Abstract
Rapid establishment of canopy coverage decreases soil evaporation relative to transpiration, improves water use efficiency and light interception, and increases soybean competitiveness against weeds. The objective of this study was to identify genomic loci associated with canopy coverage (CC). Canopy coverage was evaluated using a panel of 373 MG IV soybean genotypes that was grown in five environments. Digital image analysis was used to determine canopy coverage two times (CC1 and CC2) during vegetative development approximately 8 to 16 days apart for each environment. After filtration for quality control, 31,260 SNPs with a minor allele frequency (MAF) ≥ 5% were used for association mapping with the FarmCPU model. Analysis identified significant SNP-canopy coverage associations including 36 for CC1 and 56 for CC2. Five SNPs for CC1 and 11 SNPs for CC2 were present in at least two environments. The significant SNP associations likely tagged 33 (CC1) and 50 (CC2) different quantitative trait loci (QTLs). Eleven putative loci were identified in which chromosomal regions associated were coincident for CC1 and CC2. Candidate genes identified using these significant SNPs included those with reported functions associated with growth, developmental, and light responses. Favorable alleles from significant SNPs may be an important resource for pyramiding genes to improve canopy coverage and for identifying parental genotypes for use in breeding programs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Genome-wide association analysis is an alternative approach to traditional quantitative trait loci (QTL) mapping of bi-parental populations and is widely used in plant and human genetics (Nordborg and Tavaré 2002; Risch and Merikangas 1996). Genotyping diverse lines provides thousands of single-nucleotide polymorphisms (SNPs) across the genome, which allows fine mapping of complex trait variation down to the nucleotide level by exploiting historical recombination events (Zhu et al. 2008). The main advantages of genome-wide association studies (GWAS) over traditional linkage mapping include increased mapping resolution, reduced research time, and greater allele number (Yu et al. 2006).
In soybean, GWAS have identified chromosomal regions associated with seed protein and oil concentrations (Hwang et al. 2014), carotenoids (Dhanapal et al. 2015a), carbon 13 to 12 ratio (Dhanapal et al. 2015b; Kaler et al. 2017a), oxygen 18 to 16 ratio (Kaler et al. 2017a), canopy wilting (Kaler et al. 2017b), agronomic traits (Wen et al. 2014), and ureide concentration (Ray et al. 2015) as well as other studies. GWAS in soybean are likely to increase due to recent genotyping of more than 19,000 accessions of the USDA-ARS Soybean Germplasm collection that provided over 50,000 SNP markers. The complete SNP marker dataset is available at SoyBase (www.soybase.org).
Early establishment of a closed canopy is beneficial for several reasons. A faster closed canopy increases water use efficiency by enhancing transpiration relative to soil evaporation (Purcell and Specht 2004; Rebetzke et al. 2007; Richards et al. 2007; Slafer et al. 2005). By reducing the water lost through soil evaporation, quick canopy establishment ensures that more soil moisture is available for photosynthesis and biomass production that otherwise would be lost to evaporation.
A second advantage offered by rapid establishment of canopy coverage is improved canopy solar radiation interception, which is an important factor determining crop growth and yield (Edwards and Purcell 2005; Edwards et al. 2005; Liebisch et al. 2015). The interception of radiation by the canopy provides the energy required for physiological processes including photosynthesis and transpiration (Liebisch et al. 2015). Capacity of the crop canopy to intercept solar radiation determines yield, which depends on the available leaf area, structure, and its efficiency (Gifford et al. 1984).
Rapid establishment of canopy closure also increases soybean competiveness, especially for weeds (Bussan et al. 1997). Herbicide-resistant weeds are becoming a large problem in crop production (Green and Owen 2011), and this problem is mainly due to the high selection pressures imposed by widespread use of mono-herbicide culture (Shaner 1995). As the number of herbicide-resistant weeds increase, there is a need for alternative and sustainable approaches to weed management. Faster canopy development can suppress early-season weeds (Fickett et al. 2013; Jannink et al. 2000, 2001), and rapid canopy development can, therefore, serve as a cultural control method to suppress weed growth by increasing soybean competiveness.
There are differences in competitiveness among soybean genotypes due to how quickly the canopy is established (Place et al. 2011a, b) that appear to be associated with individual seed weight. Place et al. (2011b) divided seed lots of three cultivars into five seed weight classes that ranged from 50 to 326 mg seed−1. Canopy coverage at 3 weeks after emergence for all genotypes increased with increasing individual seed weight. Larger seed classes improved competitiveness by increasing plant height and petiole length. Small-seeded, natto soybean types were generally slower to establish a canopy compared to large-seeded, tofu type soybean (Place et al. 2011a).
