Abstract
Gibberellin-sensitive dwarfing gene Rht18 was mapped in two durum wheat recombinant inbred lines (RIL) populations developed from crosses, Bijaga Yellow/Icaro and HI 8498/Icaro. Rht18 was mapped within genetic interval of 1.8 cM on chromosome 6A. Simple sequence repeat (SSR) markers S470865SSR4, barc37 and TdGA2ox-A9 specific marker showed co-segregation with Rht18 in Bijaga Yellow/Icaro population consisting 256 RILs. Effect of Rht18 on plant height was validated in HI 8498/Icaro RIL population which segregated for Rht18 and Rht-B1b. Rht-B1b from HI 8498 showed pleiotropic effect on plant height and coleoptile length, on the other hand, Rht18 did not show effect on coleoptile length. The SSR and SNP markers linked to Rht18 were also validated by assessing their allelic frequency in 89 diverse durum and bread wheat accessions. It was observed that 204 bp allele of S470865SSR4 could differentiate Icaro from rest of the wheat accessions except HI 8498, suggesting its utility for selection of Rht18 in wheat improvement programs. Rht18 associated alleles of TdGA2ox-A9, IAW4371 and IAW7940 were absent in most of the tall Indian local durum wheat and bread wheat, hence could be used to transfer Rht18 to bread wheat and local durum wheat. SSR marker barc3 showed high recombination frequency with Rht18, though it showed allele unique to Icaro. Since semidwarf wheat with GA-sensitive dwarfing genes are useful in dry environments owing to their longer coleoptile, better emergence and seedling vigor, Rht18 may provide a useful alternative to widely used GA-insensitive dwarfing genes under dry environments.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat consumption worldwide is estimated to surpass 817 million tonnes by 2030. To meet the estimated demand, production would need to increase by 23-45% from the current production level. This target has to be achieved despite limited resources, change in climatic conditions and decreasing acreage under wheat cultivation in dry environments. Present increase in wheat production has been achieved through introduction of semidwarfing genes and selection for grain yield. Semidwarf plants do not lodge under high fertility conditions, exhibit greater harvest index and thereby contribute to increase in grain yield. Therefore, introduction of dwarfing genes in cereal crops was crucial to increase grain yield during the green revolution (Hedden 2003). However, gibberellin-insensitivity conferred by semidwarfing genes Rht-B1b and Rht-D1b not only affects culm elongation but also many other gibberellic acid (GA)-dependent developmental processes such as α-amylase production in germinating seeds (Bhagwat and Bhatia 1994), root elongation (Bai et al. 2013), coleoptile length (Botwright et al. 2001) and leaf expansion (Appelford and Lenton 1991). These pleiotropic effects of GA-insensitive semidwarfing genes can lead to reduction in yield.
In low-precipitation dry land wheat-growing regions, deep seed placement is a better option to obtain sufficient moisture to initiate germination. Earlier studies have shown that dwarfing genes Rht-B1b or Rht-D1b exhibit strong negative correlation with coleoptile length and seedling vigor (Rebetzke et al. 2001; Richards 1992). The Rht-B1b and Rht-D1b wheat cultivars with a short coleoptile have difficulty in emerging from deep sowing, particularly when sown at a depth more than 4 to 5 cm, which can result in poor seedling establishment. In contrast, wheat cultivars with GA-sensitive dwarfing gene and long coleoptiles emerge with higher frequency especially when sown deep or where stubble has been retained (Allan et al. 1962; Rebetzke et al. 2005). Field studies have demonstrated that GA-sensitive semidwarf wheats emerge significantly better at 11 cm sowing depth than GA-insensitive semidwarf wheats. In wheat genotypes with rht (tall) and Rht8, longer coleoptile showed positive association with greater number of emerged seedlings, greater seedling area and seedling biomass, and negative association with shallower crown depth (Rebetzke et al. 2007). Comparisons between tall and semidwarf wheat varieties showed that reduced plant height due to GA-sensitive dwarfing genes is associated with increased grain yield (Rebetzke and Richards 2000), thus have potential for improving wheat establishment through greater coleoptile length and seedling vigor. GA-sensitive dwarfing gene Rht13 was found to be associated with reduced peduncle length with significantly greater biomass, yield, harvest index, grain number and spike number (Rebetzke et al. 2011). Another GA-sensitive dwarfing gene Rht12 substantially reduced plant height without altering seedling vigor and significantly increased spikelet fertility in common wheat under favorable sowing environment (Chen et al. 2013). In evaluation of breeding potential of GA-sensitive Rht18 from tetraploid wheat in the background of common wheat, Rht18 moderately reduced plant height and increased harvest index without affecting seedling vigor, root growth and coleoptile length (Yang et al. 2015). Considering these advantages, GA-sensitive dwarfing genes are needed for diversification of reduced height genes in wheat breeding programs to facilitate access to soil moisture at deeper sowing in dry environments.
