to a 14 cM region on chromosome 18 using composite interval mapping. The locus explained between 26 and 40% of the phenotypic variation suggests the presence of a major locus and additional unidentified minor quantitative trait loci (QTL). A new symbol qRag-G is proposed to designate this new locus that will be useful for the development of durable aphid-resistant soybean cultivars.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glycine max (soybean) is one of the two most important field crops cultivated in the USA, with about 90 million acres planted in 2017 (NASS 2018). The soybean aphid, Aphis glycines Matsumura (Hemiptera: Aphididae), was first found in the USA in 2000 and has become one of the most economically important pests threatening soybean production in the Midwestern states (Hartman et al. 2001). Severe infestations of aphids in soybean fields can result in leaf curling, wilting, and premature leaf drop (DiFonzo and Hines 2002). Yield losses in soybean attributed to the aphid were reported to be greater than 50% in Minnesota (Ostlie 2002), and significant losses of seed oil content have also been observed (Riedell and Catangui 2006). In addition to feeding damage, aphids can transmit plant viruses including alfalfa mosaic virus, soybean dwarf virus, and soybean mosaic virus to soybean (Iwaki et al. 1980; Hartman et al. 2001; Hill et al. 2001).
Host plant resistance is one of the most environmentally friendly and economically sustainable approaches to reducing pest damage to crop plants. In 2006, two cultivars “Dowling” and “Jackson” were identified as having strong resistance to the soybean aphid in Illinois (biotype 1). A single dominant gene, Rag1 (resistant to Aphis glycines), was mapped to chromosome 7 of the soybean genome (Hill et al. 2006a, 2006b). In Ohio, Rag1, was defeated by a new virulent aphid (biotype 2) (Kim et al. 2010b). Rag2, a single dominant gene from PI 243540, was mapped to chromosome 13 and conferred resistance against both biotypes 1 and 2 (Mian et al. 2008). Rag3, a single dominant gene, was mapped on chromosome 16 from PI 567543C (Zhang et al. 2010) and Rag4, a recessive gene on chromosome 13, each confers resistance to soybean aphids in Michigan (Zhang et al. 2009). Rag5 was also mapped on chromosome 13 and provides antixenosis resistance to biotypes 1 and 2 (Jun et al. 2012). More recently, additional resistance loci have been identified and new aphid biotypes have also been discovered. Rag6 and Rag3c from a Glycine soja accession were fine-mapped to loci on chromosomes 8 and 16, respectively (Zhang et al. 2017). These new loci conferred resistance to soybean aphids were found in Michigan that defeated both Rag1 and Rag2, revealing new virulent biotypes.
Currently, there are four characterized biotypes of the soybean aphid (Alt and Ryan-Mahmutagic 2013). However, these biotypes were identified before Rag gene introgressed cultivars were deployed, suggesting pre-existing virulence diversity or a rapid capacity of aphids to adapt to new resistant plants (Michel et al. 2011). Yet, the evolution of soybean aphid virulence is still unclear. Genetic analysis of aphid populations has not revealed a correlation to geographic location or biotype (Wenger and Michel 2013). More recently, genetic differences among biotypes were associated with a microbial endosymbiont Buchnera aphidicola (Cassone et al. 2015). The bacterial endosymbiont Arsenophonus also had an added benefit to virulent aphids grown on aphid-resistant soybeans (Wulff and White 2015; Enders and Miller 2016). Microbiomes of aphids include obligate and facultative symbionts, with the latter exhibiting more rapid genome evolution and horizontal transfer between host species (Oliver et al. 2010). Therefore, continual identification of aphid-resistance genes is necessary to keep pace with aphid virulence acquisition.
PI 606390A is a maturity group (MG) IV soybean accession collected from Vietnam that was previously identified as exhibiting resistance to soybean aphid biotypes 1, 2, and 3 (Bansal et al. 2013). In the current study, we also observed strong resistance to aphid biotype 4 in this line. The objective of this study was to map host plant resistance QTL in PI 606390A for soybean aphid biotypes 3 and 4 to provide new resources for the development of cultivars with broad and more durable resistance against the aphid.
