Abstract
Diabetes mellitus is one of the top ten causes of death worldwide, accounting for 6.7 million deaths in 2021, and is one of the most rapidly growing global health emergencies of this century. Although several classes of therapeutic drugs have been invented and applied in clinical practice, diabetes continues to pose a serious and growing threat to public health and places a tremendous burden on those affected and their families. The strategy of reducing carbohydrate digestibility by inhibiting the activities of α-glucosidase and α-amylase is regarded as a promising preventative treatment for type 2 diabetes. In this study, we investigated the dual inhibitory effect against two polysaccharide hydrolytic enzymes of flavonoid derivatives from an in-house chemical database. By combining molecular docking and structure–activity relationship analysis, twelve compounds with docking energies less than or equal to − 8.0 kcal mol−1 and containing required structural features for dual inhibition of the two enzymes were identified and subjected to chemical synthesis and in vitro evaluation. The obtained results showed that five compounds exhibited dual inhibitory effects on the target enzymes with better IC50 values than the approved positive control acarbose. Molecular dynamics simulations were performed to elucidate the binding of these flavonoids to the enzymes. The predicted pharmacokinetic and toxicological properties suggest that these compounds are viable for further development as type 2 diabetes drugs.
Graphical abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Diabetes mellitus (DM) is a metabolic disease, characterized by a chronically elevated level of blood sugar and caused by an impairment of insulin excretion and/or a decrease in insulin efficiency [1]. According to the report by International Diabetes Federation (IDF) in 2021, about 537 million adults are living with DM and is estimated to rise to 783 million people by the year 2045, making it one of the most prevalent chronic diseases [2]. People with uncontrolled DM are prone to serious implications, including cardiovascular, kidney, eye, and infectious diseases [3]. Type II diabetes (T2DM) is the most common type of DM, accounting for over 90% of all diagnosed DM cases in the world [2, 4]. T2DM is associated with the deficiency of insulin (insulin resistance) and therefore, multiple anti-diabetic agents aiming to increase the level of insulin, enhance the sensitivity of insulin on organs receptors, or decrease the level of blood sugar have been established [5], which could be categorized into subfamilies such as biguanides (metformin), sulfonylureas, meglitinides, thiazolidinediones, glucagonlike-peptide-1 receptor agonist (GLP1-RA), dipeptidyl peptidase IV inhibitors (DPP-4i), sodium-glucose transporter-2 inhibitors (SGLT-2i), and α-glucosidase inhibitors [1, 5].
A promising approach for the management of T2DM is by inhibiting α-glucosidase and α-amylase, the two key enzymes which are responsible for the hydrolyzation and digestion of polysaccharide chains before being absorbed into the bloodstream, thereby reducing the level of blood sugar [6, 7]. Currently available α-glucosidase inhibitors include acarbose, miglitol, and voglibose, of which the first drug has been launched two decades ago [6]. Different from voglibose and miglitol, acarbose also demonstrates a strong inhibition against α-amylase, which could explain its strong efficacy in maintaining postprandial blood glucose levels [8]. Moreover, α-glucosidase inhibitors have also been proven to increase the sensitivity of insulin, thereby releasing the stress on the islet β-cells and slowing down the progression of T2DM [9]. Although having been proven to be well tolerated because of their localized action in the intestine, gastrointestinal side effects such as flatulence, abdominal pain, and diarrhea are the main drawbacks that make this class unpopular in recent years [9]. This raises the need for the exploration and development of new α-glucosidase and α-amylase dual-target inhibitors with minimal adverse drug reactions and better efficacy.
α-Amylase (E.C.3.2.1.1) is a digestive enzyme that cleaves starch at α-1,4-linked glucose molecules to produce a range of shorter polysaccharide chains [10]. In humans, α-amylase exists in two isoforms, which could be found in saliva and pancreatic fluid [11]. Both isoforms are composed of 496 amino acids and could be divided into three different domains (A, B, and C), with the catalytic triad (Asp197, Glu233, and Asp300) responsible for the cleaving function of the enzymes located in the A domain [12]. On the other hand, α-glucosidase is a group of small-intestinal brush border-bound hydrolyzing enzymes that cleave disaccharides and oligosaccharides into monosaccharides such as glucose [11]. This group consists of two complexes that work concurrently in the digesting pathway of starch, namely Maltase-glucoamylase (MGAM) and Sucrase-isomaltase (SI) [13], with MGAM (maltase—E.C.3.2.1.20) is the most well-studied enzyme [14]. In humans, the MGAM-containing chain is composed of duplicated catalytic domains, N-terminal MGAM (NtMGAM; residues 1-868) and C-terminal MGAM (CtMGAM; residues 955-1867) [15]. NtMGAM domain has a shallow substrate-binding pocket which allows only 2 sugar subsites whereas the addition of 21 residues in CtMGAM allows this isozyme to cleave a longer chain [14]. In this study, we focused on the NtMGAM, of which, a range of acidic and basic residues (Asp203, Asp327, Asp443, Arg526, Asp571, Asp542, His600) are responsible for the activity of the enzymes [14].