GA-responsive height reducing genes such as Rht4 (2BL), Rht5 (3BS), Rht8 (2DS), Rht9 (5AL), Rht12 (5AL) and Rht13 (7BS) have been mapped and DNA markers linked to these genes reported in bread wheat (Ellis et al. 2005). However, there are very few reports on genetic studies of GA-sensitive dwarfing genes in durum wheat. Out of the 22 dwarfing genes reported in wheat 16 are sensitive to GA, of which five genes (Rht14, Rht15, Rht16, Rht18 and Rht19) are available in durum wheat (Konzak 1987). These induced Rht mutants have better emergence potential, since their coleoptile length and gibberellic acid (GA) sensitivity is unaltered. In a genetic mapping study using three F2 populations, Rht14, Rht16, and Rht18 appeared to be allelic and linked to barc3 on chromosome 6A (Haque et al. 2011). The genes were mapped at 11.7 to 28.0 cM distance from barc3. Using a marker at such a long distance would result in to very high chances of false positive selection in breeding programs. The present study was undertaken with the following objectives: (1) mapping of GA-sensitive dwarfing gene Rht18 in durum wheat and development of closely linked simple sequence repeat (SSR) and single nucleotide polymorphism (SNP) for efficient selection and deployment of Rht18 in durum wheat breeding. (2) Validation of newly identified markers in diverse wheat genetic backgrounds to assess their usefulness in wheat improvement programs.
Materials and methods
Plant material
Icaro (Ic; PI503555) is a GA-sensitive dwarf mutant obtained from Cv. Anhinga, carrying semidominant dwarfing gene Rht18, with long coleoptile and high breeding value (Konzak 1987; Maluszynski and Szarejko 2003). It was crossed with two Indian durum wheat cultivars Bijaga Yellow (BY) and HI 8498 (HI) and two sets of recombinant inbred line (RIL) populations (BY/Ic-256 RILs and HI/Ic-134 RILs) were developed by single-seed descent method. Bijaga Yellow is a tall durum wheat cultivar released in 1965 for cultivation under rainfed conditions. HI 8498 is a cultivar carrying GA-insensitive Rht-B1b, with semidwarf plant type and short coleoptile. It was released in 1999 for cultivation under high fertility conditions. BY/Ic population was used for mapping Rht18 while HI/Ic population was mainly used to validate the effect of Rht18 on plant height in the background of Rht-B1b. A set of 89 wheat accessions composed of durum cultivars, local durums and bread wheat cultivars was also included in the study to test allelic variation of markers linked to Rht18.
Field trials and phenotype analysis
The RIL populations were planted at experimental farm of Agharkar Research Institute, Pune (180 31′ N, 730 55′ E) during regular crop seasons of 2013–14 (date of sowing November 17, 2013) and 2014–15 (date of sowing November 07, 2014). Fields were irrigated after 42 days from sowing when most of the RILs were at booting stage. Plant height was determined as the measurement of the main tiller from soil surface to the top of the ear (excluding awns) at maturity. Coleoptile length was measured for ten seedlings of each genotype as per method described by Li et al. (2011). In brief, ten similarly sized seeds from each RIL were placed side by side in a straight line, 0.5 cm apart with the germ end downward on the germination paper. The germination paper was folded, rolled and placed vertically in a container with nutrient solution (Bai et al. 2013). Coleoptile length was measured at emergence of first leaf through coleoptile (Zadoks scale 10) and expressed as a mean of 10 measurements. Similar experiment was conducted to test the response of Bijaga Yellow, HI 8498 and Icaro to exogenous gibberellic acid (GA3, 100 μM) in nutrient solution. Control seedlings were treated with nutrient solution only. Response to gibberellic acid was measured in terms of elongation of coleoptile at Zadoks scale 10. The differences in coleoptile length due to GA3 treatment and dwarfing genes were tested for significance by Student’s t- test.