Purcell (2000) described a method of analyzing digital images that offers a simple and effective way to determine canopy coverage. Canopy coverage was measured as a fraction of green pixels relative to the total number of pixels in an image, and canopy coverage was approximately equivalent to the fraction of radiation intercepted. Digital-image analysis provides an inexpensive and rapid way of measuring canopy coverage over other methods of estimating radiation interception (Campillo et al. 2008; Fiorani et al. 2012). The analysis of digital images has been widely accepted as a high-throughput method for determining canopy coverage or the fraction of solar radiation intercepted (De Bruin and Pedersen 2008; Edward et al. 2005; Gaspar and Conley 2015; Xavier et al. 2017).
Canopy coverage is a quantitative trait that is influenced by genotype, environment, and their interaction (Xavier et al. 2017). The complexity of this trait arises from the segregation of alleles at many chromosomal regions, each with small effects on the phenotype, and interacting with other alleles and with the environment (Tuberosa et al. 2007). Therefore, investigation of genetic control of canopy coverage may be used to improve crop performance by selecting and pyramiding favorable loci associated with faster establishment of the canopy into elite cultivars (Xavier et al. 2017).
To date, Xavier et al. (2017) have conducted the only genetic evaluation of canopy coverage in soybean. They phenotyped the soybean nested association mapping (SoyNAM) population for canopy coverage with a relatively small set of markers (4077 SNPs) and found six genomic regions that were associated with canopy coverage. The SoyNAM panel was developed from crossing 40 genotypes to one hub parent, and from each cross, 140 RILs were developed. Although the SoyNAM panel was selected for diversity, it is likely that these 41 genotypes may be somewhat limited in capturing the wide diversity of phenotypes that would impact canopy coverage. In the present research, 31,260 polymorphic SNPs were utilized for GWAS of canopy coverage, and canopy coverage was assessed on a panel of 373 diverse maturity group (MG) IV accessions. The objective of this study was to use GWAS to explore the genetic variation of canopy coverage present within this panel by identifying significant loci associated with this trait.
Materials and methods
Germplasm collection and field experiments
A panel of 373 MG IV soybean accessions was selected from the USDA-ARS Soybean Germplasm Collection based on genetic diversity and agronomic characteristics (www.ars.grin.gov). Our selection criteria were based on relatively high yield, relatively good lodging and shattering scores, and diverse geographic distribution. A thorough description of genotype selection criteria was provided by Dhanapal et al. (2015b).
Field experiments were conducted in five environments including the Main Arkansas Agricultural Research Center, Fayetteville, AR (FY; 36° 9′N, 94° 17′W), in 2016 on a Captina silt loam; Pine Tree Research Station, Colt, AR (PT; 35° 7′N, 90° 55′W), in 2016 on a Calloway silt loam; Rohwer Research Station, Rohwer, AR (RH; 33° 48′N, 91° 17′W), in 2016 on a Sharkey silty clay; Salina, KS (SA; 38° 70′N, 97° 60′W), in 2016 on a Hord silt loam; and at the Rice Research and Extension Center, Stuttgart, AR (ST; 34° 47′N, 91° 51′W), in 2015 on a Crowley silt loam. Planting dates were 8 June 2015 for ST, 23 May 2016 for RH, 1 June 2016 for FY, 2 June 2016 for PT, and 15 June 2016 for SA. Seeds were planted at a density of 37 m−2 at a 2.5-cm depth. At ST, plots were 4.57 m long and two rows wide with 0.76-m row spacing. At FY, single row plots were 5.48 m in length with a 0.76-m row spacing. At SA, there were two-row plots that were 3.65 m in length with a 0.76-m row spacing. At PT and RH, seeds were sown with a drill (19-cm row spacing) and plots were 1.52 m wide and 4.57 m long. At each environment, the experiment was conducted as a randomized complete block design with two replications.
For each environment, soil water deficit was estimated for each day beginning at planting as described by Purcell et al. (2007). Potential evapotranspiration (Eto) for a given day was determined using a modified Penman-Monteith approach (Allen et al. 1998) and multiplied by the fraction of radiation intercepted by the crop, which served as a crop coefficient (equivalent to canopy coverage). Estimated soil-water deficits were cumulated and adjusted with rainfall additions as needed.
