Abstract
The soybean aphid (Aphis glycines Matsumura), an invasive species, has posed a significant threat to soybean [Glycine max (L.) Merr.] production in North America since 2001. Use of resistant cultivars is an effective tactic to protect soybean yield. However, the variability and dynamics of aphid populations could limit the effectiveness of host-resistance gene(s). Gene pyramiding is a promising way to sustain host-plant resistance. The objectives of this study were to determine the prevalent aphid biotypes in Michigan and to assess the effectiveness of different combinations of aphid-resistance genes. A total of 11 soybean genotypes with known resistance gene(s) were used as indicator lines. Based on their responses, Biotype 3 was a major component of Michigan aphid populations during 2015–2016. The different performance of Rag-“Jackson” and Rag1-“Dowling” along with the breakdown of resistance in plant introductions (PIs) 567301B and 567324 may be explained by Biotype 3 or an unknown virulent biotype establishing in Michigan. With the assistance of flanking markers, 12 advanced breeding lines carrying different aphid-resistance gene(s) were developed and evaluated for effectiveness in five trials across 2015 to 2017. Lines with rag1c, Rag3d, Rag6, Rag3c + Rag6, rag1b + rag3, rag1c + rag4, rag1c + rag3 + rag4, rag1c + Rag2 + rag3 + rag4, and rag1b + rag1c + rag3 + rag4 demonstrated strong and consistent resistance. Due to the variability of virulent aphid populations, different combinations of Rag genes may perform differently across geographies. However, advanced breeding lines pyramided with three or four Rag genes likely will provide broader and more durable resistance to diverse and dynamic aphid populations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soybean [Glycine max (L.) Merr.] is one of the most important crops in North America because of its multiple uses as an animal feed, cooking oil, biofuel, and human protein source. In 2016, the USA ranked first in world soybean production (117.3 million metric tons) with 55.3 million metric tons exported (SoyStats 2016). However, soybean production in North America has been threatened by the soybean aphid, Aphis glycines Matsumura (Hemiptera: Aphididae), an invasive species native to Asia (Wu et al. 2004).
Soybean aphid has aggressively dispersed to all major soybean producing areas in the USA and Canada (Ragsdale et al. 2011) since its discovery in southern Wisconsin in 2000 (Alleman et al. 2002). The direct aphid stylet-feeding on plant sap is the most prominent damage that can cause up to 40% soybean yield loss (Ragsdale et al. 2007). Under heavy infestations, soybean foliage can be stunted, wrinkled, distorted, and wilted; yield components, such as seed size and number, are also reduced (Wu et al. 2004). Transmissions of plant viruses by soybean aphids lead to further yield loss in soybean production (Hill et al. 2001; Clark and Perry 2002). In addition, honeydew secreted by aphids promotes growth of sooty mold on leaves, impairing soybean photosynthesis by blocking sunlight and causing additional yield loss (Malumphy 1997; Lemos Filho and Paiva 2006).
Currently, insecticides are widely used to manage soybean aphids. However, this control method increases production cost, the risk of environmental contamination, and the mortality of beneficial insects (e.g., natural enemies and pollinators) (Ohnesorg et al. 2009; Lundin et al. 2015). The formation of insecticide resistance in soybean aphid populations is also an increasing concern. A more cost-effective and environmentally friendly way to managing soybean aphids is to utilize the native host-plant resistance present in soybean germplasm. Extensive screening of different soybean germplasm pools has identified ~ 30 plant introductions (PIs) and cultivars with antibiosis (affecting insect biology or reproduction) or antixenosis (non-preference) resistance (Hill et al. 2004; Li et al. 2004; Yang et al. 2004; Mensah et al. 2005; Hesler et al. 2007; Mian et al. 2008a; Fox et al. 2014).
Despite the high number of PIs and cultivars identified as resistant to soybean aphid, many share same resistance genes or alleles; this might be due to the genetic bottleneck of soybean in North America (Hyten et al. 2006). Aphid-resistance QTLs identified in North America are designated as Rag (Resistance to Aphis glycines); different resistance alleles have been uncovered at six loci, Rag1 to Rag6. The dominant antibiosis-resistant Rag1/Rag (Hill et al. 2006a, b; Li et al. 2007), the recessive antibiosis-resistant rag1c (Zhang et al. 2009), and rag1b (Bales et al. 2013) were mapped to chromosome 7 between markers Satt463 and Satt567. Additionally, Rag1 was fine-mapped to a 115-kb interval between markers SNPKS9-3 and SNPKS5 (Kim et al. 2010a). The dominant Rag2 (Mian et al. 2008b; Hill et al. 2009) and Rag5 (Jun et al. 2012) were mapped to a genomic region between Satt334 and Sct_033 on chromosome 13, but they confer different resistance modality (antibiosis vs. antixenosis) (Michel et al. 2010; Jun et al. 2012). Rag2 later was refined to a 54-kb interval between markers SNP46169.7 and SNP21A (Kim et al. 2010b). Aphid resistance in 20 PIs is associated with Rag2, indicating Rag2 may be a major aphid-resistance source in the USDA soybean germplasm collection (Fox et al. 2014). The recessive antibiosis rag4 was mapped to a different location (between Satt649 and Satt348) on chromosome 13 (Zhang et al. 2009). Jun et al. (2013) identified two major QTLs (QTL_13_1 and QTL_13_2) near Rag2 and rag4, and a minor QTL (QTL_6_1) on chromosome 6; these three QTLs suggested PI 567324 has oligogenic antixenosis resistance to soybean aphids. Six aphid-resistance QTLs/alleles were detected in a region between markers Satt285 and Satt654 on chromosome16, and designated Rag3 (antixenosis), Rag3b (antibiosis), rag3 (antibiosis), Rag3c (antibiosis), Rag3d (antibiosis), and Rag3e (antixenosis) (Zhang et al. 2010, 2013; Bales et al. 2013; Du 2016; Zhang et al. 2017a). Additionally, Rag3c was delimited to a 150-kb interval between markers Gm16-3 and Gm16-5 (Zhang et al. 2017b). The antibiosis-resistance gene Rag6 was refined to a 49-kb interval between markers Gm08-15 and Gm08-17 on chromosome 8 (Zhang et al. 2017a, b).
