Abstract
Wx is considered to be the most important gene controlling eating and cooking qualities and pasting properties in rice (Oryza sativa L.). In this study, a recombinant inbred line population derived from indica rice parents differing in apparent amylose content (AAC) was used to detect quantitative trait loci (QTLs) for ten grain quality parameters for rice quality improvement. QTL mapping was performed on the whole population and on two sub-populations based on Wx genotypes. A total of 29 QTLs were found in the whole population. Ten QTLs for 7 traits were detected in the two sub-populations, four of which (qPRO3.1, qPV9, qHPV9, and qCS7) were also detected in the whole population, whereas the other six were QTLs with minor effects that might be covered by the Wx locus. Besides the Wx locus with the largest effect on AAC and most pasting properties, there were another six QTL clusters contributing to grain quality located on chromosomes 2, 3, 5, 6 and 9. It was also found that some QTLs for peak viscosity, breakdown and consistency were closely linked to rice grain shape related QTLs on chromosome 3. A QTL cluster on chromosome 9 for peak viscosity, hot paste viscosity and cold paste viscosity was detectable in the whole population, which was close to the isoamylase 3 (ISA3) locus. A QTL cluster for both peak time and pasting temperature on chromosome 6 was near to the starch synthase I locus, and was potentially a new QTL with minor effect for peak time and pasting temperature. These findings will promote better understanding of the genetic regulation of rice eating and cooking qualities.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice (Oryza sativa L.) is one of the most important staple foods, and is consumed by more than 50 % of the world’s population. As the largest proportion of milled rice, the starch component in the endosperm is a key factor in determining the eating qualities of table rice and the processing qualities of rice flours. The cooking and eating qualities (ECQs) of rice are largely determined by some physicochemical characteristics of the starch in the endosperm: apparent amylose content (AAC), gel consistency and gelatinization temperature (GT) and pasting properties (Bao 2012).
The AAC, gel consistency and most Rapid Visco Analyzer (RVA) pasting viscosity properties are mainly controlled by the Wx gene which encodes granule-bound starch synthase I (GBSSI) responsible for amylose synthesis in endosperm (Bao et al. 2000, 2002). In non-glutinous rice, the rice Wx locus has two functional alleles, Wx a and Wx b. Most japonica varieties with the Wx b allele, containing 18 % or less amylose content, have the sequence AGTTATA at the putative leader intron 5′ splice site, while most indica varieties with the Wx a allele, containing a higher proportion of amylose content, have the sequence AGGTATA (Bligh et al. 1998; Hirano et al. 1998). Other single nucleotide polymorphisms (SNPs) in exons 6 and 10 cause amino acid substitutions, resulting in different AAC and pasting viscosity (Larkin and Park 2003). In glutinous rice, the wx gene contains a 23-bp duplication in exon 2, resulting in a premature translation termination codon, and loss of the function GBSSI (Wanchana et al. 2003).
In earlier studies on the inheritance of rice RVA profiles, they are found to be predominantly controlled by a single gene, with partial control by minor-effect genes with various additive effects, and are closely linked with the inheritance of amylose content (Gravois and Webb 1997; Larkin et al. 2003; Bao et al. 2000). As the Wx gene is the main factor in determining most RVA properties in non-glutinous accessions, many studies have then focused on the detection of minor-effect quantitative trait loci (QTLs) for RVA profiles based on the similar genetic background. With a doubled haploid population derived from two similar intermediate-AAC parents, Bao et al. (2000) detected two QTLs for setback (SB) and consistency (CS) located close to the starch branching enzyme 1 locus on chromosome 6 and a QTL for both hot paste viscosity (HPV) and breakdown (BD) on chromosome 2 that was close to the starch branching enzyme 3 locus. Using a recombinant inbred line (RIL) population derived from two indica cultivars with similar amylose content, Yan et al. (2014) identified 66 minor QTLs for rice ECQs. In an F3 progeny population derived from two parents with similar high AACs, Traore et al. (2011) showed that the Waxy exon 10 SNP was associated with the proportion of soluble to insoluble apparent amylose and most RVA pasting measurements. In glutinous varieties, association mapping studies have been carried out to detect minor-effect QTLs influencing RVA properties which are expected to be irrespective of the effect of Wx. Han et al. (2004) found that polymorphisms in both SBE1 and SBE3 loci accounted for ∼70 % of the observed variations in HPV and cold paste viscosity (CPV), and for 40 % of the observed variations in peak viscosity (PV) and CS. Yan et al. (2011) genotyped 17 starch synthesis-related genes (SSRG) of 118 glutinous accessions and did association analysis with starch paste viscosity parameters; they found that pullulanase (PUL) was the main QTL responsible for most RVA properties. Xu et al. (2013) found that soluble starch synthase IIa (SSIIa) and starch synthase I (SSI) loci were responsible for most RVA properties of waxy rice. However, whether the population derived from parents with normal AAC could be used to detect minor QTLs for RVA pasting viscosity needs to be further addressed.
