Abstract
Anthocyanins have several biological functions in plants and are beneficial to human health. To elucidate the metabolic profile of anthocyanins and determine the genetic basis controlling anthocyanin accumulation in zicaitai (Brassica rapa L. ssp. chinensis var. purpurea), we conducted anthocyanin profile characterization and quantitative trait locus (QTL) analysis. Seventeen anthocyanin compounds were identified as cyanidin glycosides in zicaitai. A genetic linkage map based on 200 F2 lines was constructed using 161 insertion/deletion markers. Total anthocyanin content (TAC) was determined by pH differential spectrophotometry for the F2 lines. Using the map and phenotypic data, a major QTL which explained 56.7 % of phenotypic variation was identified for TAC on chromosome A09. Two genes, BrEGL3.1 and BrEGL3.2, as syntenic orthologs of AtEGL3 encoding basic helix–loop–helix transcription factors in this QTL region, are candidate genes for a key role in the control of anthocyanin accumulation in zicaitai.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Anthocyanin pigments are an important type of flavonoid compound and exhibit a wide range of biological functions in plants. These include serving as attractants for pollinators and seed dispersers, and protecting plants against abiotic and biotic stresses (Harborne et al. 1999; Andersen and Markham 2006). Anthocyanins have beneficial roles in human health, and are potentially protective factors against cancer and heart disease (Lam et al. 2009; van Poppel et al. 1999). These beneficial health properties are partially attributed to their strong antioxidant capacity (Yamasaki et al. 1997). Their bioavailability and activity varies widely depending on chemical structure and content (Prior and Wu 2006). Zicaitai (Brassica rapa L. ssp. chinensis var. purpurea) has purple petioles and flower stalks, and is a representative purple variety of B. rapa vegetables. The purple pigments of zicaitai have been identified as anthocyanins (Podsędek 2007). Although several studies have characterized anthocyanins in different Brassica species, they have only investigated either B. oleracea, including cauliflower and cabbage (Lo Scalzo et al. 2008), or B. juncea, including mustard (Mobin and Khan 2007) and red mustard greens (Lin et al. 2011).
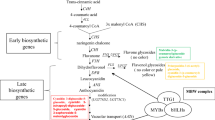
Studies on the genetic or molecular mechanisms of anthocyanin biosynthesis and accumulation have been performed in many kinds of plants (Holton and Cornish 1995). The biosynthetic pathways of anthocyanins are well characterized, and corresponding genes have been isolated from various plants (Broun 2005; Koes et al. 2005; Grotewold 2006). In B. rapa, Burdzinski and Wendell (2007) mapped the anthocyaninless (anl) locus to chromosome A09 in a rapid-cycling B. rapa line using an F2 population. Hayashi et al. (2010) reported that anthocyanin pigmentation in purple turnip (B. rapa ssp. rapa) is controlled by a single dominant gene, and mapped a novel locus (Anp) regulating anthocyanin pigmentation on chromosome A07, based on a doubled haploid (DH) population from a cross between purple turnip and Chinese cabbage (B. rapa L. ssp. pekinensis). Microarray analysis on green and red Chinese cabbage identified anthocyanin-specific genes that likely play regulatory roles in anthocyanin production, and anthocyanin-related transcription factors that may be active in the pigmentation signaling pathway (Kim et al. 2010, 2011). Liu et al. (2013b) mapped the pur gene, controlling purple leaf color, to the end of chromosome A03 using an F2 population constructed from a Chinese cabbage line and a purple pakchoi (B. rapa L. ssp. chinensis) line and proposed that this phenotype was controlled by a dominant gene with an additive effect. In these studies, qualify distances of the closest flanking markers were too far from the target genes to determine candidate genes. Results from previous research indicate that the genetic or molecular mechanisms of anthocyanin biosynthesis and accumulation are complicated, and candidate genes are not identical in different purple varieties of B. rapa.
In this study, the anthocyanin profile was determined in zicaitai using a liquid chromatography–mass spectrometry (LC–MS) technique. We analyzed the total anthocyanin content (TAC) in zicaitai leaves using an F2 population derived from a cross between zicaitai and caixin (B. rapa L. ssp. parachinensis). We mapped this trait to a 1.9-Mb region on chromosome A09. Two syntenic orthologs of AtEGL3, BrEGL3.1 and BrEGL3.2, encoding basic helix–loop–helix transcription factors, are candidate genes for controlling anthocyanin accumulation in zicaitai.