The biggest concern of employing host-plant resistance is the breakdown of single resistance genes by virulent biotypes. To date, four different soybean aphid biotypes have been discovered in North America. Biotype 1 is avirulent to all Rag genes (Hill et al. 2004). Biotype 2 can reproduce on soybean plants with Rag1 (Kim et al. 2008). Biotype 3 readily colonizes soybeans with Rag2; it also reproduces on soybeans with Rag1 in choice tests (Hill et al. 2010). A recent multi-year study reported that the occurrence of soybean aphid biotypes was highly variable across both locations and years in the Midwestern USA (Cooper et al. 2015). The variability and dynamics of aphid populations could limit the durability of effectiveness of a single resistance gene. In this study, PI 567541B (a natural pyramid of rag1c/rag4) and PI 567598B (a natural pyramid of rag1b/rag3) demonstrated the widest spectrum of resistance to aphids across locations and years (Cooper et al. 2015). Similarly, other studies showed that soybean lines with artificial pyramids of Rag1/Rag2 had significantly lower aphid colonization than lines with the Rag1 or Rag2 gene alone (Wiarda et al. 2012; McCarville et al. 2014). However, Alt and Ryan-Mahmutagic (2013) reported a new soybean aphid biotype, Biotype 4, capable of colonizing PI 567541B, PI 567598B, and soybean lines with the pyramid of Rag1/Rag2. There are likely more virulent biotypes not yet discovered. Therefore, integrating cultivars with multiple resistance genes, particularly with different modes of action, is important to achieve a broader and more durable resistance against different aphid populations.
The Soybean Breeding and Genetics Program at Michigan State University (MSU) has identified seven soybean accessions carrying resistant alleles at four resistance loci, including Rag1, Rag3, Rag4, and Rag6 (Bales et al. 2013; Zhang et al. 2009, 2010, 2013; Zhang et al. 2017a; Du 2016). Zhang et al. (2017b) refined Rag6 to a 49-kb interval between markers Gm08-15 and Gm08-17, and Rag3c to a 150-kb interval between markers Gm16-3 and Gm16-5. Fine mapping studies of five other aphid-resistance QTLs (rag1b, rag1c, rag3, Rag3d, and rag4) refined their genomic locations and identified closely linked SNP markers (unpublished data). With the assistance of these SNP markers, a pool of improved soybean germplasm with different combinations of aphid-resistance genes was developed. The objectives of this study were to (1) assess the introgression of aphid-resistance gene(s) using the Illumina Infinium SoySNP6K iSelect BeadChip, (2) determine the prevalence of soybean aphid biotypes in Michigan, and (3) assess the effectiveness of different Rag gene combinations against Michigan aphid populations.
Materials and methods
Plant materials
A total of 11 resistant soybean genotypes, including “Jackson,” LD05-16060 (Rag1-“Dowling”), PI 243540, PI 567543C, PI 567585A, PI 567597C, PI 567598B, PI 567541B, PI 567301B, E08934 (derived from G. soja 85-32) (Zhang et al. 2017a), and PI 567324, were used as indicator lines to screen for aphid biotypes in field-cage trials during the summers of 2015 and 2016. LD05-16060 was an advanced breeding line carrying the Rag1 gene from “Dowling” and was developed by Dr. Brian Diers at University of Illinois Urbana-Champaign (UIUC).
In total, 12 advanced breeding lines (Table 1) carrying different Rag gene(s) were developed through marker-assisted selection (MAS) with markers flanking the initial-mapped or fine-mapped regions (Li et al. 2007; Hill et al. 2009; Kim et al. 2010a, b; Zhang et al. 2017a, b; unpublished data). LD05-16657a with Rag1 and LD08-12430a with Rag2 were developed by Dr. Brian Diers at UIUC while “E” lines were developed at MSU in East Lansing, Michigan, with different combinations of rag1b, rag1c, Rag2, Rag3c, Rag3d, rag3, rag4, and Rag6 (Hill et al. 2009; Zhang et al. 2009; Bales et al. 2013; Du 2016; Zhang et al. 2017a) (Table 1). E00003 has been consistently susceptible to Michigan aphids over the years (Zhang et al. 2017a, b), and it served as a susceptible check in this study.