Development of SNP and SSR markers
Since Icaro has been reported as sensitive to exogenous GA3 (Konzak 1987) and also showed response to exogenous GA3 at Zadoks scale 10 in the present study (Supplementary Fig. S1; Table S1); it was hypothesized that Icaro may be carrying altered GA biosynthesis or GA inactivation pathway. Gibberellin 2-oxidase-A9 (TaGA2ox-A9) was reported on chromosome 6A with putative function of inactivation of active GA (Pearce et al. 2015), therefore, genomic DNA sequence of this gene was used to design PCR primers to amplify GA2ox-A9 in Bijaga Yellow, Icaro and HI 8498. A DNA fragment of 5.1 kb comprising complete ORF as well as 5′ and 3′ regulatory regions was amplified using primer pair TdGA2ox-A9F/TdGA2ox-A9R and HiFi HotStart polymerase, Kapa Biosystems. Amplicon was ligated in pSC-A-amp/kan vector and transformed into SoloPack competent cells using Strataclone PCR Cloning Kit, Agilent Technologies, USA. Four clones for each insert were sequenced on ABI PRISM® 3100-Avant Genetic Analyzer. GA2ox9 homoeologue from A-genome (TdGA2ox-A9) was verified by comparing it with three scaffolds containing sequences of A, B and D homoeologues of GA2ox9 kindly shared by Dr. Andrew Phillips, Rothamsted Research, Harpenden, UK. DNA Sequence polymorphism observed at −182 bp (C/GT) in 5′ regulatory region of TdGA2ox-A9 was targeted to develop co-dominant marker differentiating between alleles of TdGA2ox-A9 in Bijaga Yellow and Icaro. Primers were designed by following strategy reported earlier for development of SNP-based co-dominant markers for disease resistance genes in rice (Ramkumar et al. 2015). The primers were used to map TdGA2ox-A9 in segregating RILs. Nucleotide sequences of TdGA2ox-A9 derived from Bijaga Yellow, Icaro and HI 8498 were submitted to GenBank (Accession numbers KX163067, KX163068, and KX163069). The scaffold 470865-6AL containing GA2ox-A9 was further searched for presence of SSR using SSR identification tool (SSRIT) available at http://archive.gramene.org/db/markers/ssrtool. Total four putative SSRs were identified and tested for polymorphism. On the basis of their map position, six SNP markers were selected from high density consensus SNP map of tetraploid wheat (Maccaferri et al. 2015) and IWGSC wheat genome sequence repository (available at https://wheat-urgi.versailles.inra.fr/Seq-Repository/Genes-annotations) to test their putative linkage with Rht18. PCR markers specific to these SNPs were designed by keeping SNP at 3′ end of either forward or reverse primer (Supplementary Table S2) as described by Ellis et al. (2002).
Mapping of Rht18
Genomic DNA was extracted from tender leaves of RILs by a modified Cetyltrimethylammonium bromide (CTAB) method (Rogers and Bendich 1985). Since Rht18 was assigned to chromosome 6A in earlier report (Haque et al. 2011), a total of 62 SSR markers from chromosome 6A were tested for polymorphism between parental genotypes. Polymorphic markers were used initially to genotype a subset of 139 RILs and framework map was generated for chromosome 6A using MapMaker version 3.0 (Lander et al. 1987). Considering near centromere position of Rht18 locus, eight markers flanking Rht18 were used to genotype BY/Ic population of 256 RILs to obtain further resolution in the map. HI/Ic population was also segregating for Rht-B1, therefore, a locus specific marker for Rht-B1 (Ellis et al. 2002) from chromosome 4B was used for genotyping and single-marker regression analysis was carried out to estimate effect of Rht-B1 on plant height and coleoptile length. Map of chromosome 6A was drawn using the program MapChart 2.1 (Voorrips 2002). Markers linked to Rht18 locus were tested on 89 diverse durum and bread wheat accessions to study their allelic variation in the germplasm. Markers flanking to Rht18 were tested on 89 diverse wheat genotypes to determine frequency of their alleles in germplasm.