Canopy coverage determination
Canopy coverage was determined by analyzing digital images that were taken of the canopy with a camera mounted on a monopod (Purcell 2000). Digital images were taken at 160 cm above the plots at a resolution of 1280 × 960 pixels. For phenotypic variation of canopy coverage at ST, FY, and SA, images consisted of a single row, and at PT and RH, images were taken above the center of the plots and were composed of seven rows. For digital images of single rows, care was taken to ensure that the image was centered directly over the row and that the image frame extended from the center line between the two adjacent rows. At the times these measurements were made, leaves from adjacent rows did not cross the center line between plots. The first set of pictures (CC1) was taken 23 to 28 days after emergence when plants were between V2 and V3. A second set of pictures (CC2) was taken 8 to 16 days later between V4 and V7. Plant densities were measured in a 1-m section of row, and results indicated no significant effect of genotype on the plant density. Weeds potentially interfere with canopy cover estimates, and plots were weed free before taking pictures. Digital images were analyzed using SigmaScan Pro (v.5.0, SPSS, Inc., Chicago, IL) with a macro that utilizes batch analysis (Karcher and Richardson 2005). Software measured the number of green pixels of each image as a fraction of the total pixel count in the frame. SigmaScan Pro has selectable ranges of hue and saturation values for identifying green pixels in images. For each environment, the settings for the hue and saturation were determined for several representative images prior to batch analysis. Typical ranges that selectively identified green leaves without soil background were from 50 to 100 for hue and 0 to 100 for saturation.
Place et al. (2011b) reported a strong association of early canopy coverage and individual seed weight. Hence, average phenotypic data of individual seed weight (i.e., the average single seed weight based upon 100 seeds) from 3 years (2014, 2015, and 2016) were included in this study to assess a possible association of canopy coverage with individual seed weight at the genomic level.
Statistical analysis
Data were tested for heterogeneity of variance by checking the residual plots, which did not show any heterogeneity of variance. Genotype and environment were treated as fixed effects and replication within environment was considered a random effect for analysis of variance (ANOVA). The PROC GLIMMIX procedure (α = 0.05) of SAS 9.4 (SAS Institute Inc., Cary, NC, USA 2013) was used for ANOVA with a model suggested by Bondari (2003):
where μ is the total mean, G i is the genotypic effect of the ith genotype, E j is the effect of the jth environment, (GE) ij is the interaction effect between the ith genotype and the jth environment, Bk(ij) is the effect of replications within the jth environment, and ε ijk is a random error following \( N\left(0,{\sigma}_e^2\right) \). Analysis of variance was independently applied to CC1 and CC2. Descriptive statistics and Pearson correlation analysis for canopy coverage were performed using the PROC UNIVARIATE and PROC CORR procedures (α = 0.05) of SAS version 9.4, respectively. Broad sense heritability on an entry-mean basis was calculated as
where \( {\sigma}_G^2 \) is the genotypic variance, \( {\sigma}_{GE}^2 \) is the genotype by environment variance, \( {\sigma}_{\varepsilon}^2 \) is the residual variance, k is the number of environments, and r is the number of replications. The PROC VARCOMP of SAS 9.4 with the restricted maximum likelihood estimation (REML) method was used to estimate the above variance components.
For association analysis, the best linear unbiased prediction (BLUP) values were estimated using the PROC GLIMMIX procedure by environment and across all environments (AAE). The BLUP values were used because they reduce the effects of replication (within environment) and the effects of environment (when conducted across multiple environments).
Genotyping and association analysis
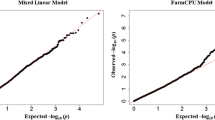
Data for a total of 42,509 SNP markers of all 373 genotypes were obtained from the Illumina Infinium SoySNP50K iSelect SNP Beadchip (Song et al. 2013), which are available at SoyBase (www.soybase.org). Genotype quality controls were applied by eliminating monomorphic markers, markers with minor allele frequency (MAF) < 5%, and markers with missing rate higher than 10%. The remaining missing markers in a set of 31,260 SNPs were imputed using a LD-kNNi method, which is based on a k-nearest-neighbor genotype (Money et al. 2015) and then used in association analysis. Population structure is a confounding factor in GWAS that induces false associations. Commonly, the mixed linear model (MLM) is used to reduce these false associations. However, these adjustments also compromise true positive associations. As described by Kaler et al. (2017a, b), the fixed and random model Circulating Probability Unification (FarmCPU) model, developed by Liu et al. (2016), effectively controlled false positives and false negatives using this same genotype panel. Different models (e.g., compressed mixed linear model from GAPIT, GLM and MLM models from TASSEL) were compared based on their Q-Q plots. The FarmCPU model was superior in controlling the false discovery rate compared to other models evaluated. The Q-Q plot of the FarmCPU model resulted in a sharp deviation from the expected P value distribution in the tail area, indicating that false positives were adequately controlled (data not shown) in agreement with previous research (Kaler et al. 2017b). Therefore, FarmCPU was used for association analysis in the present research. Default parameters were used in FarmCPU model as recommended by the developer of FarmCPU (Liu et al. 2016) except in the change of threshold output from 0.01 to 1, which allowed all markers to be included in the output Excel file.