Flavonoid is a ubiquitous phytochemical class that comprises a C6-C3-C6 skeleton and has been widely studied for its broad bioactivity in recent years [16,17,18]. Moreover, multiple flavonoid structures have been shown to have anti-diabetic properties, via different mechanisms [11, 19, 20]. Therefore, this study aims to assess the dual inhibitory activity of our in-house flavonoid compounds against α-glucosidase and α-amylase, using both molecular modeling and experimental approaches. The entire workflow of the screening process for dual inhibitors against α-amylase and α-glucosidase has been graphically depicted in Fig. 1. Both structure-based and ligand-based drug design approaches were applied in this work. Using high-quality crystal structures of the two enzymes co-crystallizing with known inhibitors, molecular docking models were constructed for the virtual screening process. The enzyme inhibitors published in the literature allow the establishment of the structure–activity relationships (SARs) to facilitate a more efficient selection of hit compounds for inclusion in chemical synthesis and in vitro experiments. Finally, the flavonoid derivatives identified as dual enzyme inhibitors were further investigated using molecular dynamics (MD) simulations and absorption, distribution, metabolism, excretion, and toxicity (ADMET) evaluation.
Materials and methods
Molecular docking
The crystal structures of α-glucosidase and α-amylase obtained from the Protein Data Bank (PDB) [21] were used to build molecular docking models. Currently, several three-dimensional (3D) structures of α-glucosidase from various species are available in the PDB. In this current work, the human intestinal α-glucosidase structure co-crystallized with acarbose imaged by X-ray diffraction method with a resolution of 1.90 Å was used (PDB code: 2QMJ) [14]. Acarbose is one of the well-known α-glucosidase inhibitors that has been used for the treatment of T2DM [22]. For α-amylase, the structure of human pancreatic enzyme co-crystallized with myricetin inhibitor captured by X-ray diffraction with 1.20 Å resolution was used (PDB ID: 4GQR) [23]. Myricetin is a member of the flavonoid class of polyphenolic compounds and has been reported to inhibit α-amylase with an IC50 of 30.2 μM [23].
After downloading from the PDB, the enzyme structures in *.pdb format were processed using the AutoDock Tools 1.5.6rcl program. First, water molecules and unnecessary structural components were removed, after which polar hydrogen atoms were added and Kollman charges were assigned to the protein. Finally, the structure of each enzyme was converted into *.pdbqt format. Molecular docking models were defined by the Grid box function of the AutoDock Tools software. For α-glucosidase, the size and coordinates of the grid box were determined based on the coordinate of the acarbose and residues in the enzyme catalytic cavity (Asp327, Asp542, His600, Arg526, Asp443, and Asp203). Likewise, the grid box for α-amylase was established based on the placement of the myricetin and residues in the binding site (Asp197, Glu233, Asp300, Trp59, and Tyr62). The receiver operating characteristic (ROC) curve [24] and the re-docking approach were used to validate the molecular docking models for each enzyme. A docking model is considered reliable if the root-mean-square deviation (RMSD) value between the co-crystallized ligand and its re-docked conformation is less than 2 Å [25]. In the ROC curve assessment method, active and inactive compounds were docked to the enzymes. Active flavonoid compounds were collected from previous studies (detailed in the SAR analysis section). Inactive decoys were generated from active molecules using the DUD-E tool (http://dude.docking.org) [26]. The discriminant ability of molecular docking models was reflected by the ROC curve plotted using the Screening Explorer tool (http://stats.drugdesign.fr) [27]. The capability of discrimination between active and inactive compounds of the docking models was assessed using Area Under the ROC curve (AUC-ROC), in which the AUC-ROC value equals 0.5 meaning random discrimination. The higher the AUC-ROC value, the better model’s performance [28].