Statistical analysis
The differences in phenotype due to GA3 treatment and dwarfing genes were tested for significance by Student’s t- test. Genotypic (σ 2 g ) and phenotypic variances (σ 2 p ) were estimated as σ 2 g = (MS v − MS e )/r and σ 2 p = σ 2 g + σ 2 e , respectively, and were used further to calculate heritability (h 2 = σ 2 g /σ 2 p ), where r is number of replications, MS v and MS e are mean sum of square for genotype and residual error, respectively. Relationships between the traits were examined by Pearson correlation coefficient.
Results
Phenotype evaluation
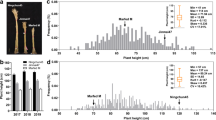
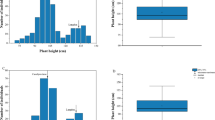
Significant differences were observed between plant height of Bijaga Yellow (102.8 to 105.3 cm), Icaro (57.9 to 64.3 cm) and HI 8498 (74.2 to 74.8 cm) over two wheat-growing seasons. Detail data in parental genotypes and two RIL populations over two seasons is presented in Table 1. Very high broad-sense heritability was observed for plant height (0.95 to 0.97). In both the RIL populations, plant height measured across 2 years showed significant correlation (P < 0.01; Supplementary Table S3). Coleoptile length showed significant correlation with plant height in HI 8498/Icaro population. Though correlation was observed between coleoptile length and plant height (2013–14) in Bijaga Yellow/Icaro, the magnitude of correlation coefficient was low. Analysis of variance showed that genotype, environment and their interaction (G × E) had significant effect on plant height (P < 0.01) in both the populations (Supplementary Table S4), however, high broad-sense heritability values suggested that genotype had major contribution to variation in plant height. Bi-modal distribution for plant height was observed in BY/Ic population, suggesting that plant height is governed by a single major locus in the population (Fig. 1a). RILs with plant height less than 85 cm were classified as dwarf lines and RILs with plant height greater than 85 cm were considered tall. BY/Ic population consisted of 97 dwarf lines and 159 tall lines. Average plant height of dwarf lines was 64 cm while that of tall lines was 98 cm over two seasons. Plant height was skewed towards semidwarf phenotype in HI/Ic population because of presence of two dwarfing genes in the population (Fig. 1b). All the internodes in Bijaga Yellow were significantly longer than that of HI 8498 and Icaro. Peduncle and second internode showed significant differences for length among Bijaga Yellow, Icaro and HI 8498 (Supplementary Fig. S2). However, lengths of third and fourth internodes of Icaro and HI 8498 were not significantly different. Contrasting values for coleoptile length were observed in Bijaga Yellow (10.2 ± 0.8 cm), HI 8498 (6.7 ± 1.0 cm) and Icaro (9.1 ± 0.6 cm). Coleoptile length in BY/Ic and HI/Ic RILs ranged from 7.1 to 14.2 cm and 5.5 to 14.8 cm with mean 10.3 and 8.8 cm, respectively (Fig. 1c; Table 1). Application of exogenous GA3 (100 μM) showed 46.2% increase in coleoptile length in Icaro, suggesting its sensitivity towards GA3. Bijaga Yellow also showed moderate sensitivity towards GA3 application (Supplementary Fig. S1; Table S1).
Development of SNP and SSR markers linked Rht18
Three SNP markers out of six could differentiate between BY and Ic, while only two (IWA4371 and IWA7940) could give reproducible amplification in RIL population. Nucleotide sequence of 4.2 kb was derived for putative TdGA2ox-A9 from Bijaga Yellow, Icaro and HI 8498 (GenBank accessions KX163067, KX163068 and KX163069). Derived sequence comprised of three exons of 489, 324 and 231 bp separated by two introns of 814 and 677 bp. Upstream 5′ (1388 bp) and downstream 3′ (314 bp) regions were also partially sequenced. Sequence alignment of Bijaga Yellow and Icaro showed sequence variation at −182 bp (C/GT) in 5′ regulatory region and single nucleotide polymorphism (SNP) in exon 1 (+438 bp) as well as exon 3 (+2435 bp). SNP in exon 3 region resulted in substitution of amino acid proline (Bijaga Yellow) by serine (Icaro). A co-dominant PCR marker targeting sequence variation at −182 bp in 5′ regulatory region could differentiate between alleles of TdGA2ox-A9 present in Bijaga Yellow and Icaro (Fig. 2). The marker was subsequently used for genotyping of RILs and mapping TdGA2ox-A9. A PCR primer pair targeting SNP observed between HI 8498 and Icaro at +2196 bp was also developed but it failed to give consistent amplification due to A/T rich flanking region. Four putative SSRs were identified from the scaffold 470865-6AL containing TdGA2ox-A9. However, only one SSR S470865SSR4 with trinucleotide repeat (GTA) n showed polymorphism between Bijaga Yellow (186 bp) and Icaro (204 bp) that was used further for linkage mapping.