Although FarmCPU controlled the false positives and false negatives, we still used a stringent threshold value (− log10(P) ≥ 3.5) to declare a significant association between SNPs and canopy coverage. This level of stringency was used to limit the number of associations to those QTLs likely contributing biologically significant effects. This threshold value is more stringent than that reported in other soybean association mapping studies of soybean (Dhanapal et al. 2015a, b; Hao et al. 2012; Hwang et al. 2014; Zhang et al. 2015). We also eliminated any SNP from consideration if the allelic effect was less than the standard error of CC1 or CC2 phenotypic measurements. A threshold value of P ≤ 0.05 was used to identify significant SNPs present in more than one environment but only if representative SNPs met a lower association of P ≤ 0.0003 in at least one other environment.
Candidate gene identification
Candidate genes were considered when they were within ± 10 kb of a SNP with a significant association. This distance was chosen because it approximates the average distance between SNPs (18 kb). Candidate genes and their associated functional annotation and biological function were determined using Glyma1.1, Glyma1.0, and NCBI RefSeq gene models in SoyBase (www.soybase.org) with consideration for those that may have an association with growth, developmental, and light responses.
Results
Phenotype statistics
There was a broad range of CC1 and CC2 values observed within each environment. Overall all 373 accessions, the range (i.e., maximum value–minimum value) of CC1 values was 0.27 (FY), 0.17 (PT), 0.45 (RH), 0.30 (SA), and 0.23 (ST) (Table 1). For CC2, the range was 0.40 (FY), 0.28 (PT), 0.62 (RH), 0.29 (SA), and 0.50 (ST). Analysis of variance indicated that genotype, environment, and their interaction had significant effects (P ≤ 0.05) on CC1 and CC2. Correlations of canopy coverage between environments for CC1 or CC2 were significant and positive (0.10 ≤ r ≤ 0.39) except for the correlation between RH and ST for CC1 (r = − 0.01) (Table 2). Correlations between CC1 and CC2 within environments averaged 0.59 and ranged from 0.76 (FY) to 0.30 (PT) (Table 2). Broad sense heritability by environment on an entry-mean basis indicated low-to-moderate heritability for CC1 and CC2 (0.21 ≤ H2 ≤ 0.54) (Table 1). When considered over all environments, H2 was 0.58 (CC1) and 0.51 (CC2).
For silt-loam soils in our study (FY, ST, PT, and SA), irrigation is typically recommended when soil-moisture deficits exceed 35 mm, and for the clay soil at RH, irrigation is recommended when soil moisture deficits exceed 50 mm (Purcell et al. 2007). Using these irrigation thresholds as indicators of stress, the week prior to CC1, soil moisture was adequate for ST, RH, and SA but was limiting for FY (3 out of 7 days) and PT (7 out of 7 days). Between CC1 and CC2, soil moisture was adequate at ST every day but limiting at FY (3 out of 14 days), PT (5 out of 15 days), SA (9 out of 11 days), and RH (5 out of 9 days). The differences in soil-moisture availability among environments and between CC1 and CC2 likely contributed to the G × E effect.