DNA extraction and the Illumina Infinium SoySNP6K iSelect BeadChip genotyping analyses to assess the effectiveness of MAS
Leaf tissue was collected from a seedling of each advanced breeding line. Genomic DNA from each sample was extracted using the modified CTAB protocol described by Kisha et al. (1997), and genotyped using the Illumina Infinium SoySNP6K iSelect BeadChip (Illumina, San Diego, CA), which consists of 5403 single nucleotide polymorphisms (SNPs) selected from the Illumina Infinium SoySNP50K iSelect BeadChip (Song et al. 2013). The genome-wide SNP distribution of the Illumina Infinium SoySNP6K iSelect BeadChip was visualized with R (R Development Core Team 2016) (Supplementary Fig. 1). Genotypes were called using the program GenomeStudio (1.9.4 version, Illumina, San Diego, CA). Each SNP was coded based on the standard codes for nucleotides derived from the International Union of Pure and Applied Chemistry. The quality of each SNP was checked as previously reported (Yan et al. 2010). SNPs with call rate < 80% across all samples were removed from the dataset. The genome-wide SNP data of each advanced breeding line was compared to that of the original aphid-resistance-gene(s) donor, mined from the public SoySNP50K iSelect BeadChip data on SoyBase (Grant et al. 2010) except for E12901. Graphic representation of genomic regions of interest from each sample was drawn with the program FlapJack (Milne et al. 2010). SNP markers that are monomorphic between the original donor line and the elite parental line were filtered. At each SNP of the advanced breeding line, the allele same as that of the original donor was assigned with the black color, and the alternative allele was assigned with the gray color.
Evaluation for soybean aphid resistance
Indicator lines and the advanced breeding lines were evaluated in choice tests in field-cage trials (Mensah et al. 2005) during the summers of 2015 and 2016. All the lines were planted in a randomized complete block design with three replications in a 12.2 × 18.3 m aphid- and predator-proof polypropylene cage (Redwood Empire Awning Co., Santa Rosa, CA) on the Agronomy Farm of MSU, East Lansing, Michigan. In each replication, 15 seeds from each line were planted in a single 60 cm long plot with 60 cm row spacing.
The advanced breeding lines were also evaluated in the greenhouse choice tests (Mensah et al. 2005) in the Plant Sciences greenhouse at MSU during fall 2015, spring 2016, and spring 2017. Eight seeds from each line were planted in a 125-mm deep, 105-mm-diameter plastic pot. All the lines were arranged in a randomized complete block design with three replications. The greenhouse was maintained at 26/15 °C day/night with supplemental light (14 h/day) provided by sodium vapor lights.
Soybean aphids were collected from multiple locations across Michigan in the early summer of each testing year and maintained on susceptible soybean plants (E00003) in field cages or the greenhouse. In each trial, each plant was artificially infested with two wingless aphids at the soybean V2 stage (Fehr and Cavinese 1977). Each plant was visually rated for aphid resistance using a 0–4 scale (Mensah et al. 2005) when the susceptible check reached rating of 3.0 (usually 3 weeks after the initial infestation). Criteria of the 0–4 scale are as follows: 0 = no aphids; 0.5 = fewer than 10 aphids; 1 = 11–100 aphids; 1.5 = 101–150 aphids; 2 = 151–300 aphids; 2.5 = 301–500 aphids; 3 = 501–800 aphids, leaves and stems are covered with aphids, leaves appear slightly curly and shiny; 3.5 = more than 800 aphids, the plant appears stunted with curled yellow leaves, the plant is covered with few cast skins, no sooty mold; 4 = more than 800 aphids, the plant appears stunted with severely curled yellow leaves, the plant is covered with cast skins and sooty mold (Mensah et al. 2005). A damage index (DI) for each replication of each line was calculated as DI (%) = ∑ (rating value × no. of plants in the category) / (4 × total no. of plants) × 100 (Mensah et al. 2005). The DI ranged from 0% (no infestation) to 100% (most severe infestation). In each trial, the average DI of each line from three replications was analyzed with one-way analysis of variance (ANOVA) at a significance level of 0.05 followed by paired wise comparisons using the PROC GLM function in SAS 9.4 (SAS Institute, Cary, NC). Lines with DI less than 37.5% were considered as aphid resistant (Zhang et al. 2017a, b).
Results and discussion
Data from the Illumina Infinium SoySNP6K iSelect BeadChip verified the successful introgressions of all targeted aphid-resistance genes
As shown in Fig. 1, genomic regions inherited from the original donor are indicated with the black color whereas genomic regions from the elite germplasm are presented in gray color. Targeted aphid-resistance genes with their published genomic locations (Glyma.Wm82.a1) were listed for each advanced breeding line. Unpublished fine-mapped regions of some Rag genes (including rag1b, rag1c, rag3, rag4) were indicated with rectangle boxes. When inspecting the regions of interest, all targeted aphid-resistance genes were successfully integrated into these advanced breeding lines, which verified the different Rag gene combination in each of the advanced breeding lines. The original genome-wide SNP data of each advanced breeding line along with E12901 (the donor of Rag6 and Rag3c) were presented in Supplementary Table 1 and shared publicly.