Materials and methods
Plant materials for UFLC-PDA-ESI/MS/MS and mapping population
Zicaitai DH line ZCT095 was used as the female parent and Caixin DH line L58DH as the male parent for construction of an F2 population. ZCT095 accumulates purple pigments in the petioles (Fig. 1a), whereas the leaves and petioles of L58DH are green (Fig. 1b). An F2 population consisting of 200 plants was constructed from a cross between these two parental lines. Leaves from parental and F2 lines were harvested 50 days after sowing, and tissues immediately freeze-dried for future anthocyanin extraction and DNA isolation.
Sample treatment and anthocyanin extraction
Anthocyanin extraction was performed in accordance with the method described by Lin and Harnly (2007), with slight modifications. Freeze-dried leaf material (~0.2 g) was extracted in 30 ml of acidified methanol (0.1 % HCl, v/v) by ultrasonication with an FS30 Ultrasonic sonicator (40 kHz, 100 W) (Fisher Scientific, Pittsburgh, PA, USA) for 30 min at room temperature (<35 °C). The acidified extraction solvent was necessary to prevent degradation of the anthocyanins (Longo and Vasapollo 2006). The slurry mixture was centrifuged (Centrifuge 5810R, Eppendorf, Germany) at 5,000 rpm for 15 min at 4 °C. The supernatant was collected and concentrated using a rotary evaporator (RV10, IKA, Germany) at 35 °C. The residue was reconstituted in 2 ml or 10 ml acidified water (0.1 % formic acid, v/v), and filtered through 0.22-μm filters for ultra-fast liquid chromatography (UFLC)–ultraviolet (UV) triple quadrupole linear ion trap mass spectrometry (Q-Trip), time-of-flight (TOF) mass spectrometry, or pH differential spectrophotometry analysis.
Anthocyanin profile characterization
The chromatographic system, which was set up in accordance with the method described by Lin et al. (2011) and Ferreres et al. (2008) with modifications, consisted of an UFLC system (Prominence™ UFLC, Shimadzu, Japan) equipped with an ultraviolet detector. Chromatographic separation was achieved using a Shim-pack XR-ODS column (75 mm × 2.0 mm i.d., 2.2 μm particle size; Shimadzu, Japan) at a flow rate of 0.25 ml/min. The column oven temperature was set at 40 °C. The mobile phase was a binary solvent system consisting of solvent A (formic acid 0.1 % v/v in water) and solvent B (acetonitrile). The gradient conditions were as follows: 5–15 % B from 0 to 25 min, maintained at 15 % B for 20 min; 15–18 % B from 45 to 60 min; 18–95 % B from 60 to 70 min, maintained at 95 % B for 2 min; and 5 % B from 72 to 75 min for equilibration of the column for the next run. The sample injection volume was 5 μl. Chromatograms were recorded at 520 nm for anthocyanins. The analytes were identified using a triple quadrupole linear ion trap mass spectrometer (AB SCIEX 5500 Qtrap™, USA) equipped with a turbo-spray ion source. Product ion spectra (MS2 and MS3) were acquired using enhanced product ion (EPI) and MS3 profile scans in positive ionization modes. High-resolution accurate mass measurements were made using a 6540 Agilent quadrupole/time-of-flight mass spectrometer (Q-TOF, Agilent Technologies, USA) equipped with an electrospray ionization (ESI) source. Mass spectrometer acquisition was operated in the positive ion mode with accurate mass ranging from 100 to 1,700 m/z at resolution 100,000.
pH differential spectrophotometry
The pH differential spectrophotometry method was performed in accordance to that reported by (Lee et al. 2005), with slight modifications, to determine total anthocyanin content (TAC) in parental and F2 lines. Absorbance was measured at 520 and 700 nm. The TAC was expressed as cyanidin-3-glucoside (cyd-glu, molar extinction coefficient 26,900 l/cm mol, molecular weight 449.2 g/mol). The units for TAC were mg/g dry weight (DW) of detected samples. Measurements of TAC in all samples were replicated three times.
Molecular marker assays
DNA from parents and F2 individuals was isolated from freeze-dried mature leaves, as described by Wang et al. (2005).