Genome-wide association analysis
Genome-wide association analysis identified 36 SNPs significantly associated with CC1 at level of − log10 (P) ≥ 3.5; P ≤ 0.0003 (Table S1 and Fig. S1) in at least one of the five environments. Of these 36 SNPs, six were significant in at least two environments. Significant SNPs that were present in a linkage disequilibrium block on the same chromosome were considered as one locus. Thus, the 36 significant SNPs comprised 33 putative QTLs (Table S1). There were 56 significant SNPs associated with CC2 at a level of − log10(P) ≥ 3.5; P ≤ 0.0003 (Table S2 and Fig. S2) in at least one environment. Eleven SNPs of these 56 were significant in at least two environments. These 56 significant SNPs comprised 47 putative QTLs (Table S2). The allelic effect (fractional change in canopy coverage for the major compared to the minor allele) for these significant SNPs for CC1 ranged from − 0.050 to 0.068 (Table S1) and for CC2 ranged from − 0.048 to 0.086 (Table S2). The positive sign indicates that the minor allele was associated with increased canopy coverage. These allelic effects are in comparison to the standard errors of measurement for CC1 and CC2 of 0.004 and 0.001, respectively. The list of all 33 significant loci for CC1 and 47 significant loci for CC2, their corresponding MAF, major or minor allele, allelic effect, R2 value, and common environments where SNPs were significant are listed in Tables S1 and S2, respectively. Table 3 shows the list of significant SNPs associated with CC1 and CC2 that were present in more than one environment, common to both CC1 and CC2, and coincident with previously reported QTLs for canopy coverage (Xavier et al. 2017). Association analysis of canopy coverage averaged across all environments identified 11 SNPs and 12 SNPs significantly associated with CC1 and CC2, respectively, at level of − log10 (P) ≥ 3.5; P ≤ 0.0003 (Table S5).
Individual seed weight averaged over 3 years for these 373 genotypes was significantly (P ≤ 0.05) correlated with CC1 and CC2 (0.14 ≤ r ≤ 0.41) at 7 of the 10 possible combinations of CC1/CC2 and environment (Table 2). Analysis identified 52 SNPs significantly associated with average individual seed weight at a significance level of − log10 (P) ≥ 3.5; P ≤ 0.0003 (data not shown). Nine chromosomal regions were identified, where QTLs associated with canopy coverage and average seed weight were coincident (six with CC1 (Table S1) and three with CC2 (Table S2)). These associations were important because previous studies showed a positive relationship between individual seed weight and canopy coverage (Place et al. 2011a, b). Identifying the common regions would be useful for further research.
Candidate gene identification
Based on the location of SNPs with significant associations, nearby genes (within ± 10 kb of the SNP) were identified that may have association with growth, developmental, and light response. There were 36 genes near SNPs tagging CCI and 56 genes near CC2. Of these genes, 19 (CC1) and 21 (CC2) had SNPs with significant associations within the gene. The remaining genes were present within ± 10 kb of the respective SNPs. Based on their biological function of growth, developmental, and light response that were reported in SoyBase (www.soybase.org), 13 genes for CC1 and 19 genes for CC2 are potential candidate genes for establishing faster canopy coverage.
Discussion
There was wide phenotypic variation of canopy coverage for both CC1 and CC2 within each environment, which is important for dissecting complex traits through association mapping (McCarthy et al. 2008). That there were significant positive correlations between environments for CC1 and CC2, and moderate heritability indicates that canopy coverage can be improved, which has implications for increasing transpiration relative to soil evaporation, light interception, and competitiveness for weeds.
Some of the variability between CC1 and CC2 within environments and among canopy coverage measurements across environments may be related to soil-moisture availability prior to measurements. Drought decreases leaf area development and leaf expansion rate (Clauw et al. 2015; Manandhar et al. 2017; Tardieu and Tuberosa 2010). The week before CC1 measurements there were no soil-moisture limitations at ST, RH, or SA, but soil-moisture deficits exceeded the threshold for irrigation for 3 or 7 days before CC1 at FY and PT, respectively. Between CC1 and CC2 measurements, ST was the only environment in which daily soil-moisture deficits were above the irrigation threshold.
Of the nine genomic regions common to both average seed weight and canopy coverage, eight had the same directional allelic effect (Tables S1 and S2). For example, a genomic region on Gm01, where both canopy coverage and average seed weight were coincident, there was a positive allelic effect for canopy coverage of 0.02 (Table S1) and this region also had a positive allelic effect for average seed weight of 0.05 g seed−1. Both these regions were located within the coding region of a gene, Glyma01g42890, which codes a JUMONJI domain-containing protein with a biological function associated with meristem growth (Table S3). Interestingly, the SNP associated with canopy coverage in this region was present in more than one environment (Table S1). Similarly, two coincident genomic regions on Gm05 and Gm16 also had positive allelic effects for both canopy coverage and average seed weight. These regions were located close to genes, Glyma05g02130 and Glyma16g25880, which have biological functions involved with response to light stimulus (Table S3).