Materials and methods
Aphid colonies
Aphis glycine biotype 3 was originally collected in a greenhouse at the OARDC in Wooster, OH. The colony has been maintained asexually in the Department of Plant Pathology at the OARDC campus. A. glycine biotype 4 was originally collected in Arlington, WI, in 2013. The colony has been asexually maintained by Dr. Andy Michel in the Department of Entomology at the OARDC in Wooster, OH. Colonies were maintained in growth chambers or rearing rooms at temperatures between 22 and 24 °C, with a photosynthetically active radiation of 330 mol m−2 s−1 for 15 h daily and 60–70% relative humidity (Hill et al. 2004).
Mapping population and soybean aphid resistance assessment
HR09-018 is a high-yielding experimental line developed by USDA-ARS in Wooster, OH, that is highly susceptible to the soybean aphid. PI 243540 is a maturity group IV soybean plant introduction from Japan that carries a single dominant gene Rag2 conferring resistance to soybean aphid biotypes 1 and 2 (Mian et al. 2008). PI 606390A is a maturity group IV soybean plant introduction from Vietnam that is resistance to soybean aphid biotypes 1, 2, and 3 (Bansal et al. 2013). Rag1/Rag2 (Rag1/2) stack is an experimental line homozygous for both aphid-resistance loci (Wiarda et al. 2012). Soybean genotypes were planted into 13.9 cm diameter × 10.5 cm deep pots with four plants per pot and two replicates per genotype in a completely randomized design. Pots were infested with five wingless biotype 3 or biotype 4 adult aphids per plant at the V1 stage in choice tests then evaluated for aphid resistance by counting the number of aphids per plant at 14 days post infestation. Each soybean aphid biotype experiment was separated by space on different greenhouse tables.
A population of 108 F6:7 recombinant inbred lines (RILs) derived from a cross of a USDA experimental line HR09-018 × PI 606390A advanced by single seed decent and the two parental lines were used in this study. The parents and the F6:7 families were assessed for their responses to soybean aphid biotypes 3 and 4 in choice tests in greenhouses in Wooster, OH, using methods and conditions previously described by Jun et al. (2012). Soybean genotypes were planted into 13.9 cm diameter × 10.5 cm deep pots with four plants per pot. Each plant was infested with five wingless adult aphids at the V1 stage in choice tests using a completely randomized design. Biotype 3 aphid resistance was evaluated 4 weeks post infestation from (B3rep1) May 24th to June 21st and (B3rep2) October 4th to November 1st in 2017. In initial screening, plants with high levels of biotype 4 susceptibility became sooty with black mold between 3 and 4 weeks after infestation (data not shown); thus, biotype 4 aphid resistance was evaluated 3 weeks post infestation from (B4rep1) August 16th to September 6th, 2017, and (B4rep2) December 24th, 2017, to January 17th, 2018. Each line was evaluated for aphid resistance by counting the number of aphids per plant.
Molecular marker genotyping and linkage analysis
The construction of the genetic map was carried out using all 108 RILs and the two parental lines. DNA was extracted from the first trifoliate leaves using DNeasy Plant Minikit (Qiagen). SNP genotyping was performed at the Genome Center, University of California, Davis, using the Illumina Infinium BARCSoySNP6K v2 BeadChip. Marker genotypes were analyzed for quality using the GenomeStudio software version 2.0 (Illumina Inc., San Diego, CA). Unique polymorphic SNPs were filtered for missing data and heterozygosity (< 10%). The genetic map was constructed with 1847 unique SNPs (31% of the BeadChip) using Kosambi’s mapping function in JoinMap4 (Van Ooijen 2006). Linkage group assignment was called with a logarithm of odds (LOD) threshold of 4.0 and a maximum recombination frequency of 0.4.