The two parental lines were re-sequenced to a depth of 5× using the Illumina sequencing platform service provided by the BGI Tech Solutions Co., Ltd. The genome sequence of B. rapa was retrieved from BRAD (http://brassicadb.org/brad/) and used as the reference sequence. Genome-wide detection of short insertion/deletion length polymorphisms (InDels, <5 bp) and the development of InDel markers were performed as described by Liu et al. (2013a). In total, 300 pairs of InDel primers uniformly distributed (~2,000 kb) across the whole genome were designed to construct the linkage map. Each 20-μl polymerase chain reaction (PCR) mixture contained 2 μl PCR buffer, 1.6 μl dNTPs (2.5 mM each), 0.4 μl Taq DNA polymerase (2.5 U/μl), 2 μl DNA template (50 ng/μl), 0.6 μl forward primer (10 μM), 0.6 μl reverse primer (10 μM), and 12.8 μl ddH2O. The reaction mixture was incubated in a thermal cycler (9700, ABI, USA) at 94 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 45 s, with a final extension at 72 °C for 7 min. The PCR products were separated by electrophoresis on 8 % polyacrylamide gel running at 160 V for 1.5 h, and then subjected to silver staining for band statistics (Bassam et al. 1991).
Construction of linkage map
The linkage map was constructed with JoinMap 4.0 (http://www.kyazma.nl/) using a minimum logarithm of odds (LOD) score of 4.0 (Van Ooijen 2006). The linkage groups (LGs) were determined using a LOD threshold ≥4.0 and a maximum recombination fraction of 0.4. The Kosambi mapping function was used to obtain genetic distances between markers (Kosambi 1944).
Statistical analysis and quantitative trait locus (QTL) mapping
The software MapQTL 5.0 (Van Ooijen 2004) was used to detect QTLs by both interval mapping and multiple QTL mapping (MQM) methods. In regions containing putative QTLs, markers with the highest LOD values were taken as co-factors. The LOD thresholds for QTL significance were determined by permutation tests (1,000 replications) in MapQTL with a genome-wide significance level of P = 0.05 for significant linkages; LOD = 3.0 was used as the significance threshold based on the permutation test. Frequency distribution and ANOVA analysis of TAC was performed using the SPSS version 12.0.1 statistical package (SPSS Inc., Chicago, IL, USA).
Results
Anthocyanin profile analysis in zicaitai
Several anthocyanin groups (e.g., cyanidin, pelargonidin, and delphinidin) exist in the plant kingdom (Tanaka et al. 2008). To characterize the anthocyanin profile in zicaitai and caixin, we performed UFLC-UV-Q-trip/TOF MS analysis on the parental lines. In total, 17 anthocyanin compounds were detected in zicaitai ZCT095 (Fig. 2a; Table 1), while no anthocyanin was detected in caixin L58DH (Fig. 2b). Identification of anthocyanins in extracts was based on chromatographic behavior, accurate molecular masses obtained by Q-TOF mass spectrometry, characteristic MS/MS fragmentation product ions obtained by Q-Trip mass spectrometry, UV spectra, and comparison with scientific publications (Mazza and Miniati 1993; Wu and Prior 2005; Lin et al. 2011). All 17 anthocyanins were different forms of cyanidin glycosides, with glycosyl groups at both the 3- and 5-positions. The glycosyl groups at the 3-position were di/triglucosides; these were acylated by coumaroyl, sinapoyl, caffeoyl, and feruloyl. Glucoside was the only glycosylated modification at the 5-position, and some of the anthocyanins were acylated by malonyl. Therefore, all 17 anthocyanins were different acylated cyanidin 3-di/triglucosides-5-glucosides. The use of UFLC-UV-Q-Trip and UFLC-Q-TOF analysis generated a comprehensive metabolic profile of anthocyanins in zicaitai (Table 1).
Liquid chromatography chromatograms (520 nm) of a Zicaitai ZCT095 and b Caixin L58DH anthocyanin extracts. Horizontal axis shows retention time (min); vertical axis gives the strength of the chromatographic peak response (AU/uV). Labels correspond to compounds shown in Table 1
Quantitative variation in total anthocyanin content
The TAC of the parents and F2 lines was determined by pH differential spectrophotometry. The TAC of the female parental line ZCT095 and the male parental line L58DH was 0.8 mg/g DW and 0.0 mg/g DW, respectively. The purpleness of plants and plant extracts from the F2 population varied widely in comparison to the purple or green seen in the parental lines (Fig. 3a); this indicated that anthocyanin accumulation was under multigenic control in zicaitai leaves. The TAC of the F2 population ranged from 0.0 to 0.78 mg/g DW, and the average TAC was 0.19 mg/g DW. As the variation among F2 lines for TAC was between that of the parental lines, it indicated that this trait showed inter-parent variations and that alleles with positive effects for anthocyanin accumulation might only exist in the female parental line. The TAC of the F2 population showed a bias distribution lower than the parent, at approximately 67 % lower than the average level.