For CC1, 19 major alleles out of 41 were linked with an increase in canopy coverage (positive value of allelic effect indicates that the major allele was associated with an increase in canopy coverage) (Table S1). One SNP on Gm08 that had the largest positive allelic effect (0.07) was present within the coding region of a gene, Glyma08g13160, which codes a chaperone binding protein that has a biological function associated with photosynthesis (Tables S1 and S3). A SNP on Gm16 that had the second largest positive allelic effect (0.05) was also present within the coding region of a gene, Glyma16g25880, which codes a root phototropism protein that has a biological function involved with response to light stimulus (Tables S1 and S3). Out of 41 SNPs, minor alleles of 22 were associated with an increase in canopy coverage (negative value of allelic effect indicates that minor allele was associated with an increase in canopy coverage) (Table S1). One SNP on Gm02 that had a large negative allelic effect (− 0.03) was present within the coding region of a gene, Glyma02g40960, which codes an early growth response protein (Tables S1 and S3).
For CC2, there were 38 out of 56 SNPs for which the major allele associated with an increase in canopy coverage (Table S2). A SNP on Gm18 that had the largest positive allelic effect (0.09) was present within ± 5 kb range of a gene, Glyma18g00530, which codes a DNA repair protein (RAD50) that has a biological function involved with meristem structural organization (Tables S2 and S4). Out of 56 SNPs, the minor alleles of 18 were associated with an increase in canopy coverage (Table S2). One SNP on Gm09 with the largest negative allelic effect (− 0.05) was present within ± 10 kb range of a gene, Glyma09g30370, which codes a protein functioning as a glutamine synthetase clone R1 that has a biological function involved with leaf senescence (Tables S2 and S4).
Xavier et al. (2017) identified seven SNPs associated with canopy coverage, but two of the SNPs on Gm10 were close to one another, which they considered as one QTL. They, therefore, reported six QTLs for canopy coverage using the SoyNAM population. Location of the CC1 and CC2 chromosomal regions identified in this study were compared with QTLs reported by Xavier et al. (2017) for canopy coverage. Likewise, we compared genomic regions of CC1 and CC2 to see if they were coincident (Table 3 and Fig. 1). Four out of six QTLs reported by Xavier et al. (2017) were located close to genomic regions that were associated with CC1 and CC2 in this study (Table 3 and Fig. 1). Twelve putative loci on Gm02 (2), Gm06, Gm07, Gm09 (3), Gm11, Gm16 (3), and Gm20 were identified, where chromosomal regions were associated with both CC1 and CC2 (Table 3 and Fig. 1). These chromosomal regions have candidate genes with a direct function associated with response to auxin, response to gibberellic acid, meristem growth, light-regulated protein, early growth response protein, and response to light intensity (Tables S3 and S4). These putative loci may indicate the stability and importance for improving faster canopy coverage and may highlight the important regions of the genome for further investigations.
Conclusions
A high marker density of 31,260 SNPs with a MAF ≥ 5% was used in this study for association mapping of canopy coverage at two dates (CC1 and CC2) in five environments. There were 36 significant SNPs associated with CC1 and 56 significant SNPs associated with CC2 at a significance level of − log10 (P) ≥ 3.5. Five significant SNPs for CC1 and 11 SNPs for CC2 were present in at least two environments. The 36 SNPs for CC1 and 56 SNPs for CC2 likely tagged 33 and 50 QTLs, respectively. Four different putative loci were located within four genomic regions that were previously reported (Xavier et al. 2017) as QTLs for canopy coverage. Twelve putative loci were identified, where chromosomal regions associated with CC1 and CC2 were coincident. Several of these loci were close to or within genes related to growth and development. We identified significant SNPs that were present in more than one environment, and where chromosomal regions associated with both CC1 and CC2 were found within the same genomic location. This information may be useful in pyramiding alleles for faster canopy coverage and for improving radiation interception, WUE, crop growth, and soybean competitiveness.