Statistical and mapping analysis
The soybean genotype mean of aphid number data from each biotype and experiment were analyzed separately. Data normalization was performed using the square root (sqrt) of the aphid number. Analysis of variance (ANOVA) and broad-sense heritability estimates were performed in R using the package (Rcmdr) R commander (Fox, 2005). Composite interval mapping (CIM) was conducted using the Haley-Knott regression in R using the package (Rqtl) (Broman et al. 2003). The genome-wide LOD threshold at 5% significance level was determined using a 1000 permutation test for each experiment individually. The phenotypic variation explained by each locus was calculated from the value at the QTL peak indicated by CIM.
Results
Evaluation of aphid resistance
The numbers of soybean aphid biotype 3 on HR09-018, PI 243540 (Rag2), PI 606390A, and Rag1/2 stack plants ranged from 33.75 to 162.25 at 2 weeks post infestation (wpi), whereas the numbers of biotype 4 aphids ranged 52.75–378.75 in choice tests at 2 wpi (Table 1). Analysis of variance of the mean number of aphids indicated that the main effects of aphid biotype and soybean genotype were both significant (P = 1.05 × 10−8 and P = 8.58 × 10−10, respectively). A significant interaction between aphid biotype × soybean genotype was also detected (P < 1.05 × 10−4). PI 606390A was more resistant to soybean aphid biotype 3 than HR09-018 and PI 243540 and was more resistant to biotype 4 than HR09-018, PI 243540, and Rag1/2 stack (Tukey’s test, P < 0.05).
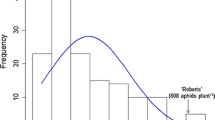
In the experiments to characterize the genetics of aphid resistance in PI 606390A, significantly more biotype 3 (P < 0.0018) and biotype 4 (P < 0.00013) aphids were found on the susceptible parent HR09-018 than the resistance parent PI 606390A. The frequency distributions of aphid numbers for the RILs population were continuous, but not normal, and were skewed toward the resistant parent (Fig. 1). Broad-sense heritability estimates ranged from 0.69 to 0.78 for biotype 3 aphid numbers and from 0.85 to 0.91 for biotype 4 numbers (Table 2). In addition, correlation coefficients between biotypes 3 and 4 in the summer (0.45; B3rep1 and B4rep1) and in the autumn (0.55; B3rep2 and B4rep2) were significant (P < 0.001) (Table 2).
Linkage analysis and QTL mapping
Genotyping with the Infinium BARCSoySNP6K BeadChip revealed 2696 polymorphic SNPs (44.9%) between HR09-018 and PI 606390A that segregated in the RIL population. Across all markers and individuals, genotypic frequencies were consistent with a F6 population (AA, 47.6%; AB, 3.3%; BB, 49.1%). A total of 1847 unique SNPs were mapped into 22 linkage groups where markers from chromosomes 14 and 19 mapped to two linkage groups each. The total length of the map was 2497.94 cM with a mean linkage group length of 113.54 cM. Average marker interval ranged from 0.78 to 3.97 cM with an overall mean of 1.47 cM (Table 3); however, two large gaps (> 20 cM) were observed on chromosome 1. Seven SNPs were mapped to linkage groups that did not correspond to previously determined chromosomal assignments (Supplemental Data 1).