Construction of the genetic linkage map
A total of 1,228 InDel primer pairs, comprising 928 previously designed (Wang et al. 2011; Liu et al. 2013a) and 300 newly developed (Supplementary Table 1) for this study, were screened for polymorphisms between the parental lines. Seventy-two of the previously developed InDel primer pairs and 107 newly developed ones showed polymorphisms between parents. The 179 polymorphic InDel markers were relatively evenly distributed on the B. rapa genome, with 12, 14, 17, 18, 19, 18, 15, 11, 35, and 16 on chromosomes A01–A10 respectively, and a further four located on unassigned scaffolds. All polymorphic primers returned reliable PCR products, and could distinguish parental and heterologous genotypes in the F2 population. These markers were then used to genotype the F2 mapping population. A total of 161 InDel markers were assigned to 10 linkage groups (LGs); these covered a genetic distance of 1,314.3 cM, with an average internal length of 8.16 cM between two adjacent markers (Fig. 4). The linkage groups were anchored to their corresponding reference chromosomes in accordance with the physical positions of the markers.
Genetic linkage map based on a population of 200 F2 lines of B. rapa. Marker loci are listed to the right, and recombination distances (cM) to the left of each linkage group. Marker IDs listed in this linkage map omit “BrID” in front of the ID number; for example, “120013” and “90271” represent “BrID120013” and “BrID90271”, respectively, in Supplemental Table S1 and previous studies (Wang et al. 2011)
Identification of a QTL controlling total anthocyanin concentration in zicaitai
In total, two QTLs associated with this trait were identified that satisfied the threshold LOD of >3. A major QTL was identified on chromosome A09 with a LOD value of 22.95, and explained 56.7 % of TAC variation in the F2 population (Fig. 5). The flanking markers of this major QTL were BrID11259 and BrID11861. This major QTL showed a positive additive effect, indicating that the purple parent ZCT095 at this locus mostly increased the anthocyanin accumulation. A minor QTL was observed on chromosome A07 between InDel markers BrID120533 and BrID120549 with a LOD value of 6.10, and 16.3 % of TAC variation was explained by this QTL which showed a negative additive effort opposite to the major one. Taken together, the major QTL on chromosome A09 might play a key role in the anthocyanin accumulation of zicaitai.
The major QTL on chromosome A9 for total anthocyanin content in zicaitai. a Red and green lines indicate the LOD value and contribution rate of the QTL, respectively. b Position of flanking markers surrounding the major QTL on chromosome A09; the red line indicates the peak position of the QTL. (Color figure online)
Identification of candidate genes
The genetic distance between the two flanking markers of the major QTL identified on chromosome A09 was 8.7 cM; this corresponds to a physical distance of 1.9 Mb. There are 274 annotated genes distributed in this region. As B. rapa is closely related to Arabidopsis thaliana, comparative genomic analysis between B. rapa and A. thaliana allowed genes involved in the anthocyanin biosynthesis pathway of B. rapa to be identified on a genome-wide level (Guo et al. 2014). We evaluated the 274 predicted genes within the QTL interval to identify potential genes involved in the anthocyanin biosynthesis pathway. Two candidate genes, BrEGL3.1 (Bra027796) and BrEGL3.2 (Bra027653), which are syntenic orthologs of AtEGL3, were located in this region.