The GT and most thermal properties are mainly determined by the ALK gene (Wang et al. 2007; Bao 2012). Map-based cloning revealed that ALK encodes SSIIa (Gao et al. 2003). Two functional SNPs in SSIIa were detected at +4198 bp (G/A) and +4330 bp (GC/TT) (Bao et al. 2006a). The SNP at +4198 bp (G/A) is crucial for SSIIa activity; the enzyme is inactive when it is an A SNP regardless of which GC/TT allele is present at +4330 bp (Nakamura et al. 2005), but it is a rare allele in the natural population (Bao et al. 2006a; Bao 2012). The GC/TT polymorphism can differentiate rice varieties with high and intermediate GT (GC allele) from low GT (TT allele) rice varieties (Bao et al. 2006a).
Besides the ECQs mainly influenced by starch components, rice storage proteins also play a role in determining rice qualities (Likitwattanasade and Hongsprabhas 2010). Common rice generally contains about 7 % protein (Xie et al. 2008). The main storage protein in rice is oryzenin, which is composed of subunits that are linked by both intra- and inter-molecular disulfide bonds. Early in cooking, proteins affect the amount of water the rice absorbs, and the availability of water will determine the texture of the cooked rice (Martin and Fitzgerald 2002). It was found that the higher protein content rice displays higher stickness (Chrastil 1994) and hardness (Lyon et al. 1999) than low protein content rice. Proteins also influence viscosity curves, both through binding water and through the agency of a network linked by disulfide bonds (Martin and Fitzgerald 2002).
In this study, an RIL population derived from two non-waxy parents differing in AAC was used to detect QTLs for ten ECQ-related traits. Our strategy was to divide the population into two sub-populations based on Wx genotype, so that new QTLs could be identified in these sub-populations which might be covered by the effect of Wx in the whole population. The objective of this study was to find some new minor-effect QTLs for ECQ-related traits which were covered by Wx genes.
Materials and methods
Materials
A mapping population consisting of 234 RILs at F8 and F9 derived from a cross between M201 and JY293 (Xu et al. 2015) was used in the study. In 2012 and 2013, seeds were sown in late May and harvested in late September at the experimental farm of Zhejiang University, Hangzhou, China.
After being air-dried and stored at room temperature for 2 months, the rice samples were stored in the cooling rooms at 4 °C until use. The samples were milled to white rice using a Satake rice machine (Satake Corp.) and then ground to flour in a Cyclone sample mill (UDY Corp., Fort Collins, CO, USA), and then sieved through a 100-mesh sieve.
AAC and protein content
Determination of AAC was carried out using the iodine staining method (Bao et al. 2006b). The absorbance of the solution was measured at 620 nm against the blank solution using a spectrophotometer. AAC was calculated using a standard curve made from rice samples with known AAC. Protein content was measured by the Kjeldahl method using a Kjeltec-Foss 70 2400 Auto-analyzer using 5.95 as the nitrogen-to-protein conversion factor.