A single genomic region on the genetic map was significantly associated with soybean aphid biotype 3 and 4 resistance using composite CIM (Fig. 2). Fourteen markers in the interval between 92 and 100 cM on chromosome 18 were associated with biotype 3 resistance in B3rep1 (Table 4). Among these markers, Gm18_56839033_A_C had the most significant association, explaining 40.35% of the phenotypic variation (LOD 14.54) of the aphid resistance (Table 4). In B3rep2, biotype 3 resistance was associated with six markers between 98 and 104 cM on chromosome 18. A composite marker, c18.loc100, had the highest LOD score (8.36) and explained 26.43% of the phenotypic variation (Table 4). The same ten markers in an interval between 98 and 106.54 cM on chromosome 18 were associated with soybean aphid biotype 4 resistance in B4rep1 and B4rep2 (Table 4). The most significant marker for B4rep1 was Gm18_57832929_C_A, while a composite marker c18.loc102 had the highest LOD score (12.13) for B4rep2. Each marker explained 32.53 and 35.31% of the phenotypic variation in each of the soybean aphid biotype 4 screening, respectively (Table 4). Estimates of the additive effect ranged between − 16.61 and − 66.43 aphids per plant (Supplemental Fig. S1).
Discussion
Host plant resistance in PI 606390A to soybean aphid biotypes 1, 2, and 3 was first reported by Bansal et al. (2013). We developed a RIL population from a cross between HR09-018 (aphid susceptible) and PI 606390A (aphid resistant) to map soybean aphid resistance. In an initial study, choice tests were performed in the greenhouse using biotype 3 and biotype 4 aphids on HR09-018, PI 243540 (Rag2), PI 606390A, and a Rag1/2 stack line. Soybean aphid biotype 3 numbers on PI 243540 in this study were consistent with results (aphid numbers = 100–200, 2 wpi) from Bansal et al. (2013). We found that, under the same environmental conditions, biotype 4 numbers on Rag1/2 were larger than those for biotype 3. Greater biotype 4 numbers than biotype 3 numbers on a different Rag1/2 line was also observed by Ajayi-Oyetunde et al. (2016). A significant interaction of aphid biotype × soybean genotype was observed which indicates that the soybean aphid biotypes were biologically different. Moreover, PI 606390A was more resistant to biotype 4 aphids than HR09-018, PI 243540, and Rag1/Rag2 (Table 1).
A linkage map was constructed using 1847 SNP markers distributed across 22 linkage groups (Table 3). Overall, the linkage map corresponded well with the Soybean Consensus Map 4.0 for the total length of the genetic map (2537 cM) (Hyten et al. 2010) and a more recent and improved high-resolution map (2346 cM) (Lee et al. 2015a). Markers from chromosomes 14 and 19 were each mapped to two independent linkage groups. Splitting of chromosomes into multiple linkage groups can be the result of poor marker coverage and/or large regions of heterochromatin with reduced recombination rates. In Williams 82, heterochromatic regions were recently shown to extend from 9.7 to 43.7 Mb and 8.9 to 34.3 Mb on chromosomes 14 and 19, respectively (Song et al. 2016). In addition, several markers mapped to linkage groups comprised markers from a different chromosome (Supplemental data 1). This erroneous linkage may be due to sequence variation between the reference genome (Williams 82) and the parental germplasm or possible hybridization of the oligonucleotides to paralogous sequences (Schmutz et al. 2010).
To identify QTLs associated with soybean aphid biotype 3 and 4 resistance, we used a F6:7 population derived from the cross of HR09-018 × PI 606390A. Aphid numbers on test plants were lower in experiments run in the late fall (B3rep2 and B4rep2) compared with those carried out in the summer (B3rep1 and B4rep1) regardless of biotype, consistent with reduced rates of aphid population doubling at sub-optimal temperatures (McCornack et al. 2004). In both tests of soybean responses to aphid biotypes 3 and 4, the frequency distribution of aphid numbers was continuous and skewed toward lower aphid numbers suggesting polygenic resistance (Fig. 1). However, the moderate correlation of aphid numbers on individuals in the summer (B3rep1 vs. B4rep1; 45%) and late fall (B3rep2 and B4rep2; 55%) indicated the presence of a single gene or tightly linked genes conferring resistance to both biotypes 3 and 4. Resistance to both aphid biotypes mapped to an overlapping 14 cM long region of chromosome 18 in PI 606390A. Across all trials, the locus explained was between 26 and 40% of the phenotypic variation; however, the lowest R2 value was observed in the fall biotype 3 screening (B3rep2) when aphid counts were the lowest. Further research is needed to test whether resistance to both biotypes is conferred through a single gene or two tightly linked genes. Moreover, the locus explained between 26 and 40% of the phenotypic variation suggesting additional QTL may be present that requires a larger population size to elucidate.