We compared re-sequencing data from the parental lines to the Chiifu-401-42 reference genome to reveal sequence variations in these two genes between the parents. For BrEGL3.1, we found two InDels and 34 single-nucleotide polymorphisms (SNPs). The two InDels, and 18 of the 34 SNPs, were located in non-coding region (introns). The 16 SNPs located in the coding region (exons) comprised six synonymous and 10 nonsynonymous mutations, resulting in eight amino acid substitutions (Supplementary Fig. 1a). Of the eight amino acid substitutions, one (Leu180Phe) was located in the MYB interacting region (MIR), five (Ser226Lys, Ala342Thr, Ser379Asn, Lys394Thr, Ile409Val) were located in the domain of interaction with WD40, and two (Asp490Glu and Ser529Arg) were located in the carboxy-terminal end (C-ter), while there were no variations in the basic helix–loop–helix (bHLH) domain. Three substitutions (Ser226Lys, from polarity without charge to polarity with positive; Ala 342Thr, from nonpolarity to polarity without charge; Lys394Thr, from polarity with positive to polarity without positive) in the domain of interaction with WD40 and one (Ser529Arg, from polarity without charge to polarity with positive) located in the carboxy-terminal end (C-ter) caused changes in the chemicophysical properties of amino acid residues. The sequence variations of BrEGL3.2 between the parental lines were relatively less than those of BrEGL3.1, and included two InDels and 10 SNPs. The two InDels were located in non-coding regions, while the 10 SNPs were all located in coding sequences, leading to two amino acid substitutions (Gly233Glu and Leu352Phe) which were both in the domain of interaction with WD40 (Supplementary Fig. 1b), and the substitution of Gly233Glu changed the chemicophysical property of the amino acid residue from polarity without charge to polarity with negative. The amino acid substitutions of BrEGL3.1 and BrEGL3.2 between the parents were mainly located in the domain of interaction with WD40 proteins, while there were no amino acid substitutions in the bHLH domain. For BrEGL3.1, there was one amino acid substitution in the MYB-interacting region, five in the domain of interaction with WD40 proteins, and two in the C-ter region. Of these amino acid substitutions, three in the domain of interaction with WD40 proteins and one in the C-ter region showed different chemicophysical properties. The two amino acid substitutions between parents of BrEGL3.2, one of which had identical characteristics, were both in the domain of interaction with WD40.
Discussion
Cyanidin glucosides are the major anthocyanins in zicaitai
Anthocyanin compounds are the sugar-conjugated forms of anthocyanidins, and are widely dispersed throughout the plant kingdom. The chemical structure of the anthocyanin determines its stability, color intensity, and potential biological activity. The commonest anthocyanins are pelargonidin, cyanidin, delphinidin, peonidin, petunidin, and malvidin, with cyanidin being the most common in Brassica crop species (Tatsuzawa et al. 2006; Scalzo et al. 2008; Lin et al. 2011). In Brassica, anthocyanins are complex, glycosylated with one to five sugar moieties, and acylated with different hydroxycinnamic acids (Podsędek 2007; Moreno et al. 2010). Until now, no study has comprehensively analyzed anthocyanin composition in zicaitai, a species with high accumulation of anthocyanin in the petiole.
In this study, despite the lack of commercial available standards, the accuracy of the mass measurement of MSn fragmentation analysis by Q-trap and TOF MS, together with UV/visible absorption spectra, allowed identification of the anthocyanin composition with high reliability. A total of 17 cyanidin glycosides were identified. The glycosylated and acylated properties revealed the chemical structural characteristics of these anthocyanins in zicaitai (Table 1), and lay a foundation for further investigations into the genetic mechanisms and genes that control anthocyanin pigmentation in B. rapa.
Purple pigmentation is a quantitative trait in zicaitai
The genetic mechanism controlling purple pigmentation in Brassica is complex. Previous studies revealed that the purple characteristics in rapid-cycling B. rapa, purple turnip, purple pakchoi, and purple cauliflower (B. oleracea var. botrytis) were qualitative traits controlled by different single genes, and that purple is dominant over green (Burdzinski and Wendell 2007; Hayashi et al. 2010; Chiu et al. 2010). In this study, we investigated the purple characteristics of an F2 population derived from crossing zicaitai and caixin. TAC showed continuous variation in the F2 population, indicating that the genetic mechanism of anthocyanin accumulation in zicaitai was under multigenic control. This is in contrast to the previously reported single gene control observed in purple turnip, purple pak choi, rapid-cycling B. rapa, and purple cauliflower. This indicates that anthocyanin accumulation in plants is complicated. The different anthocyanin compounds may involve specific gene control, and for some compounds it is necessary to activate a whole branch of the biosynthesis pathway. Future studies of different B. rapa purple crops will helpfully clarify gene function in each step of the anthocyanin biosynthesis pathway.
The major QTL is a novel locus regulating anthocyanin pigmentation in B. rapa
A major QTL controlling anthocyanin accumulation in zicaitai was mapped to chromosome A09. In previous studies, three loci were reported to control anthocyanin pigmentation in different B. rapa varieties, namely anl on chromosome A09 in rapid-cycling B. rapa (Burdzinski and Wendell 2007), Anp on chromosome A07 in purple turnip (Hayashi et al. 2010), and Pur on the end of chromosome A03 in purple pak choi (Liu et al. 2013a, b). Using flanking simple sequence repeat markers of anl, we compared the position of locus anl with the major QTL identified in this study. The anl locus was located within the 20,696,245–23,679,772 region of chromosome A09, while the newly identified major QTL was located in the 6,068,769–8,028,904 interval on A09, a physical distance far from anl. We therefore assume that this QTL is a novel locus regulating anthocyanin accumulation in B. rapa.