Pasting viscosity profiles
The pasting property of rice flour was determined by using a Rapid Visco Analyzer (RVA-3, Newport Scientific, Warriewood, Australia). According to the method of AACC61-02, 3.0 g (12 % m.b.) of rice flour was placed an aluminum canister, then 25 g of distilled water was added. The heating profile was set as follows: (1) the temperature is held at 50 °C for 1 min; (2) the temperature is linearly ramped up to 95 °C until 4.8 min; (3) the temperature is held at 95 °C until 7.3 min; (4) the temperature is linearly ramped down to 50 °C at 11.1 min and (5) held at 50 °C until 12.5 min.
Four primary parameters could be obtained from the pasting curve, peak viscosity (PV), hot paste viscosity (HPV), cool paste viscosity (CPV) and pasting time (PTime). Three secondary parameters were calculated from primary parameters: breakdown viscosity (BD = PV − HPV), setback viscosity (SB = CPV − PV) and consistency viscosity (CS = CPV − HPV). The viscosity parameters were measured in Rapid Visco Units (RVU). Pasting temperature (PTemp) was determined according to the method proposed by Bao (2008).
Molecular linkage map construction and QTL mapping
One hundred and eighty markers, comprising 161 SSR (simple sequence repeat), ten InDel (insertion–deletion), three STS (sequence tagged site), two CAPS (cleaved amplified polymorphic sequence) and four dCAPS (derived cleaved amplified polymorphic sequence) markers, were used to construct a linkage map. Linkage map construction and QTL mapping were carried out by QTL IciMapping v3.3 (http://www.isbreeding.net/software/?type=detail&id=14). A minimum log10-likelihood ratio (LOD) score of 2.5 was used to declare the presence of a putative QTL in a given genomic region, and the variation explained and additive effect of each QTL was also calculated. QTL nomenclature followed that described by McCouch et al. (1997).
Statistical analysis
All the statistical analyses were carried out with the SAS program v9.1 (SAS Institute Inc, Cary, NC, USA). Proc Corr was used to examine correlations between these traits. Analysis of variance (ANOVA) was carried out to using the general linear model procedure (Proc Glm) and Duncan’s new multiple-range test was used to determine significant differences.
Results
Phenotypic variation in parents and the RIL population
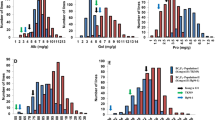
In both years, significant phenotypic differences were detected between the two parents for AAC and protein content (P < 0.01), where JY293 had higher AAC but lower protein content (Table 1). Most parameters for RVA profiles showed significant differences between the two parents. The PV, HPV, CPV, SB, CS, PTime and PTemp of JY293 were significantly higher than those of M201 in both years (Supplementary Fig. 1). The BD of JY293 was significantly lower than M201 in both years. The RIL population showed transgressive segregations in both directions for all the traits. The average values of AAC, PTime and PTemp were significantly higher in 2012 than those in 2013 (P < 0.05); the other parameters showed no significant differences between the 2 years.
Correlation analysis of all ECQ and RVA parameters
The coefficients of correlation of the same trait or parameter ranged from 0.52 for PV to 0.83 for AAC between 2012 and 2013 (P < 0.001) (Supplementary Table 1). In both years, AAC was positively correlated with HPV, CPV, SB and CS, and was negatively correlated with BD. Protein content was negatively correlated with PV, HPV, CPV, BD and CS in 2013, and was negatively correlated with AAC, PV and BD and positively correlated with PTemp in 2012. Except for AAC and SB, PV was positively correlated with all RVA parameters, and was negatively correlated with protein content in both years. HPV and CPV were correlated with all parameters, except for PTemp in 2012, and were correlated with AAC and all RVA parameters, expect for protein content, PTime and PTemp in 2013. BD was positively correlated with PV and PTime, but was negatively correlated with all the other parameters except for PTemp in both years. SB was correlated with AAC, HPV, CPV, BD and CS in both years. CS was correlated with all parameters in both years. PTime was correlated with PV, BD, CS and PTemp in both years, but was also correlated with HPV and CPV in 2012, and was correlated with SB in 2013. PTemp was correlated with PV, CS and PTime in both years, and was correlated with protein and CPV in 2012.