Graphic representation of genomic region(s) of interest for each advanced breeding line. Genomic regions inherited from the original donor(s) of the aphid-resistance gene(s) are presented in black while genomic regions from the susceptible elite background are presented in gray. Targeted aphid-resistance genes with their published genomic locations are listed for each advanced breeding line. Unpublished fine-mapped regions of some Rag genes (including rag1b, rag1c, rag3, rag4) are indicated with rectangle boxes. The genomic locations are according to Glyma.Wm82.a1 on SoyBase (Grant et al. 2010)
Indicator lines suggested Biotype 3 and undescribed virulent biotype(s) prevailing in Michigan
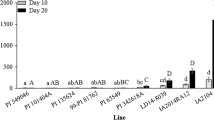
In both the 2015 and 2016 field-cage trials, LD05-16060 (Rag1), PI 243540, PI 567301B, and PI 567324 were heavily colonized by aphids collected from Michigan fields and their DIs (ranging from 61.7 to 79.2%) were not significantly different from the susceptible check, E00003 (DIs ~ 79.2 to 83.3%) (Fig. 2 and Table 2). PI 567585A was moderately resistant in 2016 (DI of 43.3%), although it performed better in 2015 (Fig. 2 and Table 2). The remaining soybean genotypes including “Jackson” showed strong resistance (DIs ranging from 12.5 to 33.3%) to the same aphid populations in both field trials (Fig. 2 and Table 2).
“Dowling”(Rag1) and “Jackson”(Rag) were reported as overcome by Biotype 2 in both choice and no-choice tests (Kim et al. 2008). Biotype 3 aphids readily colonized Rag2 soybeans in choice and no-choice tests as well as Rag1 soybeans in choice tests (Hill et al. 2010). Alt and Ryan-Mahmutagic (2013) discovered a new biotype, Biotype 4, capable of colonizing PI 567541B and PI 567598B. In our study, the Rag1 (LD05-16060) and Rag2 (PI 243540) lines were readily colonized by aphids; in contrast, the Rag line (“Jackson”), PI 567541B, and PI 567598B maintained strong resistance. This suggests that Biotype 3 aphids were a major component of the collected aphid populations in Michigan during 2015 and 2016.
The response of “Jackson” to Biotypes 3 or 4 is unknown as it was not included in the previous aphid biotype studies by Hill et al. (2010) and Alt and Ryan-Mahmutagic (2013). In our study, “Jackson” performed differently than LD05-16060 (carrying Rag1-“Dowling’) in both years; it showed a strong resistance in 2015 and a very strong resistance in 2016 whereas LD05-16060 was consistently as susceptible as E00003. In a regional investigation conducted by Cooper et al. (2015), “Jackson” was characterized as resistant in multiple states (SD, IA, MI, and OH) whereas “Dowling” was susceptible in all ten participating states in the year of 2010. Zhang et al. (2017a) also observed that “Jackson’ was resistant whereas “Dowling” was susceptible in Michigan during 2010. Combining the evidences from Cooper et al. (2015) and Zhang et al. (2017a), the different reactions of these two varieties to aphid populations in some years (2010, 2015, and 2016) suggested that Rag and Rag1 themselves are likely different, despite being mapped to a similar genomic region (Li et al. 2007). They could be allelic at a same locus or different QTLs located closely. “Jackson” showed strong resistance to aphid populations that were primarily Biotype 3 in our field trials during 2015 and 2016, which suggests Biotype 3 is likely not able to overcome the resistance in “Jackson.” Further study on the response of “Jackson” to Biotype 3 is needed to exam this hypothesis. It is also possible that the different performance of Rag1 and Rag in the present study was due to an undescribed aphid biotype capable of colonizing Rag1 but not Rag soybeans. Single clones of Michigan aphids will be tested on “Dowling” and “Jackson” to explore this possibility.
Mian et al. (2008a) reported that PI 567301B had strong antixenosis resistance to Biotypes 1 and 2, controlled by a major QTL (Rag5) and a minor QTL on chromosome 8 (Jun et al. 2012). Similarly, Mian et al. (2008a) reported that PI 567324 showed moderate antixenosis resistance to Biotype 1 and strong resistance to Biotype 2, contributed by QTL13_1 mapped closely to Rag2, QTL13_2 mapped closely to rag4, and a minor QTL_6_1 on chromosome 6 (Jun et al. 2013). Jun et al. (2013) suggested that the oligogenic resistance in PI 563724 would provide broader and more durable aphid resistance compared to lines with a single aphid-resistance gene. However, in our field trials during 2015 and 2016, both PI 567301B and PI 563724 were heavily colonized by aphids. Although the reaction of these PIs to other biotypes has not been tested, their high damage indices (ranging from 68.3 to 75%) in our study could be explained by their susceptibility to Biotype 3 aphids which appeared to predominate the aphid population in 2015 and 2016; it also could be due to an undescribed virulent biotype in Michigan. PI 567301B and PI 563724 will be tested with Biotype 3 and/or single clones isolated from Michigan aphid populations to further investigate the hypotheses.