BrEGL3.1 and BrEGL3.2 are candidate genes controlling anthocyanin accumulation in zicaitai
Transcriptional regulation of structural genes is a major mechanism by which anthocyanin biosynthesis is regulated in plants. R2R3-MYB and bHLH transcription factors and WD40 proteins represent the three major families of anthocyanin regulatory proteins (Paz-Ares et al. 1987; Chandler et al. 1989; Ludwig and Wessler 1990; de Vetten et al. 1997; Quattrocchio et al. 1999). They form regulatory complexes that activate expression of anthocyanin structural genes (Goff et al. 1992; Grotewold et al. 2000). In Arabidopsis thaliana, bHLH transcription factors playing important roles in anthocyanin biosynthesis regulation include TT8 (TRANSPARENT TESTA 8), GL3 (GLABROUS 3), and EGL3 (ENHANCER OF GLABRA 3) (Nesi et al. 2000; Zhang et al. 2003). Of these, EGL3 has a major role in the regulation of anthocyanin biosynthesis in A. thaliana, with activation of DFR and ANS/LDOX predominantly due to EGL3 (Gonzalez et al. 2008). Alteration of the bHLH transcription factor In1 causes an increase in red pigment production in maize aleurone (Burr et al. 1996). Knockout of bHLH Rc function through frame-shifting changes the color of seed pericarps from red to white in rice (Sweeney et al. 2006). An insertion of a DNA transposon into the bHLH regulatory gene bHLH2 alters the color of common morning glory (Ipomoea tricolor) flowers to pale pigmented flowers (Park et al. 2007). Two syntenic orthologs of AtEGL3, BrEGL3.1 and BrEGL3.2, were located within the QTL region identified in this study. The re-sequencing data revealed that there were eight non-synonymous substitutions of BrEGL3.1 in the purple parent ZCT095. The protein sequence of BrEGL3.1 from the male line L58DH was the same as that from Chiifu-401-42, the Chinese cabbage inbred line used for whole-genome sequencing. None of the non-synonymous substitutions of BrEGL3.1 and BrEGL3.2 was in the bHLH domain, while three amino acid substitutions with variant chemicophysical properties of BrEGL3.1 in the domain of interaction with WD40 proteins may influence the interaction between BrEGL3.1 and WD40 as well as the stability of the ternary transcriptional complex for activation of anthocyanin biosynthetic structure genes. These results indicate that BrEGL3.1 may be responsible for enhanced anthocyanin biosynthesis in ZCT095. As there is another syntenic ortholog within the QTL interval, it will be interesting to know whether the two copies are functionally diversified. Studies combining gene expression profiling with anthocyanin metabolic profiling of accessions with different colors, and fine mapping based on a larger F2 population, will be conducted to clarify the genetic mechanisms underlying anthocyanin accumulation in zicaitai.
References
Andersen OM, Markham KR (2006) Flavonoids: chemistry, biochemistry and applications. CRC Press, Taylor & Francis, Boca Raton. 397-398
Bassam BJ, Caetano-Anolles G, Gresshoff PM (1991) Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 196(1):80–83
Broun P (2005) Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr Opin Plant Biol 8(3):272–279
Burdzinski C, Wendell DL (2007) Mapping the anthocyaninless (anl) locus in rapid-cycling Brassica rapa (RBr) to linkage group R9. BMC Genet 8(1):64
Burr FA, Burr B, Scheffler BE, Blewitt M, Wienand U, Matz EC (1996) The maize repressor-like gene intensifier1 shares homology with the r1/b1 multigene family of transcription factors and exhibits missplicing. Plant Cell 8(8):1249–1259
Chandler VL, Radicella JP, Robbins TP, Chen J, Turks D (1989) Two regulatory genes of the maize anthocyanin pathway are homologous: isolation of B utilizing R genomic sequences. Plant Cell 1(12):1175–1183
Chiu LW, Zhou X, Burke S, Wu X, Prior RL, Li L (2010) The purple cauliflower arises from activation of a MYB transcription factor. Plant Physiol 154(3):1470–1480
de Vetten N, Quattrocchio F, Mol J, Koes R (1997) The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev 11(11):1422–1434
Ferreres F, Valentão P, Pereira JA, Bento A, Noites A, Seabra RM, Andrade PB (2008) HPLC-DAD-MS/MS-ESI screening of phenolic compounds in Pieris brassicae L. reared on Brassica rapa var. rapa L. J Agric Food Chem 56(3):844–853