Mapping of Rht18
Since Rht18 is located on chromosome 6A, a total of 62 SSR markers on 6A were tested for polymorphism among parents. Out of these, 18 polymorphic SSRs were used to develop genetic linkage map of chromosome 6A initially in a set of 139 RILs of BY/Ic population. No recombinants were observed between Rht18, gwm82 and barc37 (Fig. 3). S470865SSR4 and TdGA2ox-A9 specific marker developed in present study also showed co-segregation with Rht18. Both the SNP markers (IAW4371 and IAW7940) showed only one recombinant with Rht18 in 139 RILs. The genetic linkage distance between Rht18 and barc3 was 9.4 cM with 14 recombinants. To resolve this cluster further, additional 117 RILs form the same population were also used for mapping. Genotype data for 8 markers flanking Rht18 was produced for a total of 256 RILs, which showed no recombinants between Rht18, TdGA2ox-A9, S470865SSR4 and barc37. This cluster was flanked by barc118 and IWA4371 within an interval of 1.8 cM. In HI/Ic population barc118 showed five recombinants with Rht18 among RILs carrying wild allele of Rht-B1. Unexpectedly, TdGA2ox-A9, S470865SSR4 and barc37 showed monomorphic alleles in Icaro and HI 8498, therefore, could not be mapped in HI/Ic population. Regression analysis showed that Rht18 did not have significant effect on coleoptile length BY/Ic and HI/Ic populations, while Rht-B1 showed significant pleiotropic effect on coleoptile length and plant height in HI/Ic population. Additive effect showed that Rht18 reduced plant height by 16.95 cm in BY/Ic RILs. Allelic variation of markers S470865SSR4, barc118, TdGA2ox-A9, IAW4371, IAW7940 and barc3 linked to Rht18 locus were tested on 89 diverse durum and bread wheat accessions (Supplementary Table S5). It was observed that 204 bp allele of S470865SSR4 could differentiate Icaro from rest of the wheat accessions except HI 8498. Similarly, barc3 showed 217 bp allele which was also unique to Icaro. Rht18 associated alleles of TdGA2ox-A9, IAW4371 and IAW7940 were absent in most of the tall Indian local durum and bread wheat.
Discussion
Semidwarf wheats with GA-sensitive dwarfing genes are known to be useful in dry environments owing to their longer coleoptile, better emergence and seedling vigor, thus, may provide a useful alternative to widely used GA-insensitive dwarfing genes under dry environments. Therefore, it is important to develop markers for GA-sensitive dwarfing genes, which would subsequently facilitate development of wheat genotypes better adapted to semiarid conditions by marker-assisted breeding. This study was undertaken to characterize GA-sensitive dwarfing gene Rht18 at molecular level to enable its exploitation in wheat breeding program. In BY/Ic and HI/Ic RIL populations, Rht18 was mapped within interval of 1.8 cM flanked by barc118 and IWA4371, moreover, TdGA2ox-A9, S470865SSR4 and barc37 showed no recombinants with Rht18 in a population of 256 RILs (Fig. 3). Results showed that SSR and SNP markers reported in the present study are more closely linked to Rht18 than barc3 reported earlier (Haque et al. 2011). Therefore, S470865SSR4, barc37, TdGA2ox-A9 and IWA4371 assure more precise selection of Rht18 as compared to earlier reported marker. The identified markers were tested on 89 diverse durum and bread wheat accessions to survey their allele frequency in different genetic backgrounds. It was observed that S470865SSR4 could discriminate between Icaro and all the other accessions except HI 8498, therefore, can be used to transfer Rht18 in diverse wheat genetic backgrounds (Supplementary Table S5). Icaro specific allele of SNP marker IWA4371 and TdGA2ox-A9 was absent in most of the tall Indian local durum as well as bread wheat carrying Rht-B1b allele, however, their frequency was high in durum cultivars. The results suggested that S470865SSR4 can be useful to select donor allele in diverse genetic backgrounds, while IWA4371 and TdGA2ox-A9 specific markers will have limited use in the background of local durums and bread wheat. Although barc3 showed an allele unique to Icaro, comparatively large linkage distance was observed between barc3 and Rht18 in present study as well as earlier report (Haque et al. 2011), limits its utilization in marker-assisted selection for Rht18 due to higher chances of false positives.