Linkage map construction and QTL mapping
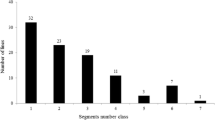
A linkage map was constructed with 180 markers, with chromosomes 1, 2 and 8 each containing two linkage groups. All these markers covered a total of 1950.28 cM of the whole genome, with an average interval of 10.83 cM between adjacent markers. A total of 29 QTLs were identified for all of the ten traits involved in ECQs, distributed on eight chromosomes (Table 2; Fig. 1). Of these QTLs, only nine could be identified in both years, indicating that most QTLs detected in this study were easily affected by environment. At least one QTL and as many as six QTLs were detected for different traits.
QTLs detected for rice RVA properties and ECQs (2012 and 2013). The QTLs in the box were detected in both years. QTLs with underlines were detected in 2012, while QTLs without underlines were detected in 2013. Chr chromosome, PVE percentage of variance explained, Add additive effect; see text for other abbreviations
Apparent amylose content
Two QTLs were detected for AAC. One was in 2012 and two in 2013, collectively accounting for 76.71 and 75.24 % of phenotypic variation, respectively (Table 2). The QTL with the largest effect detected in both years was the Wx locus on chromosome 6, which was closely related to the SSR marker of Wx (RM190). The other QTL on chromosome 5 was only detected in 2013 and accounted for 5.64 % of the phenotypic variation.
Protein content
There were altogether five QTLs for protein content detected in 2012 and 2013 (Table 2). None of them was detected in both years, but qPRO3.1 detected in 2012 was close to qPRO3.2 detected in 2013 on chromosome 3. The other three QTLs were located on chromosomes 1, 2 and 4, respectively.
Peak viscosity
Three main-effect QTLs for PV were detected in both years (Table 2). On chromosome 3, one QTL was detected in both years which explained the largest part of phenotypic variation, i.e. 11.98 % in 2012 and 15.86 % in 2013. The other two QTLs detected only in one year were located on chromosome 2 and chromosome 9.
Hot paste viscosity and cool paste viscosity
In total we detected four QTLs for HPV in 2012 and 2013 (Table 2). Three QTLs detected in 2012 and two in 2013 collectively explained 51.2 and 54.3 % of the total phenotypic variation, respectively. The QTL (qHPV6) detected in both years corresponded to the Wx locus. Two QTLs detected in 2012 were located on chromosomes 3 and 9; the QTL detected in 2013 was on chromosome 3.
Two QTLs for CPV were localized to the same position as those for HPV and the other two QTLs were also linked with the same markers (Table 2). qHPV9 and qCPV9 were QTLs contributed by M201, and increased HPV and CPV by 10.44 and 14.31 RVU, respectively, in 2012.
Breakdown, setback and consistency
Two QTLs were detected for BD in the 2 years (Table 2). The main-effect QTL for BD in both years was also located close to the Wx locus, accounting for 64.5 and 59.6 % of the total phenotypic variation, respectively. The other QTL detected in 2012 could explain 5.64 % of phenotypic variation.
Two QTLs were detected for SB in the 2 years on chromosomes 5 and 6, collectively explaining 79 and 71.8 % of phenotypic variation (Table 2). The QTL detected in both years with the largest effect was at the Wx locus. qSB5, flanked by RM249 and RM18450, was located to the same site as the QTL detected for AAC in 2013.
A total of four QTLs were well mapped for CS on chromosomes 2, 3, 6 and 7. Two QTLs were consistently detected in both years, and could collectively account for 62 and 61.4 % of the total phenotypic variation in 2012 and 2013, respectively. The QTL with the largest effect was the Wx locus. qCS2, flanked by RM423 and RM6375, was also a QTL for protein content in 2013. Another QTL detected in 2012 was located on chromosome 7.
Pasting time and pasting temperature
Only one QTL was detected for PTime in both years (Table 2). The QTL was flanked by RM584 and RM6917, and accounted for 7.9 and 10.9 % of the total phenotypeic variation in 2012 and 2013, respectively.