Goff SA, Cone KC, Chandler VL (1992) Functional analysis of the transcriptional activator encoded by the maize B gene: evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev 6(5):864–875
Gonzalez A, Zhao M, Leavitt JM, Lloyd AM (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53(5):814–827
Grotewold E (2006) The genetics and biochemistry of floral pigments. Annu Rev Plant Biol 57:761–780
Grotewold E, Sainz MB, Tagliani L, Hernandez JM, Bowen B, Chandler VL (2000) Identification of the residues in the Myb domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc Natl Acad Sci USA 97(25):13579–13584
Guo N, Cheng F, Wu J, Liu B, Zheng S, Liang J, Wang X (2014) Anthocyanin biosynthetic genes in Brassica rapa. BMC Genomics 15:426
Harborne JJB, Baxter H, Moss GP (1999) Phytochemical dictionary: a handbook of bioactive compounds from plants, 2nd edn. CRC Press, Boca Raton, pp 361–363
Hayashi K, Matsumoto S, Tsukazaki H, Kondo T (2010) Mapping of a novel locus regulating anthocyanin pigmentation in Brassica rapa. Breed Sci 60:76–80
Holton TA, Cornish EC (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7(7):1071
Kim C, Park S, Kikuchi S, Kwon S, Park S, Yoon U, Park D, Seol Y, Hahn J, Park S (2010) Genetic analysis of gene expression for pigmentation in Chinese cabbage (Brassica rapa). BioChip J 4(2):123–128
Kim C, Kim J, Kikuchi S, Choi J, Kim Y, Park H, Seol Y, Park D, Hahn J, Kim Y (2011) Computational identification of Chinese cabbage anthocyanin-specific genes. Biochip J 5(2):184–192
Koes R, Verweij W, Quattrocchio F (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10(5):236–242
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12(1):172–175
Lam TK, Gallicchio L, Lindsley K, Shiels M, Hammond E, Tao XG, Chen L, Robinson KA, Caulfield LE, Herman JG (2009) Cruciferous vegetable consumption and lung cancer risk: a systematic review. Cancer Epidemiol Biomark Prev 18(1):184–195
Lee J, Durst RW, Wrolstad RE (2005) Determination of total monomeric anthocyanin pigment content of fruit juices, beverages, natural colorants, and wines by the pH differential method: collaborative study. J AOAC Int 88(5):1269–1278
Lin LZ, Harnly JM (2007) A screening method for the identification of glycosylated flavonoids and other phenolic compounds using a standard analytical approach for all plant materials. J Agric Food Chem 55(4):1084–1096
Lin LZ, Sun J, Chen P, Harnly J (2011) UHPLC-PDA-ESI/HRMS/MSn analysis of anthocyanins, flavonol glycosides, and hydroxycinnamic acid derivatives in red mustard greens (Brassica juncea Coss Variety). J Agric Food Chem 59(22):12059–12072
Liu B, Wang Y, Zhai W, Deng J, Wang H, Cui Y, Cheng F, Wang X, Wu J (2013a) Development of InDel markers for Brassica rapa based on whole-genome re-sequencing. Theor Appl Genet 126(1):231–239
Liu J, Wang W, Zhang D, Yu S, Zhang F, Zhao X, Yu J, Lu G (2013b) Primary mapping of pur, a gene controlling purple leaf color in Brassica rapa. Acta Agric Boreal Sin 28(1):49–53 (in Chinese)
Lo Scalzo R, Genna A, Branca F, Chedin M, Chassaigne H (2008) Anthocyanin composition of cauliflower (Brassica oleracea L. var. botrytis) and cabbage (B. oleracea L. var. capitata) and its stability in relation to thermal treatments. Food Chem 107(1):136–144
Longo L, Vasapollo G (2006) Extraction and identification of anthocyanins from Smilax aspera L. berries. Food Chem 94(2):226–231
Ludwig SR, Wessler SR (1990) Maize R gene family: tissue-specific helix-loop-helix proteins. Cell 62(5):849–851
Mazza G, Miniati E (1993) Anthocyanins in fruits, vegetables, and grains. CRC Press, Boca Raton
Mobin M, Khan NA (2007) Photosynthetic activity, pigment composition and antioxidative response of two mustard (Brassica juncea) cultivars differing in photosynthetic capacity subjected to cadmium stress. J Plant Physiol 164(5):601–610
Moreno DA, Pérez-Balibrea S, Ferreres F, Gil-Izquierdo Á, García-Viguera C (2010) Acylated anthocyanins in broccoli sprouts. Food Chem 123(2):358–363
Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L (2000) The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12(10):1863–1878