Additive effect showed that Rht18 reduces plant height by 16.95 cm in BY/Ic population, which is in agreement with earlier results where GA-sensitive dwarfing genes were reported to reduce plant height in common wheat (Chen et al. 2013; Rebetzke and Richards 2000; Rebetzke et al. 2012). Rht-B1b from HI 8498 showed pleiotropic effect on plant height and coleoptile length, on the other hand, Rht18 did not show any significant effect on coleoptile length (Table 2). Therefore, Rht18 may help in better seedling establishment due to longer coleoptile under limited moisture conditions, thereby improving crop stand and higher grain yield than wheat carrying Rht-B1b. The additive effect on plant height reduction is shared by Rht18 and Rht-B1b in HI/Ic population, however, no significant difference for plant height was observed between dwarf RILs carrying either Rht18 or Rht-B1b (Supplementary Fig. S3; Table S6). This suggests that Rht18 has dwarfing effect comparable to that of Rht-B1b. Moderate height reducing effect of Rht18 on plant stature, without any negative effect on coleoptile length and root growth in the background of Chinese winter wheat has also been reported recently (Yang et al. 2015).
Chromosomal region of Rht18 in Icaro coincides to the QTL for plant height, coleoptile length, and leaf width identified earlier in bread wheat cross between Chuan-Mai18 and Vigour18 (Spielmeyer et al. 2007). A consistent QTL for plant height was identified near centromere on chromosome 6A in meta-QTL study conducted using three bread wheat doubled haploid populations (Griffiths et al. 2012). In durum wheat Rht14 and Rht16 were also reported to be allelic to Rht18 and mapped near barc3 on chromosome 6A (Haque et al. 2011). All these reports in bread and durum wheat suggest the presence of a major height reducing gene on chromosome 6A.
TdGA2ox-A9 specific marker showed perfect association with variation in plant height due to Rht18 in BY/Ic population, however, the marker showed high frequency of donor allele in durum cultivars and could not be considered as diagnostic marker. Nevertheless, the marker could be used to map TdGA2ox-A9 on chromosome 6A and its map position coincides with Rht18 locus as demonstrated in present study. Overexpression of OsGA2ox9 and PvGA2ox9b was shown to be associated with reduction in plant height along with increase in tiller number and root length in rice and switch grass (Lo et al. 2008; Wuddineh et al. 2015). Similar results were also reported in wheat, wherein overexpression of bean GA2ox yielded gibberellin-deficient dwarf and semidwarf phenotype in wheat transformants (Hedden and Phillips 2000). However, linkage between Rht18 and TdGA2ox-A9 identified in present study is indirect evidence, therefore, needs to be studied further on a comparatively larger mapping population for fine mapping or by comparing Icaro with its wild-type parent Anhinga for expression of the gene and its effect on levels of active gibberellins in developing stem.
Conclusion
Effect of Rht18 on plant height was shown in two durum wheat recombinant inbred lines populations. Rht-B1b from HI 8498 showed pleiotropic effect on plant height and coleoptile length, on the other hand, Rht18 did not show effect on coleoptile length. Rht18 was mapped on chromosome 6A in a population of 256 RILs within interval of 1.8 cM. The gene showed co-segregation with newly developed co-dominant SSR marker S470865SSR4 with very rare Icaro-type allele of 204 bp, hence could be very useful for selection of Rht18 in diverse breeding material including bread and durum cultivars carrying Rht-B1b as well as tall local durum. Closely linked SNP markers were also identified that are amenable to high-throughput systems in breeding programs. These SNP markers are more useful for selection of Rht18 in the background of bread wheat and local durum.