References
Allen RG, Pereira LS, Raes D, Smith M (1998) Crop evapotranspiration: guidelines for computing crop water requirements. Irrigation and Drainage Paper No. 56, Food and Agriculture Organization of the United Nations, Rome, Italy
Bondari K (2003) Statistical analysis of genotype × environment interaction in agricultural research. In: Paper SD15, SESUG. The Proceedings of the SouthEast SAS Users Group, St Pete Beach
Bussan AJ, Burnside OC, Orf JH, Ristau EA, Puettmann KJ (1997) Field evaluation of soybean (Glycine max) genotype for weed competitiveness. Weed Sci 45:31–37
Campillo C, Prieto MH, Daza C, Moñino MJ, García MI (2008) Using digital images to characterize canopy coverage and light interception in a processing tomato crop. Hort Science 43:1780–1786.35
Clauw P, Coppens F, De Beuf K, Dhondt S, Van Daele T, Maleux K, Storme V, Clement L, Gonzalez N, Inzé D (2015) Leaf responses to mild drought stress in natural variants of Arabidopsis. Plant Physiol 167:800–816
De Bruin JL, Pedersen P (2008) Soybean seed yield response to planting date and seeding rate in the upper Midwest. Agron J 100:696–703
Dhanapal AP, Ray JD, Singh SK, Hoyos-Villegas V, Smith JR, Purcell LC, King CA, Fritsch FB (2015a) Association mapping of total carotenoids in diverse soybean genotypes based on leaf extracts and high-throughput canopy spectral reflectance measurements. PLoS One 10(9):e0137213
Dhanapal AP, Ray JD, Singh SK, Hoyos-Villegas V, Smith JR, Purcell LC, King CA, Cregan PB, Song Q, Fritsch FB (2015b) Genome-wide association study (GWAS) of carbon isotope ratio (δ13C) in diverse soybean [Glycine max (L.) Merr.] genotypes. Theor Appl Genet 128:73–91
Edward JT, Purcell LC, Karcher DE (2005) Soybean yield and biomass responses to increasing plant population among diverse maturity groups. II Light interception and utilization Crop Sci 45:1778–1785
Edwards JT, Purcell LC (2005) Soybean yield and biomass responses to increasing plant population among diverse maturity groups: I Agronomic characteristics. Crop Sci 45:1770–1777
Fickett ND, Boerboom CM, Stoltenberg DE (2013) Soybean yield loss potential associated with EarlySeason weed competition across 64 site-years. Weed Sci 61:500–507
Fiorani F, Rascher U, Jahnke S, Schurr U (2012) Imaging plants dynamics in heterogenic environments. Curr Opin Biotech 23:227–235
Gaspar AP, Conley SP (2015) Responses of canopy reflectance, light interception, and soybean seed yield to replanting suboptimal stands. Crop Sci 55(1):377–385
Gifford RM, Thorne JH, Hitz WD, Giaquinta RT (1984) Crop productivity and photoassimilate partitioning. Science 225:801–808
Green JM, Owen MDK (2011) Herbicide-resistant crops: utilities and limitations for herbicide resistant weed management. J Agric Food Chem 59:5819–5829
Hao D, Cheng H, Yin Z, Cui S, Zhang D, Wang H, Yu D (2012) Identification of single nucleotide polymorphisms and haplotypes associated with yield and yield components in soybean (Glycine max) landraces across multiple environments. Theor Appl Genet 124:447–458
Hwang E, Song Q, Jia G, Specht JE, Hyten DL, Costa J, Cregan PB (2014) A genome-wide association study of seed protein and oil content in soybean. PLoS Genet 15:1
Jannink JL, Orf JH, Jordan NR, Shaw RG (2000) Index selection for weed suppressive ability in soybean. Crop Sci 40:1087–1094
Jannink JL, Jordan NR, Orf JH (2001) Feasibility of selection for high weed suppressive ability in soybean: absence of tradeoffs between rapid initial growth and sustained later growth. Euphytica 120:291–300
Kaler AS, Dhanapal AP, Ray JD, King CA, Fritsch FB, Purcell LC (2017a) Genome-wide association mapping of carbon isotope and oxygen isotope ratios in diverse soybean genotypes. Crop Sci 57:3085–3100
Kaler AS, Ray JD, King CA, Schapaugh WT, Purcell LC (2017b) Genome-wide association mapping of canopy wilting in diverse soybean genotypes. Theor Appl Genet 130:1–15
Karcher DE, Richardson MD (2005) Batch analysis of digital images to evaluate turfgrass characteristics. Crop Sci 45:1536–1539
Liebisch F, Kirchgessner N, Schneider D, Walter A, Hund A (2015) Remote, aerial phenotyping of maize traits with a mobile multi-sensor approach. Plant Methods 11:9
Liu X, Huang M, Fan B, Buckler ES, Zhang Z (2016) Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet 12(2):e1005767