Park KI, Ishikawa N, Morita Y, Choi JD, Hoshino A, Iida S (2007) A bHLH regulatory gene in the common morning glory, Ipomoea purpurea, controls anthocyanin biosynthesis in flowers, proanthocyanidin and phytomelanin pigmentation in seeds, and seed trichome formation. Plant J 49(4):641–654
Paz-Ares J, Ghosal D, Wienand U, Peterson P, Saedler H (1987) The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. The EMBO J 6(12):3553
Podsędek A (2007) Natural antioxidants and antioxidant capacity of Brassica vegetables: a review. LWT-Food Sci Technol 40(1):1–11
Prior RL, Wu XL (2006) Anthocyanins: structural characteristics that result in unique metabolic patterns and biological activities. Free Radic Res 40(10):1014–1028
Quattrocchio F, Wing J, van der Woude K, Souer E, de Vetten N, Mol J, Koes R (1999) Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell 11(8):1433–1444
Scalzo RL, Genna A, Branca F, Chedin M, Chassaigne H (2008) Anthocyanin composition of cauliflower (Brassica oleracea L. var. botrytis) and cabbage (B. oleracea L. var. capitata) and its stability in relation to thermal treatments. Food Chem 107(1):136–144
Sweeney MT, Thomson MJ, Pfeil BE, McCouch S (2006) Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 18(2):283–294
Tanaka Y, Sasaki N, Ohmiya A (2008) Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant Journal 54(4):733–749
Tatsuzawa F, Saito N, Shinoda K, Shigihara A, Honda T (2006) Acylated cyanidin 3-sambubioside-5-glucosides in three garden plants of the Cruciferae. Phytochemistry 67(12):1287–1295
Van Ooijen JW (2004) MapQTL5, software for mapping of quantitative trait loci in experimental populations. Kyazma BV, Wageningen
Van Ooijen JW (2006) JoinMap4, software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen
van Poppel G, Verhoeven DT, Verhagen H, Goldbohm RA (1999) Brassica vegetables and cancer prevention. In: Zappia V, Della Ragione F, Barbarisi A, Russo GL, Iacovo RD (eds) Advances in nutrition and cancer 2. Springer, pp 159–168
Wang X, Lou P, Bonnema G, Yang B, He H, Zhang Y, Fang Z (2005) Linkage mapping of a dominant male sterility gene Ms-cd1 in Brassica oleracea. Genome 48(5):848–854
Wang Y, Sun S, Liu B, Wang H, Deng J, Liao Y, Wang Q, Cheng F, Wang X, Wu J (2011) A sequence-based genetic linkage map as a reference for Brassica rapa pseudochromosome assembly. BMC Genomics 12(1):239
Wu X, Prior RL (2005) Systematic identification and characterization of anthocyanins by HPLC-ESI-MS/MS in common foods in the United States: fruits and berries. J Agric Food Chem 53(7):2589–2599
Yamasaki H, Sakihama Y, Ikehara N (1997) Flavonoid-peroxidase reaction as a detoxification mechanism of plant cells against H2O2. Plant Physiol 115(4):1405–1412
Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130(20):4859–4869
Acknowledgments
The work was funded by the National High Technology R&D Program of China (2012AA100101); the National Program on Key Basic Research Projects of China (The 973 Program: 2012CB113900, 2013CB127000, and 2013CB127006); the International Joint Research Grant of Ministry of Science and Technology, P.R. China (2011DFR31180); and the National Natural Science Foundation of China (NSFC Grants: 31301771, 31201628, 31201636 and 31301784); the Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, P.R. China and the Sino-Dutch Joint Lab of Horticultural Genomics Technology as well as the National Engineering Research Center for Vegetables in Beijing.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11032_2015_237_MOESM2_ESM.jpg
Supplementary Fig. S1 Amino acid sequence alignment and comparison of BrEG3.1 and BrEGL3.2 showing amino acid substitutions of the parental lines. Identical residues are highlighted on a black background, and similar residues are highlighted on a gray background (JPEG 329 kb)
Rights and permissions
About this article
Cite this article
Guo, N., Wu, J., Zheng, S. et al. Anthocyanin profile characterization and quantitative trait locus mapping in zicaitai (Brassica rapa L. ssp. chinensis var. purpurea). Mol Breeding 35, 113 (2015). https://doi.org/10.1007/s11032-015-0237-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-015-0237-1