Lines with rag1c or Rag3d or Rag6 or pyramided Rag genes showed strong and broad resistance
As shown in Fig. 3, several soybean lines with a single aphid-resistance gene were readily colonized by aphids in our study. LD05-16657a with Rag1 and LD08-12430a with Rag2 had severe aphid damages (DI ~ 66.8 to 88.3%) in all trials across 2015–2017 (Table 3), which was consistent with the performance of indicator lines, LD05-16060 (Rag1) and PI 243540 (Rag2). E11950 with rag3 showed strong resistance in all the greenhouse trials but had moderate aphid damages (DI ~ 42.2 to 60%) in the field trials (Table 3), whereas the original donor, PI 567598B, had very strong resistance in the field trials (DI ~ 12.5 to 16.7%) (Table 2). PI 567598B also had the lowest frequency (18%) of aphid colonization across 11 locations during 2008–2010 (Cooper et al. 2015). Combining the results from Cooper et al. (2015) and the present study, the pyramid of rag1b/rag3 is critical to provide soybean with broad and durable resistance.
Aphid damage indices (%) of a susceptible check (E00003) and the advanced breeding lines with different combinations of aphid-resistance gene(s) in a field-cage and greenhouse trials in 2015, b field-cage and greenhouse trials in 2016, and c a greenhouse trial in 2017. Damage indices from the field-cage trial were presented with gray bars followed by lowercase letters in a and b. Damage indices from the greenhouse trial were presented with black bars followed by uppercase letters in a and b. Within each trial, bars with same letter(s) are not significantly different at P < 0.05
E14923 with Rag6 alone was highly resistant (DI ~ 19.5 to 23.9%) to aphids across all trials during 2015–2016. However, its damage index (36.0%) in 2017 greenhouse trial was slightly below the resistance threshold (DI ~ 37.5%), and it was statistically greater than those of the remaining resistant lines (Fig. 3c and Table 3). The original donor, E08934 (Rag6 + Rag3c), and the advanced breeding line, E14902 (Rag6 + Rag3c), exhibited very strong and consistent resistance (DI ~ 12.5 to 16.7%) across all trials (Tables 2 and 3). Collectively, Rag6 alone offers a strong resistance; however, the pyramid of Rag6/Rag3c provides a stronger and more durable resistance.
E12904 with Rag3d appears to have a consistent stronger resistance compared to its original donor, PI 567585A. It displayed a strong resistance (DI ~ 12.5 to 27.4%) across all five trials during 2015–2017 (Fig. 3 and Table 3). However, PI 567585A had moderate aphid damage (DI ~ 43.3%) in 2016 field trial even though it had a lower damage index (20.8%) in 2015 field trial (Fig. 2 and Table 2). The consistent strong resistance effect of Rag3d in E12904 may be attributed to the elite genetic background; some background gene(s) may upregulate the expression of Rag3d.
Across all five trials during 2015–2017, E14922 with rag1c showed a consistent strong resistance (DI ~ 12.5 to 21.7%) whereas LD05-16657a with Rag1 was consistently susceptible (DI ~ 66.8 to 72.8%) (Fig. 3 and Table 3). The strong resistance provided by rag1c alone suggested that rag1c is a different gene or allele from Rag1 even though they were mapped in close proximity (Li et al. 2007; Zhang et al. 2009; Kim et al. 2010a). This conclusion is consistent with the genotypic evidence collected by Zhang et al. (2009); the band patterns of SSR markers flanking rag1c were distinctive between PI 567541B and “Dowling.”
Among the resistant soybean genotypes tested by Cooper et al. (2015), PI 567541B and PI 567598B demonstrated the widest spectrum of resistance to aphid populations across North America during 2008–2010; the broad resistance was deduced contributed by the natural pyramids of two resistance genes in these two PIs. However, PI 567541B and PI 567598B were later found fully colonized by Biotype 4 (Alt and Ryan-Mahmutagic 2013). In our study, E14912 (rag1b + rag3 from PI 567598B) and E13369 (rag1c + rag4 from PI 567541B) showed very strong resistance across 2015 to 2017 (Fig. 3 and Table 3); however, their resistance might be limited in geographic regions that have a higher pressure of Biotype 4 or other undescribed virulent biotypes.
rag1c and rag3 are the two major genes controlling aphid resistance in PI 567541B and PI 567598B, respectively (Zhang et al. 2009; Bales et al. 2013). Additionally, Chandrasena et al. (2015) detected a significant additive × additive interaction between rag1c and rag3, contributing up to 24% of the phenotypic variation in aphid resistance. To achieve a broader and more durable resistance, additional aphid-resistance gene(s) were pyramided with rag1c + rag3. Advanced breeding line E13901 was pyramided with three aphid-resistance genes, including rag1c, rag3, and rag4. Compared to E13901, E13903 has one more aphid-resistance gene, Rag2, to provide additional resistance. E14919 has all four genes from PI 567541B and PI 567598. All these advanced breeding lines (E13901, E13903, and E14919) pyramided with multiple aphid-resistance genes had very strong and consistent resistance to aphid populations in Michigan across 2015–2017 (Fig. 3 and Table 3), and they are expected to be strong and durable when combating diverse and dynamic aphid populations across geographic regions.