Two QTLs were detected for PTemp in the 2 years. The QTL detected in both years was located to the same site as the major QTL for PTime, and explained 10.8 and 11.3 % of the total phenotypic variation in 2012 and 2013. The other QTL, only detected in 2012, was on chromosome 1 with a contribution of 6.2 % to phenotypic variation.
Multiple comparison of AAC-related traits with different Wx genotypes
According to the Wx genotype, all the RIL lines can be divided into two sub-populations: one is the same as the JY293-Wx genotype and the other is the same as the M201-Wx genotype (Table 3). In the 2 years, sub-population I (JY293-Wx genotype) had significantly higher AAC, HPV, CPV, SB and CS, but lower BD than sub-population II (M201-Wx genotype). The AAC in the same sub-populations in 2012 was significantly higher than in 2013.
QTL detection in sub-populations based on Wx backgrounds
As stated above, all the lines could be divided into two sub-populations according to the Wx genotype; QTLs for grain quality parameters were detected in the two sub-populations (Table 4). This strategy may allow more QTLs with minor effect to be detected which might be covered by the Wx locus. In the 2 years, a total of ten QTLs for seven traits were detected under the JY293-Wx and M201-Wx backgrounds (Table 4). Most of these QTLs were also detected in the whole RIL population. Under the JY293-Wx background, a total of seven QTLs were identified in 2012 for AAC, PV, HPV, SB, CS and PTemp, while none was detected in 2013, indicating that these QTLs were affected by environment. Of the seven QTLs, three QTLs for PV, HPV and CS were also detected in the whole population. Under the M201-Wx background, two QTLs were detected for protein content; one detected in 2012 was located on chromosome 5 and the other detected in 2013 was located on chromosome 3, and was located exactly to the same site as qPRO3.1 detected in the whole population in 2013. We also detected a QTL flanked by RM232 and RM15413 for PV in both 2012 and 2013.
Discussion
Genetic studies have shown that many starch physicochemical properties are controlled by Wx and SSIIa genes (Bao 2012; Wang et al. 2007). In order to discover more minor-effect QTLs contributing to ECQs, many linkage mapping studies have carried out with genetic populations derived from parents with similar amylose contents or with the same Wx genotype (Bao et al. 2002; Traore et al. 2011; Han et al. 2004; Yan et al. 2011). In addition, association studies have also been carried out with glutinous accessions in which the Wx gene is dysfunctional (Han et al. 2004; Yan et al. 2011; Xu et al. 2013). We describe an additional strategy to detect minor QTLs within sub-populations derived from parents differing in AAC content and Wx genotype. This strategy is alternative to the genetic population derived from parents with similar amylose contents or with the same Wx genotype. The difference is that we artificially created two sub-populations with the same Wx genotype. The advantage of this strategy is that we can compare QTLs detected in the whole population and in two sub-populations.
To confirm whether our strategy could identify more minor-effect QTLs, we divided the whole population into two sub-populations corresponding to the JY293-Wx and M201-Wx genotypes. Within each sub-population, all lines have the same Wx genotype but they still have subtle differences in AAC, protein content and RVA pasting properties, so QTL mapping for these ten traits were carried out in two sub-populations, respectively. We detected a total of ten QTLs for seven traits in the sub-populations (Table 4). Under the JY293-Wx background (sub-population I), seven QTLs were identified in 2012 for AAC, PV, HPV, SB, CS and PTemp, but none was detected in 2013, indicating that these QTLs were also environment-responsive. Of the seven QTLs, three QTLs for PV (qPV9), HPV (qHPV9) and CS (qCS7) were also detected in the whole population (Tables 2, 4). The other four QTLs, qAAC3, qSB7, qCS3.1, and qTemp3, were specific to this sub-population. qAAC3 is close to a QTL detected by Li et al. (2014) for amylose content, which was flanked by two SSR markers RM135 and RM49. qSB7 is located to the same site as a QTL for SB detected by Hsu et al. (2014). qTemp3 is close to the QTL for peak temperature detected by Wang et al. (2007). Under the M201-Wx background (sub-population II), only three QTLs were detected, two for protein content and another QTL for PV. The QTL on chromosome 3 for protein content detected in 2013 was located exactly in the same region as qPRO3.1 detected in 2012 in the whole population. The other QTL located on chromosome 5 detected in 2012 (qPRO5) is specific to sub-population II. The QTL for PV, flanked by RM232 and RM 15413, detected in both years is specific to sub-population II, but it was also a QTL for HPV and CPV detected in the whole population in 2012. Thus, we confirmed that our strategy can identify more QTLs with minor effect for eating and cooking qualities in rice grain.