References
Allan RE, Vogel OA, Peterson CJ (1962) Seedling emergence rate of fall sown wheat and its association with plant height and coleoptile length. Agron J 54:347–350
Appelford NEJ, Lenton JR (1991) Gibberellins and leaf expansion in near-isogenic wheat lines containing Rht1 and Rht3 dwarfing alleles. Planta 183:229–236
Bai C, Liang Y, Hawkesford MJ (2013) Identification of QTLs associated with seedling root traits and their correlation with plant height in wheat. J Exp Bot 64:1745–1753
Bhagwat SG, Bhatia CR (1994) Quantitative differences in the gibberellin induced alpha amylase activity from aleurone layers of tall and semidwarf wheat cultivars. Cereal Res Commun 22:129–134
Botwright TL, Rebetzke GJ, Condon AG, Richards RA (2001) The effect of rht genotype and temperature on coleoptile growth and dry matter partitioning in young wheat seedlings. Aust J Plant Physiol 28:417–423
Chen L, Phillips AL, Condon AG, Parry MAJ, Hu Y-G (2013) GA-responsive dwarfing gene Rht12 affects the developmental and agronomic traits in common bread wheat. PLoS One 8:e62285. doi:10.1371/journal.pone.0062285
Ellis MH, Rebetzke GJ, Azanza F, Richards RA, Spielmeyer W (2005) Molecular mapping of gibberellin-responsive dwarfing genes in bread wheat. Theor Appl Genet 111:423–430
Ellis MH, Spielmeyer W, Gale KR, Rebetzke GJ, Richards RA (2002) Perfect markers for the Rht-B1b and Rht-D1b dwarfing genes in wheat. Theor Appl Genet 105:1038–1042
Griffiths S, Simmonds J, Leverington M, Wang Y, Fish L et al (2012) Meta-QTL analysis of the genetic control of crop height in elite European winter wheat germplasm. Mol Breeding 29:159–171
Haque MA, Martinek P, Watanabe N, Kuboyama T (2011) Genetic mapping of gibberellic acid-sensitive genes for semidwarfism in durum wheat. Cereal Res Commun 39:171–178
Hedden P (2003) The genes of the Green Revolution. Trends Genet 19:5–9
Hedden P, Phillips AL (2000) Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci 5:523–530
Konzak CF (1987) Mutations and mutation breeding. In: Heyne EC (ed) Wheat and wheat improvement—Agronomy Monograph no. 13, 2nd edn. American Society of Agronomy, Madison, pp 428–443
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MapMaker: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Li P, Chen J, Wu P, Zhang J, Chu C, See D, Brown-Guedira G, Zemetra R, Souza E (2011) Quantitative trait loci analysis for the effect of Rht-B1 dwarfing gene on coleoptile length and seedling root length and number of bread wheat. Crop Sci 51:2561–2568
Lo SF, Yang SY, Chen KT, Hsing YI, Zeevaart JA, Chen LJ, Yu SM (2008) A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20:2603–2618
Maccaferri M, Ricci A, Salvi S, Milner SG, Noli E, Martelli P, Casadio R, Akhunov E, Scalabrin S, Vendramin V, Ammar K, Blanco A, Desiderio F, Distelfeld A, Dubcovsky J, Fahima T, Faris J, Korol A, Massi A, Mastrangelo AM, Morgante M, Pozniak C, N’Diaye A, Xu S, Tuberosa R (2015) A high-density, SNP-based consensus map of tetraploid wheat as a bridge to integrate durum and bread wheat genomics and breeding. Plant Biotechnol J 13:648–663
Maluszynski M, Szarejko I (2003) Induced mutations in the Green and Gene Revolutions. In: Tuberosa R, Phillips RL, Gale M (eds) Proceedings of the international congress bologna, Italy, ‘in the wake of the double helix: from the Green Revolution to the gene revolution’. ©2005 Avenue media, Bologna, pp 403–425
Pearce S, Huttly AK, Prosser IM, Li Y, Vaughan SP, Gallova B, Patil A, Coghill JA, Dubcovsky J, Hedden P, Phillips AL (2015) Heterologous expression and transcript analysis of gibberellin biosynthetic genes of grasses reveals novel functionality in the GA3ox family. BMC Plant Biol 15:130. doi:10.1186/s12870-015-0520-7