Conclusion
The utilization of host-plant resistance is an effective way to control soybean aphids. However, the aphid resistance provided by Rag1 soybeans, PI 243540 (Rag2), PI 567301B (Rag5), and PI 567324 (Rag2’ + rag4’ + QTL_6_1) was overcome by aphids in our field trials during 2015 and 2016. The high damage indices of PI 567301B and PI 567324 could be explained by their susceptibility to Biotype 3 aphids which appeared to be prevalent in our field trials. In contrast to the susceptibility of Rag1 soybeans, “Jackson” maintained strong resistance in the field trials during 2015 and 2016. Coupled with the similar evidences from Cooper et al. (2015) and Zhang et al. (2017a), Rag1 and Rag are likely different loci or alleles, which may be distinguished by Biotype 3. In addition, it is possible that an undescribed virulent biotype prevalent in our field trials caused the susceptibility of PI 567301B and PI 567324 and the different responses from Rag1 soybeans and “Jackson.” Biotype 3 and single isolates of Michigan aphids will be tested on these soybean genotypes to further exam the hypotheses.
Advanced breeding lines with single aphid-resistance genes, such as rag1c, Rag3d, and Rag6, showed very strong resistance to Biotype 3 across trials during 2015–2017. The strong resistance provided by rag1c suggested that it is a different locus or allele from Rag1 even though they were mapped closely. According to a regional study by Cooper et al. (2015), soybean aphids have a high degree of virulence diversity in North America, which means the effectiveness of a single aphid-resistance gene is likely limited by soybean aphid virulence variability.
Advanced breeding lines pyramided with two aphid-resistance genes, such as rag1b + rag3, rag1c + rag4, and Rag3c + Rag6, demonstrated strong resistance in Michigan. Although Biotype 3 dominated in our trials, there is variability in soybean aphid populations from year-to-year across the Midwest, and undescribed biotypes are likely yet to be identified. Lines with multiple Rag genes, such as rag1c + rag3 + rag4, rag1c + Rag2 + rag3 + rag4, and rag1b + rag1c + rag3 + rag4, likely will provide broader and more durable resistance to diverse and dynamic aphid populations. The advanced breeding lines with different combinations of Rag genes developed in this study are significant resources for breeders to develop varieties to combat different aphid populations across many geographies.
References
Alleman RJ, Grau CR, Hogg DB (2002) Soybean aphid host range and virus transmission efficiency. In: Proceedings of Wisconsin Fertilizer, Aglime, and Pest Management Conference. Madison. http://alfi.soils.wisc.edu/extension/wcmc/2002proceedings/Alleman-Conf-2002.pdf
Alt J, Ryan-Mahmutagic M (2013) Soybean aphid biotype 4 identified. Crop Sci 53(4):1491–1495. https://doi.org/10.2135/cropsci2012.11.0672
Bales C, Zhang G, Liu M, Mensah C, Gu C, Song Q, Hyten D, Cregan P, Wang D (2013) Mapping soybean aphid resistance genes in PI 567598B. Theor Appl Genet 126(8):2081–2091. https://doi.org/10.1007/s00122-013-2120-y
Chandrasena D, Wang Y, Bales C, Yuan J, Gu C, Wang D (2015) Pyramiding 3, 1b, 4, and 1c aphid-resistant genes in soybean germplasm. Crop Sci 55(5):2108–2115. https://doi.org/10.2135/cropsci2015.02.0089
Clark AJ, Perry KL (2002) Transmissibility of field isolates of soybean viruses by Aphis glycines. Plant Dis 86(11):1219–1222. https://doi.org/10.1094/PDIS.2002.86.11.1219
Cooper SG, Concibido V, Estes R, Hunt D, Jiang GL, Krupke C, McCornack B, Mian R, O’Neal M, Poysa V, Prischmann-Voldseth D (2015) Geographic distribution of soybean aphid biotypes in the United States and Canada during 2008–2010. Crop Sci 55(6):2598–2608. https://doi.org/10.2135/cropsci2014.11.0758
Du W (2016) Map and fine map aphid resistance genes in soybean plant introduction (PI) 567597C, 567585A and 567537. Dissertation, Michigan State University, East Lansing
Fehr WR, Cavinese CE (1977) Stages of soybean development. Special Report, Agriculture and Home Economics Experiment Station, Iowa State University, 80:11. https://trove.nla.gov.au/work/18906509
Fox CM, Kim KS, Cregan PB, Hill CB, Hartman GL, Diers BW (2014) Inheritance of soybean aphid resistance in 21 soybean plant introductions. Theor Appl Genet 127(1):43–50. https://doi.org/10.1007/s00122-013-2199-1
Grant D, Nelson RT, Cannon SB, Shoemaker RC (2010) SoyBase, the USDA-ARS soybean genetics and genomics database. Nucl Acids Res 38(suppl_1):D843–D846. https://doi.org/10.1093/nar/gkp798