In the whole RIL population, we detected 29 QTLs for ten traits. Among these QTLs, the Wx gene is the most important QTL for AAC, HPV, CPV, BD, SB and CS, agreeing with previous reports (Bao et al. 2000; Hsu et al. 2014; Larkin et al. 2003; Larkin and Park 2003; Traore et al. 2011; Wang et al. 2007;). Li et al. (2014) have integrated QTLs for amylose content, finding that QTLs for AAC were located on chromosomes 1, 2, 3, 6, 7, 8, 9 and 10, so the QTL on chromosome 5 for AAC detected in 2012 might be a new locus. We detected five QTLs for protein content, but none was detected in both years, which might be caused by environmental effects. Of the five QTLs, three of them (qPRO1, qPRO3.2 and qPRO4) were also detected in previous studies (Zhang et al. 2008; Zheng et al. 2011). In correlation analysis, it was found that protein had negative correlations with PV (P < 0.001), HPV (P < 0.01), CPV (P < 0.01) and BD (P < 0.01) (Supplementary Table 1), suggesting that protein influenced ECQs by affecting starch viscosities (Martin and Fitzgerald 2002). Three QTLs for PV were detected in the 2 years. qPV3, flanked by GL3 and RM15578, was detected in both years. In this region, FLOURY ENDOSPERM 6 (FLO6)-deficient mutants also showed low PV (Peng et al. 2014). In addition, a QTL for rice grain weight (qGL3) was located in this region (Xu et al. 2015; Zhang et al. 2012). qPV9, qHPV9 and qCPV9 were located at the same site, and were also detected by Wang et al. (2007). This locus is near to the isoamylase 3 (ISA3) locus, indicating that isoamylase 3 activity might play a role in affecting PV, HPV and CPV. Almost all of the QTLs detected for HPV and CPV are located at the same sites (Table 2; Fig. 1), which might explain their significant correlation. qBD3 is flanked by GL3 and RM15578, and is close to the QTL (qGL3) affecting grain length (Xu et al. 2015; Zhang et al. 2012). qCS2 and qCS3 are located near to grain shape genes GW2 and GS3 (Fan et al. 2006; Song et al. 2007; Xu et al. 2015). These phenomena may explain why grain yield is often negatively correlated with grain quality. The QTLs qPTime6 and qTemp6 detected for PTime and PTemp, sharing the same region flanked by RM584 and RM6917, are near to the starch synthase 1 (SSI) locus, which is potentially a new QTL with minor effect for pasting time and pasting temperature.
In summary, we describe a strategy to search for more QTLs with minor effect for rice ECQs. As expected, the Wx gene has the largest effect on AAC and most pasting properties. However, there are another six QTL clusters contributing to grain quality, located on chromosomes 2, 3, 5, 6 and 9. These QTLs, detected simultaneously in the whole RIL population and in sub-populations, display minor effects on rice ECQs, which are irrespective of the effect of Wx. However, QTLs detected only in one of the sub-populations are specific to this population but are also covered by Wx. Co-location of QTLs for pasting properties at grain shape-related genes provides an answer to the question why grain yield is usually negatively correlated with grain quality. The QTL detected for PTime and PTemp, near to the SSI locus, is a potential new minor-effect QTL for pasting time and pasting temperature.