Manandhar A, Sinclair TR, Rufty TW, Ghanem ME (2017) Leaf emergence (phyllochron index) and leaf expansion response to soil drying in cowpea genotypes. Physiol Plantarum 160:201–208
McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JPA, Hirschhorn JN (2008) Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nature Rev Genet 9(5):356–369
Money D, Gardner K, Migicovsky Z, Schwaninger H, Zhong GY, Myles S (2015) LinkImpute: fast and accurate genotype imputation for non-model organisms. G3 5(11):23383–22390
Nordborg M, Tavaré S (2002) Linkage disequilibrium: what history has to tell us. Trends Genet 18(2):83–90
Place GT, Reberg-Horton SC, Dickey DA, Carter TE (2011a) Identifying soybean traits of interest for weed competition. Crop Sci 51:2642–2654
Place GT, Reberg-Horton SC, Carter TE, Smith AN (2011b) Effects of soybean seed size on weed competition. Agron J 103:175–181
Purcell LC (2000) Soybean canopy coverage and light interception measurements using digital imagery. Crop Sci 40:834–837
Purcell LC, Specht JE (2004) Physiological traits for ameliorating drought stress. In: Boema HR, Specht JE (eds) Soybeans: improvement, production, and uses. 3rd ed. Madison, WI: American Society of Agronomy. Pp, pp 569–620
Purcell LC, Edwards JT, Brye KR (2007) Soybean yield and biomass responses to cumulative transpiration: questioning widely held beliefs. Field Crop Res 101:10–18
Ray JD, Dhanapal AP, Singh SK, Hoyos-Villegas V, Smith JR, Purcell LC, King CA, Boykin D, Cregan PB, Song Q, Fritschi FB (2015) Genome-wide association study of ureide concentration in diverse maturity group IV soybean [Glycine max (L.) Merr.] accessions. G3 5(11):2391–2403
Rebetzke GJ, Ellis MH, Bonnett DG, Richards RA (2007) Molecular mapping of genes for coleoptile growth in bread wheat (Triticum aestivum L.) Theor Appl Genet 114:1173–1183
Richards RA, Watt M, Rebetzke GJ (2007) Physiological traits and cereal germplasm for sustainable agricultural systems. Euphytica 154:409–425
Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273(5281):1516–1517
SAS Institute (2013) The SAS system for Windows. Version 9.3. SAS Inst. Inc., Cary, NC
Shaner DL (1995) Herbicide resistance: where are we? How did we get here? Where are we going? Weed Technol 9:850–856
Slafer GA, Araus JL, Royo C, Garcia Del Moral LF (2005) Promising eco-physiological traits for genetic improvement of cereal yields in Mediterranean environments. Ann Appl Biol 146:61–70
Song Q, Hyten DL, Jia G, Quigley CV, Fickus EW, Nelson RL, Cregan PB (2013) Development and evaluation of SoySNP50K, a high-density genotyping array for soybean. PLoS One 8(1):e54985
Tardieu F, Tuberosa R (2010) Dissection and modelling of abiotic stress tolerance in plants. Curr Opin Plant Biol 13:206–212
Tuberosa R, Salvi S, Giuliani S, Sanguineti MC, Bellotti M, Conti S, Landi P (2007) Genome-wide approaches to investigate and improve maize response to drought. Crop Sci 47:120–141
Wen Z, Tan R, Yuan J, Bales C, Du W (2014) Genome-wide association mapping of quantitative resistance to sudden death syndrome in soybean. BMC Genomics 15:809
Xavier A, Hall B, Hearst AA, Cherkauer KA, Rainey KM (2017) Genetic architecture of phenomic-enabled canopy coverage in Glycine max. Genetics 206:1081–1089. https://doi.org/10.1534/116.198713
Yu J, Pressoir G, Briggs WH, Vroh BI, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen HJB, Kresovich S, Buckler ES (2006) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38:203–208
Zhang J, Song Q, Cregan PB, Nelson RL, Wang X, Wu J, Jiang GL (2015) Genome-wide association study for flowering time, maturity dates and plant height in early maturing soybean (Glycine max) germplasm. BMC Genomics 16:217. https://doi.org/10.1186/s12864-015-1441-4
Zhu C, Gore MA, Buckler ES, Yu J (2008) Status and prospects of association mapping in plants. Plant Genome 1:5–20
Acknowledgements
The authors gratefully acknowledge partial funding of this research from the United Soybean Board. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. The USDA is an equal opportunity provider and employer.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kaler, A.S., Ray, J.D., Schapaugh, W.T. et al. Association mapping identifies loci for canopy coverage in diverse soybean genotypes. Mol Breeding 38, 50 (2018). https://doi.org/10.1007/s11032-018-0810-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-018-0810-5