Hesler LS, Dashiell KE, Lundgren JG (2007) Characterization of resistance to Aphis glycines in soybean accessions. Euphytica 154(1–2):91–99. https://doi.org/10.1007/s10681-006-9273-6
Hill JH, Alleman R, Hogg DB, Grau CR (2001) First report of transmission of soybean mosaic virus and alfalfa mosaic virus by Aphis glycines in the new world. Plant Dis 85(5):561–561. https://doi.org/10.1094/PDIS.2001.85.5.561C
Hill CB, Li Y, Hartman GL (2004) Resistance to the soybean aphid in soybean germplasm. Crop Sci 44(1):98–106. https://doi.org/10.2135/cropsci2004.9800
Hill CB, Li Y, Hartman GL (2006a) A single dominant gene for resistance to the soybean aphid in the soybean cultivar Dowling. Crop Sci 46(4):1601–1605. https://doi.org/10.2135/cropsci2005.11-0421
Hill CB, Li Y, Hartman GL (2006b) Soybean aphid resistance in soybean Jackson is controlled by a single dominant gene. Crop Sci 46(4):1606–1608. https://doi.org/10.2135/cropsci2005.11-0438
Hill CB, Kim KS, Crull L, Diers BW, Hartman GL (2009) Inheritance of resistance to the soybean aphid in soybean PI 200538. Crop Sci 49(4):1193–1200. https://doi.org/10.2135/cropsci2008.09.0561
Hill CB, Crull L, Herman TK, Voegtlin DJ, Hartman GL (2010) A new soybean aphid (Hemiptera: Aphididae) biotype identified. J Econ Entomol 103(2):509–515. https://doi.org/10.1603/EC09179
Hyten DL, Song Q, Zhu Y, Choi IY, Nelson RL, Costa JM, Specht JE, Shoemaker RC, Cregan PB (2006) Impacts of genetic bottlenecks on soybean genome diversity. Proc Natl Acad Sci 103(45):16666–16671. https://doi.org/10.1073/pnas.0604379103
Jun TH, Mian MAR, Michel AP (2012) Genetic mapping revealed two loci for soybean aphid resistance in PI 567301B. Theor Appl Genet 124(1):13–22. https://doi.org/10.1007/s00122-011-1682-9
Jun TH, Mian MAR, Michel AP (2013) Genetic mapping of three quantitative trait loci for soybean aphid resistance in PI 567324. Heredity 111(1):16–22. https://doi.org/10.1038/hdy.2013.10
Kim KS, Hill CB, Hartman GL, Mian MA, Diers BW (2008) Discovery of soybean aphid biotypes. Crop Sci 48(3):923–928. https://doi.org/10.2135/cropsci2007.08.0447
Kim KS, Bellendir S, Hudson KA, Hill CB, Hartman GL, Hyten DL, Hudson ME, Diers BW (2010a) Fine mapping the soybean aphid resistance gene Rag1 in soybean. Theor Appl Genet 120(5):1063–1071. https://doi.org/10.1007/s00122-009-1234-8
Kim KS, Hill CB, Hartman GL, Hyten DL, Hudson ME, Diers BW (2010b) Fine mapping of the soybean aphid-resistance gene Rag2 in soybean PI 200538. Theor Appl Genet 121(3):599–610. https://doi.org/10.1007/s00122-010-1333-6
Kisha TJ, Sneller CH, Diers BW (1997) Relationship between genetic distance among parents and genetic variance in populations of soybean. Crop Sci 37(4):1317–1325. https://doi.org/10.2135/cropsci1997.0011183X003700040048x
Lemos Filho JP, Paiva ÉA (2006) The effects of sooty mold on photosynthesis and mesophyll structure of mahogany (Swietenia macrophylla King., Meliaceae). Bragantia 65(1):11–17. https://doi.org/10.1590/S0006-87052006000100003
Li Y, Hill CB, Hartman GL (2004) Effect of three resistant soybean genotypes on the fecundity, mortality, and maturation of soybean aphid (Homoptera: Aphididae). J Econ Entomol 97(3):1106–1111. https://doi.org/10.1093/jee/97.3.1106
Li Y, Hill CB, Carlson SR, Diers BW, Hartman GL (2007) Soybean aphid resistance genes in the soybean cultivars Dowling and Jackson map to linkage group M. Mol Breed 19:25–34
Lundin O, Rundlöf M, Smith HG, Fries I, Bommarco R (2015) Neonicotinoid insecticides and their impacts on bees: a systematic review of research approaches and identification of knowledge gaps. PLoS One 10(8):e0136928. https://doi.org/10.1371/journal.pone.0136928
Malumphy CP (1997) Morphology and anatomy of honeydew eliminating organs. In: Ben-Dov, Hodgson CJ (eds) World Crop Pests, vol 7. Elsevier Science B.V., Amsterdam, pp 269–274
McCarville MT, O’Neal ME, Potter BD, Tilmon KJ, Cullen EM, McCornack BP, Tooker JF, Prischmann-Voldseth DA (2014) One gene versus two: a regional study on the efficacy of single gene versus pyramided resistance for soybean aphid management. J Econ Entomol 107(4):1680–1687. https://doi.org/10.1603/EC14047
Mensah C, DiFonzo C, Nelson RL, Wang D (2005) Resistance to soybean aphid in early maturing soybean germplasm. Crop Sci 45(6):2228–2233. https://doi.org/10.2135/cropsci2004.0680
Mian MAR, Hammond RB, St Martin SK (2008a) New plant introductions with resistance to the soybean aphid. Crop Sci 48(3):1055–1061. https://doi.org/10.2135/cropsci2007.06.0357