Rag2 was identified as a single dominant locus on chromosome 13 conferring resistance to soybean aphid biotypes 1 and 2 (Mian et al. 2008); however, this locus is defeated by soybean aphid biotypes 3 and 4. Pyramiding Rag1 (Chr 7) and Rag3 (Chr 16) or Rag1, Rag2, and Rag3 is required for strong biotype 3 resistance and moderate resistance to biotype 4 (Ajayi-Oyetunde et al. 2016). In addition, five soybean PIs with resistance to biotype 4 aphids were recently characterized; although, resistance mapped to previously known Rag loci. These PIs originated from China (Hill et al. 2017). PI 606390A was collected from Vietnam, suggesting a genetically distinct resistance mechanism. Soybean aphid-resistance loci have not previously been mapped to chromosome 18. Using the convention of the Soybean Genetics Committee, the new major locus identified in PI 606390A will be designated qRag-G, standing for resistant to Aphis glycines QTL on linkage group G.
The region of chromosome 18 most strongly correlated with resistance includes a 6.18 cM segment (0.99 Mb) that encodes 121 putative genes in the Williams 82 v2 genome assembly (www.soybase.org). Of these genes, there are several defense-related genes including 10 kinases (five leucine-rich repeat receptor-like kinases, LRR-RLK) along with an AUTOPHAGY 4-RELATED gene. LRR-RLKs have been shown to function in pest perception and as initiators of pathogen/microbe/damage-associated molecular patterns (Goff and Ramonell 2007). Rag5 also maps to a region where 4 of 13 candidate genes are LRR proteins. Moreover, transcriptome analysis of near-isogenic lines with and without Rag5 indicated a suite of differentially expressed genes consistent with the host response to phloem-feeding insects (Lee et al. 2017). In addition, fine mapping of Rag1 and Rag2 has revealed the presence of nucleotide-binding site leucine-rich repeat (NBS-LRR) genes within the QTL segments that confer biotype-specific resistance to the soybean aphid (Kim et al. 2010a, 2010b).
qRag-G also overlaps several QTLs which confer biotype-specific resistance to Aulacorthum solani foxglove aphid. These QTLs were mapped in PI 366121, a Glycine soja accession (wild soybean). A major foxglove aphid-resistance locus, Raso2, also mapped in PI 366121 corresponds to the Rag1 interval on chromosome 7 (Lee et al. 2015b). The sources of Rag1, G. max cv. “Jackson” and “Dowling” were not resistant to A. solani. Thus, further research is needed to determine if Rag genes evolved from Raso genes during the domestication of G. max.
In summary, a new major locus conferring resistance to the soybean aphid biotypes 3 and 4 has been identified and mapped to chromosome 18. The qRag-G locus is independent from all other previously mapped Rag loci. SNP markers linked to qRag-G will be useful for fine mapping and marker-assisted selection of aphid resistance into high-yielding cultivars. Due to the identification of four known soybean aphid biotypes, pyramiding of qRag-G with other Rag loci may enhance the durability of soybean lines with various combinations of aphid-resistance loci.