References
Bao JS (2008) Accurate measurement of pasting temperature by the rapid visco-analyser: a case study using rice flour. Rice Sci 15:69–72
Bao JS (2012) Toward understanding the genetic and molecular bases of the eating and cooking qualities of rice. Cereal Foods World 57:148–156
Bao JS, Zheng XW, Xia YW, He P, Shu QY, Lu X, Chen Y, Zhu LH (2000) QTL mapping for the paste viscosity characteristics in rice (Oryza sativa L.). Theor Appl Genet 100:280–284
Bao JS, Wu YR, Hu B, Wu P, Cui HR, Shu QY (2002) QTL for rice grain quality based on a DH population derived from parents with similar apparent amylose content. Euphytica 128:317–324
Bao JS, Corke H, Sun M (2006a) Nucleotide diversity in starch synthase IIa and validation of single nucleotide polymorphisms in relation to starch gelatinization temperature and other physicochemical properties in rice (Oryza sativa L.). Theor Appl Genet 113:1171–1180
Bao JS, Shen SQ, Sun M, Corke H (2006b) Analysis of genotypic diversity in the starch physicochemical properties of nonwaxy rice: apparent amylose content, pasting viscosity and gel texture. Starch-Starke 58:259–267
Bligh HF, Larkin P, Roach P, Jones C, Fu H, Park W (1998) Use of alternate splice sites in granule-bound starch synthase mRNA from low-amylose rice varieties. Plant Mol Biol 38:407–415
Chrastil J (1994) Stickiness of oryzenin and starch mixtures from preharvest and postharvest rice grains. J Agric Food Chem 42:2147–2151
Fan C, Xing Y, Mao H, Lu T, Han B, Xu C, Li X, Zhang Q (2006) GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet 112(6):1164–1171
Gao ZY, Zeng DL, Cui X, Zhou YH, Yan M, Huang D, Li JY, Qian Q (2003) Map-based cloning of the ALK gene, which controls the gelatinization temperature of rice. Sci China Ser C 46:661–668
Gravois KA, Webb BD (1997) Inheritance of long grain rice amylograph viscosity characteristics. Euphytica 97:25–29
Han YP, Xu ML, Liu XY, Yan CJ, Korban SS, Chen XL, Gu MH (2004) Genes coding for starch branching enzymes are major contributors to starch viscosity characteristics in waxy rice (Oryza sativa L.). Plant Sci 166:357–364
Hirano HY, Eiguchi M, Sano Y (1998) A single base change altered the regulation of the Waxy gene at the posttranscriptional level during the domestication of rice. Mol Biol Evol 15:978–987
Hsu YC, Tseng MC, Wu YP, Lin MY, Wei FJ, Hwu KK, Hsing YI, Lin YR (2014) Genetic factors responsible for eating and cooking qualities of rice grains in a recombinant inbred population of an inter-subspecific cross. Mol Breed 34:655–673
Larkin PD, Park WD (2003) Association of waxy gene single nucleotide polymorphisms with starch characteristics in rice (Oryza sativa L.). Mol Breed 12:335–339
Larkin PD, McClung AM, Ayres NM, Park WD (2003) The effect of the Waxy locus (Granule Bound Starch Synthase) on pasting curve characteristics in specialty rices (Oryza sativa L.). Euphytica 131:243–253
Li X, Ma W, Wang Z, Guo Y, Liu F, Wu H (2014) Study on integration of QTLs related to amylose content of rice. J Northeast Agric Univ 45:8–14
Likitwattanasade T, Hongsprabhas P (2010) Effect of storage proteins on pasting properties and microstructure of Thai rice. Food Res Int 43:1402–1409
Lyon BG, Champagne ET, Vinyard BT, Windham WR, Barton FE, Webb BD, McClung AM, Moldenhauer KA, Linscombe S, McKenzie KS, Kohlwey DE (1999) Effects of degree of milling, drying condition, and final moisture content on sensory texture of cooked rice. Cereal Chem 76:56–62
Martin M, Fitzgerald M (2002) Proteins in rice grains influence cooking properties. J Cereal Sci 36:285–294
McCouch SR, Cho YG, Yano M (1997) Report on QTL nomenclature. Rice Genet Newsl 14:11–14