Mian MAR, Kang ST, Beil SE, Hammond RB (2008b) Genetic linkage mapping of the soybean aphid resistance gene in PI243540. Theor Appl Genet 117(6):955–962. https://doi.org/10.1007/s00122-008-0835-y
Michel AP, Mian MR, Davila-Olivas NH, Cañas LA (2010) Detached leaf and whole plant assays for soybean aphid resistance: differential responses among resistance sources and biotypes. J Econ Entomol 103(3):949–957. https://doi.org/10.1603/EC09337
Milne I, Shaw P, Stephen G, Bayer M, Cardle L, Thomas WT, Flavell AJ, Marshall D (2010) Flapjack—graphical genotype visualization. Bioinformatics 26(24):3133–3134. https://doi.org/10.1093/bioinformatics/btq580
Ohnesorg WJ, Johnson KD, O'Neal ME (2009) Impact of reduced risk insecticides on soybean aphid and their natural enemies. J Econ Entomol 102(5):1816–1826. https://doi.org/10.1603/029.102.0512
R Development Core Team (2016) R: A language and environment for statistical computing. R foundation for statistical computing, Vienna
Ragsdale DW, McCornack BP, Venette RC, Potter BD, MacRae IV, Hodgson EW, O’Neal ME, Johnson KD, O’Neil RJ, DiFonzo CD, Hunt TE (2007) Economic threshold for soybean aphid (Hemiptera: Aphididae). J Econ Entomol 100(4):1258–1267. https://doi.org/10.1093/jee/100.4.1258
Ragsdale DW, Landis DA, Brodeur J, Heimpel GE, Desneux N (2011) Ecology and management of the soybean aphid in North America. Annu Rev Entomol 56(1):375–399. https://doi.org/10.1146/annurev-ento-120709-144755
Song Q, Hyten DL, Jia G, Quigley CV, Fickus EW, Nelson RL, Cregan PB (2013) Development and evaluation of SoySNP50K, a high-density genotyping array for soybean. PLoS One 8(1):E54985. https://doi.org/10.1371/journal.pone.0054985
SoyStats (2016) http://soystats.com
Wiarda SL, Fehr WR, O'Neal ME (2012) Soybean aphid (Hemiptera: Aphididae) development on soybean with Rag1 alone, Rag2 alone, and both genes combined. J Econ Entomol 105(1):252–258. https://doi.org/10.1603/EC11020
Wu Z, Schenk-Hamlin D, Zhan W, Ragsdale DW, Heimpel GE (2004) The soybean aphid in China: a historical review. Ann Entomol Soc Am 97(2):209–218. https://doi.org/10.1093/aesa/97.2.209
Yan J, Yang X, Shah T, Sánchez-Villeda H, Li J, Warburton M, Zhou Y, Crouch JH, Xu Y (2010) High-throughput SNP genotyping with the GoldenGate assay in maize. Mol Breed 25(3):441–451. https://doi.org/10.1007/s11032-009-9343-2
Yang Z, Honda K, Wang S, Ma X (2004) Re-evaluation of Glycine soja germplasm from north eastern China for aphid resistance. J Jilin Agric Sci 29(5):3–6
Zhang G, Gu C, Wang D (2009) Molecular mapping of soybean aphid resistance genes in PI 567541B. Theor Appl Genet 118(3):473–482. https://doi.org/10.1007/s00122-008-0914-0
Zhang G, Gu C, Wang D (2010) A novel locus for soybean aphid resistance. Theor Appl Genet 120(6):1183–1191. https://doi.org/10.1007/s00122-009-1245-5
Zhang G, Gu C, Wang D (2013) Mapping and validation of a gene for soybean aphid resistance in PI 567537. Mol Breed 32(1):131–138. https://doi.org/10.1007/s11032-013-9857-5
Zhang S, Zhang Z, Bales C, Gu C, DiFonzo C, Li M, Song Q, Cregan P, Yang Z, Wang D (2017a) Mapping novel aphid resistance QTL from wild soybean, Glycine soja 85-32. Theor Appl Genet 130(9):1941–1952. https://doi.org/10.1007/s00122-017-2935-z
Zhang S, Zhang Z, Wen Z, Gu C, An Y, Bales C, DiFonzo C, Song Q, Wang D (2017b) Fine mapping of the aphid resistance genes Rag6 and Rag3c from Glycine soja 85-32. Theor Appl Genet 130(12):2601–2615. https://doi.org/10.1007/s00122-017-2979-0
Acknowledgments
We thank Dr. Brian Diers for providing LD05-16060, LD05-16657a, and LD08-12430a. We appreciate the funding support from the United Soybean Board, Michigan Soybean Promotion Committee, and USDA National Institute of Food and Agriculture, Hatch project 1011788.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This work complies with the current laws of the USA.
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Supplementary Fig. 1
The genome-wide SNP distribution of the Illumina Infinium SoySNP6K iSelect BeadChip visualized with R (GIF 324 kb)
High Resolution Image
(TIFF 172 kb)
Supplementary Table 1
(XLS 1425 kb)
Rights and permissions
About this article
Cite this article
Zhang, S., Wen, Z., DiFonzo, C. et al. Pyramiding different aphid-resistance genes in elite soybean germplasm to combat dynamic aphid populations. Mol Breeding 38, 29 (2018). https://doi.org/10.1007/s11032-018-0790-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-018-0790-5