References
Ajayi-Oyetunde OO, Diers BW, Lagos-Kutz DM, Hill CB, Hartman GL, Reuter-Carlson U, Bradley CA (2016) Differential reactions of soybean isolines with combinations of aphid resistance genes Rag1, Rag2, and Rag3 to four soybean aphid biotypes. J Econ Entomol 109:1431–1437
Alt J, Ryan-Mahmutagic M (2013) Soybean aphid biotype 4 identified. Crop Sci 53:1491–1495
Bansal R, Mian MAR, Michel A (2013) Identification of novel sources of host plant resistance to known soybean aphid biotypes. J Econ Entomol 106(3):1479–1485
Broman KW, Wu H, Sen S, Churchill GA (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19:889–890
Cassone BJ, Wenger JA, Michel AP (2015) Whole genome sequence of the soybean aphid endosymbiont Buchnera aphidicola and genetic differentiation among biotype-specific strains. J Genomics 3:85–94
DiFonzo C, Hines R (2002) Soybean aphid in Michigan: Update from 2001 season, Michigan State University Extension Bulletin E–2746
Enders LS, Miller NJ (2016) Stress-induced changes in abundance differ among obligate and facultative endosymbionts of the soybean aphid. Ecol Evol 6(3):818–829
Fox J (2005) The R Commander: a basic statisicals graphical user interface to R. J Stat Softw 14:1–42
Goff KE, Ramonell KM (2007) The role and regulation of receptor-like kinases in plant defense. Gene Reg Sys Biol 1:167
Hartman GL, Domier LL, Wax LM, Helm CG, Onstad DW, Shaw JT, Solter LF, Voegtlin DJ, D’Arcy CJ, Gray ME, Steffey KL, Isard SA, Orwick PL (2001) Occurrence and distribution of Aphis glycines on soybeans in Illinois in 2000 and its potential control. Online Plant Health Progress. Plant Health Progress
Hill JH, Alleman R, Hogg DB, Grau CR (2001) First report of transmission of soybean mosaic virus and alfalfa mosaic virus by Aphis glycines in the New World. Plant Dis 85:561
Hill CB, Li Y, Hartman GL (2004) Resistance of Glycine species and various cultivated legumes to the soybean aphid (Homoptera: Aphididae). J Econ Entomol 97:1071–1077
Hill CB, Li Y, Hartman GL (2006a) A single dominant gene for resistance to the soybean aphid in the soybean cultivar Dowling. Crop Sci 46:1601–1605
Hill CB, Li Y, Hartman GL (2006b) Soybean aphid resistance in soybean Jackson is controlled by a single dominant gene. Crop Sci 46:1606–1608
Hill CB, Shiao D, Fox CM, Hartman GL (2017) Characterization and genetics of multiple soybean aphid biotype resistance in five soybean plant introductions. Theor Appl Genet 130:1335–1348
Hyten DL, Choi IY, Song Q, Specht JE, Carter TE, Shoemaker RC, Hwang EY, Matukumalli LK, Cregan PB (2010) A high density integrated genetic linkage map of soybean and the development of a 1536 universal soy linkage panel for QTL mapping. Crop Sci 50:960–968
Iwaki M, Roechan M, Hibino H, Tochihara H, Tantera DM (1980) A persistent aphid borne virus of soybean, Indonesian soybean dwarf virus transmitted by Aphis glycines. Plant Dis 64:1027–1030
Jun TH, Mian MAR, Michel AP (2012) Genetic mapping revealed two loci for soybean aphid resistance in PI 567301B. Theor Appl Genet 124:13–22
Kim KS, Bellendir S, Hudson K, Hill C, Hartman G, Hyten D, Hudson M, Diers B (2010a) Fine mapping the soybean aphid resistance gene Rag1 in soybean. Theor Appl Genet 120:1063–1071
Kim KS, Hill CB, Hartman GL, Hyten DL, Hudson ME, Diers BW (2010b) Fine mapping of the soybean aphid-resistance gene Rag2 in soybean PI 200538. Theor Appl Genet 121:599–610
Lee S, Freewalt KR, McHale LK, Song Q, Jun T-H, Michel AP, Dorrance AE, Mian MAR (2015a) A high resolution genetic linkage map of soybean based on 357 recombinant inbred lines genotyped with BARCSoySNP6k. Mol Breed 35:58. https://doi.org/10.1007/s11032-015-0209-5