Ramkumar G, Prahalada GD, Hechanova SL, Vinarao R, Jena KK (2015) Development and validation of SNP-based functional codominant markers for two major disease resistance genes in rice (O. sativa L.) Mol Breeding 35:129. doi:10.1007/s11032-015-0323-4
Rebetzke GJ, Richards RA (2000) Gibberellic acid-sensitive dwarfing genes reduce plant height to increase kernel number and grain yield of wheat. Aust J Agric Res 51:235–245
Rebetzke GJ, Appels R, Morrison AD, Richards RA, McDonald G, Ellis MH, Spielmeyer W, Bonnett DG (2001) Quantitative trait loci on chromosome 4B for coleoptile length and early vigour in wheat. Aust J Agric Res 52:1221–1234
Rebetzke GJ, Bruce S, Kirkegaard JA (2005) Genotypic increases in coleoptile length improves emergence and early vigour with crop residues. Plant Soil 270:87–100
Rebetzke GJ, Ellis MH, Bonnett DG, Condon AG, Falk D, Richards RA (2011) The Rht13 dwarfing gene reduces peduncle length and plant height to increase grain number and yield of wheat. Field Crop Res 124:323–331
Rebetzke GJ, Richards RA, Fettel NA, Long M, Condon AG, Forrester RI, Botwright TL (2007) Genotypic increases in coleoptile length improves stand establishment, vigour and grain yield of deep-sown wheat. Field Crops Res 100:10–12
Rebetzke GJ, Ellis MH, Bonnett DG, Mickelson B, Condon AG, Richards RA (2012) Height reduction and agronomic performance for selected gibberellin-responsive dwarfing genes in bread wheat (Triticum aestivum L.) Field Crops Res 126:87–96
Richards RA (1992) The effect of dwarfing genes in spring wheat in dry environments II. Growth, water use and water use efficiency. Aust J Agric Res 43:529–539
Rogers SO, Bendich AJ (1985) Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol Biol 5:69–76
Spielmeyer W, Hyles J, Joaquim P, Azanza F, Bonnett D, Ellis ME, Moore C, Richards RA (2007) A QTL on chromosome 6A in bread wheat (Triticum aestivum) is associated with longer coleoptiles, greater seedling vigour and final plant height. Theor Appl Genet 115:59–66
Voorrips RE (2002) Mapchart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wuddineh WA, Mazarei M, Zhang J-Y, Poovaiah CR, Mann DGJ, Ziebell A, Sykes RW, Davis MF, Udvardi MK, Stewart CN Jr (2015) Identification and overexpression of gibberellic acid 2-oxidase (GA2ox) in switchgrass (Panicum virgatum L.) for improved plant architecture and reduced biomass recalcitrance. Plant Biotechnol J 13:636–647
Yang Z, Zheng J, Liu C, Wang Y, Condon AG, Chen Y, Hu Y (2015) Effects of the GA-responsive dwarfing gene Rht18 from tetraploid wheat on agronomic traits of common wheat. Field Crop Res 183:92–101
Acknowledgements
Authors wish to thank Dr. M. D. Bhagwat for initiating development of mapping populations and useful suggestions. Authors are grateful to Dr. Andrew Phillips, Rothamsted Research, Harpenden, UK, for providing scaffolds containing sequences of three homoeologues of TaGA2ox9. Authors wish to thank three anonymous reviewers for constructive suggestions on the manuscript. Financial support by the Science and Engineering Research Board, Department of Science and Technology, New Delhi under Start-Up Research grants for Young Scientists-SB/FT/LS-243/2012 to RP is gratefully acknowledged. The Junior Research Fellowship by Agharkar Research Institute to PV under project GEN15 is also gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The research was partially funded by the Science and Engineering Research Board, Department of Science and Technology, New Delhi (SB/FT/LS-243/2012) and the Agharkar Research Institute. The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Vikhe, P., Patil, R., Chavan, A. et al. Mapping gibberellin-sensitive dwarfing locus Rht18 in durum wheat and development of SSR and SNP markers for selection in breeding. Mol Breeding 37, 28 (2017). https://doi.org/10.1007/s11032-017-0641-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-017-0641-9