Nakamura Y, Francisco PB, Hosaka Y, Sato A, Sawada T, Kubo A, Fujita N (2005) Essential amino acids of starch synthase IIa differentiate amylopectin structure and starch quality between japonica and indica rice varieties. Plant Mol Biol 58:213–227
Peng C, Wang YH, Liu F, Ren YL, Zhou KN, Lv J, Zheng M, Zhao SL, Zhang L, Wang CM, Jiang L, Zhang X, Guo XP, Bao YQ, Wan JM (2014) FLOURY ENDOSPERM6 encodes a CBM48 domain-containing protein involved in compound granule formation and starch synthesis in rice endosperm. Plant J 77:917–930
Song XJ, Huang W, Shi M, Zhu MZ, Lin HX (2007) A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet 39:623–630
Traore K, McClung AM, Chen MH, Fjellstrom R (2011) Inheritance of flour paste viscosity is associated with a rice Waxy gene exon 10 SNP marker. J Cereal Sci 53:37–44
Wanchana S, Toojinda T, Tragoonrung S, Vanavichit A (2003) Duplicated coding sequence in the waxy allele of tropical glutinous rice (Oryza sativa L.). Plant Sci 165:1193–1199
Wang LQ, Liu WJ, Xu Y, He YQ, Luo LJ, Xing YZ, Xu CG, Zhang QF (2007) Genetic basis of 17 traits and viscosity parameters characterizing the eating and cooking quality of rice grain. Theor Appl Genet 115:463–476
Xie LH, Chen N, Duan BW, Zhu ZW, Liao XY (2008) Impact of proteins on pasting and cooking properties of waxy and non-waxy rice. J Cereal Sci 47:372–379
Xu FF, Zhang G, Tong C, Sun X, Corke H, Sun M, Bao JS (2013) Association mapping of starch physicochemical properties with starch biosynthesizing genes in waxy rice (Oryza sativa L.). J Agric Food Chem 61:10110–10117
Xu FF, Sun X, Chen YL, Huang Y, Tong C, Bao JS (2015) Rapid identification of major QTLs associated with rice grain weight and their utilization. PLoS ONE. doi:10.1371/journal.pone.0122206
Yan CJ, Tian ZX, Fang YW, Yang YC, Li JA, Zeng SY, Gu SL, Xu CW, Tang SZ, Gu MH (2011) Genetic analysis of starch paste viscosity parameters in glutinous rice (Oryza sativa L.). Theor Appl Genet 122:63–76
Yan B, Tondi Yacouba N, Chen J, Wang Y, Gao G, Zhang Q, Liu X, He Y (2014) Analysis of minor quantitative trait loci for eating and cooking quality traits in rice using a recombinant inbred line population derived from two indica cultivars with similar amylose content. Mol Breed 34:2151–2163
Zhang WW, Bi JC, Chen LM, Zheng LN, Ji SL, Xia YM, Xie K, Zhao ZG, Wang YH, Liu LL, Jiang L, Wan JM (2008) QTL mapping for crude protein and protein fraction contents in rice (Oryza sativa L.). J Cereal Sci 48:539–547
Zhang XJ, Wang JF, Huang J, Lan HX, Wang CL, Yin CF, Wu YY, Tang HJ, Qian Q, Li JY, Zhang HS (2012) Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc Natl Acad Sci USA 109:21534–21539
Zheng L, Zhang W, Chen X, Ma J, Chen W, Zhao Z, Zhai H, Wan J (2011) Dynamic QTL analysis of rice protein content and protein index using recombinant inbred lines. J Plant Bio 54:321–328
Acknowledgments
We thank Yafang Shao, Gan Zhang and Fufu Tang for their technical assistance. This work was financially supported by the Fundamental Research Funds for the Central Universities (2011FZA6011), the Natural Science Foundation of Zhejiang Province (LZ13C130001), and the Special Fund for Agro-scientific Research in the Public Interest (201103007) from the Ministry of Agriculture.
Author information
Authors and Affiliations
Corresponding author
Additional information
Feifei Xu and Chengxiao Sun have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xu, F., Sun, C., Huang, Y. et al. QTL mapping for rice grain quality: a strategy to detect more QTLs within sub-populations. Mol Breeding 35, 105 (2015). https://doi.org/10.1007/s11032-015-0296-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-015-0296-3