Lee JS, Yoo MH, Jung JK, Bilyeu KD, Lee JD, Kang S (2015b) Detection of novel QTLs for foxglove aphid resistance in soybean. Theor Appl Genet 128:1481–1488
Lee S, Cassone B, Wijeratne A, Jun T, Michel A, Mian MAR (2017) Transcriptomic dynamics in soybean near-isogenic lines differing in alleles for an aphid resistance gene, following infestation by soybean aphid biotype 2. BMC Genomics 18:472
Mccornack BP, Ragsdale DW, Venette RC (2004) Demography of soybean aphid (Homoptera: Aphididae) at summer temperatures. J Econ Entomol 97:854–861
Mian MAR, Kang ST, Beil SE, Hammond RB (2008) Genetic linkage mapping of the soybean aphid resistance gene in PI 243540. Theor Appl Genet 117:955–962
Michel AP, Mittapalli O, Mian MAR (2011) Evolution of soybean aphid biotypes: understanding and managing virulence to host-plant resistance. In Soybean-Molecular Aspects of Breeding. Edited by Sudarić A. Intech; 355–372
NASS (2018) National Agricultural Statistics Service. United States
Oliver KM, Degnan PH, Burke GR, Moran NA (2010) Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu Rev Entomol 55:247–266
Ostlie K (2002) Managing soybean aphid, University of Minnesota Extension Service, St Paul. http://www.soybeans.umn.edu/crop/insects/aphid/aphid_publication_managingsba.htm.
Riedell W, Catangui M (2006) Greenhouse studies of soybean aphid (Hemiptera: Aphididae) effects on plant growth, seed yield and composition. J Agric Urban Entomol 23:225–235
Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu S, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, Du J, Tian Z, Zhu L, Gill N, Joshi T, Libault M, Sethuraman A, Zhang X, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht J, Grimwood J, Rokhsar D, Stacey G, Shoemaker RC, Jackson SA (2010) Genome sequence of the palaeopolyploid soybean. Nature 463:178–183
Song Q, Jenkins J, Jia G, Hyten D, Pantalone V, Jackson S, Schmutz J, Cregan P (2016) Construction of high resolution genetic linkage maps to improve the soybean genome sequence assembly Glyma1.01. BMC Genomics 17:33
Van Ooijen JW (2006) JoinMap4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations. Kyazma BV, Wageningen
Wenger JA, Michel AP (2013) Implementing an evolutionary framework for understanding genetic relationships of phenotypically defined insect biotypes in the invasive soybean aphid (Aphis glycines). Evol Appl 6:1041–1053
Wiarda SL, Fehr WR, O’Neal ME (2012) Soybean aphid (Hemiptera: Aphididae) development on soybean with Rag1 alone, Rag2 alone, and both genes combined. J Econ Entomol 105(1):252–258
Wulff JA, White JA (2015) The endosymbiont Arsenophonus provides a general benefit to soybean aphid (Hemiptera: Aphididae) regardless of host plant resistance (Rag). Environ Entomol 31:1–8
Zhang G, Gu C, Wang D (2009) Molecular mapping of soybean aphid resistance genes in PI 567541B. Theor Appl Genet 118:473–482
Zhang G, Gu C, Wang D (2010) A novel locus for soybean aphid resistance. Theor Appl Genet 120:1183–1191
Zhang S, Zhang Z, Wen Z, Gu C, An Y, Bales C, DiFonzo C, Song Q, Wang D (2017) Fine mapping of the aphid resistance genes Rag6 and Rag3c from Glycine soja 85-32. Theor Appl Genet 130(12):2601–2615
Funding
This research was partially funded through a grant from the Ohio Soybean Council
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
LaMantia, J.M., Mian, M.A.R. & Redinbaugh, M.G. Genetic mapping of soybean aphid biotype 3 and 4 resistance in PI 606390A. Mol Breeding 39, 53 (2019). https://doi.org/10.1007/s11032-019-0956-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-019-0956-9