Database of flavonoid derivatives for virtual screening
In the present work, we used an in-house library containing 378 flavonoid derivatives to investigate the dual inhibition of α-glucosidase and α-amylase. The structure in SMILES form and the molecular descriptors according to Lipinski’s Rule of Five (Ro5) of the compounds in this chemical library are presented in Table S1. All 378 flavonoid compounds used for virtual screening in this study were designed to satisfy the drug-like properties without more than one violation of Lipinski’s Ro5.
The 2D structures of flavonoid derivatives were drawn using ChemDraw 19 software. After that, they were converted into 3D molecules and energy minimized using the Molecular Operating Environment (MOE) software version 2022.10 (https://www.chemcomp.com) [29] with a Gradient RMS of 0.0001 kcal mol Å2. During energy minimization, partial charges were calculated and orient − OH groups were optimized. The ligands were then saved in *.pdb format and converted to *.pdbqt format using the Open Babel software version 2.4.1 (https://openbabel.org) [30]. The ligands were then docked into α-glucosidase and α-amylase, respectively, using the AutoDock Vina 1.2.0 software [31] with the previously defined grid box. During the docking process, the exhaustiveness value was set to 8. The investigated compounds were ranked based on their Autodock Vina binding energy (ΔGdock, kcal mol−1) and their interactions with the important residues of α-glucosidase and α-amylase using the Protein–Ligand Interaction Profiler (PLIP) tools (https://plip-tool.biotec.tu-dresden.de) [32].
Structure–activity relationship analysis of known inhibitors of α-glucosidase and α-amylase
Flavonoid derivatives with α-glucosidase and α-amylase inhibitory activities were collected from the literature and used for structure–activity relationship (SAR) analysis. These known inhibitors were also docked into the two enzymes to validate the docking protocols as previously described. Simultaneously, the docking results of these compounds were combined with the SAR analysis to reveal the structural features required for a flavonoid possessing dual inhibitory activity against the enzymes.
In most of the previous reports, the positive control for α-glucosidase and α-amylase inhibition assays was acarbose. However, the IC50 values of acarbose and other inhibitors in different publications are often inconsistent. For the convenience of SAR analysis, the IC50 values of α-amylase and α-glucosidase inhibitor flavonoids were normalized to a standard IC50 value of acarbose. Particularly, we have collected 51 flavonoid inhibitors of α-glucosidase from the following original articles [33,34,35,36,37,38], and [39]. The α-glucosidase IC50 values of the compounds were standardized based on the IC50 value of acarbose at 607 μM according to the study of Proença Carina et al. [34]. Similarly, 69 flavonoids inhibiting α-amylase were collected from publications [40,41,42,43,44,45,46], and [17], with the IC50 value of acarbose used for standardization being 18.08 μM according to the research of Saleem Faiza et al. [41]. The lists of known α-glucosidase and α-amylase inhibitors are presented in Table S2 and S3, respectively, along with their coding names and standardized IC50 values. SAR analysis combined with virtual screening by molecular docking will guide the rational selection of flavonoid derivatives for inclusion in the next phase of biological testing.
Chemicals and instruments
All commercial reagents and solvents were used as received without further purification. Benzaldehyde, 2-chlorobenzaldehyde, 3-chlorobenzaldehyde, 4-chlorobenzaldehyde, 3,4-dichlorobenzaldehyde, 2-chloro-6-fluorobenzaldehyde, 3-pyridinecarboxaldehyde, 4-pyridinecarboxaldehyde, and 2,3-dimethoxybenzaldehyde were purchased from Acros Organics-Thermo Fisher Scientific (Geel-Belgium). The chemicals including 1-(4-((2-hydroxybenzyl)amino)phenyl)ethanone and 1-(4-((4-hydroxybenzyl)amino)phenyl)ethanone were obtained from Medicinal Chemistry’s Laboratory, Faculty of Pharmacy, University of Medicine and Pharmacy at Ho Chi Minh city, Vietnam. α-Glucosidase from Saccharomyces cerevisiae, p-nitrophenyl-α-D-glucopyranose (p-NPG), soluble starch (ACS reagent grade), iodine, and potassium iodide were obtained from Sigma-Aldrich (United States). α-Amylase from malt was purchased from Himedia (India). Acarbose was procured from Abcam (UK). All the other chemicals were of analytical quality.
The procedures of synthesis were monitored using TLC (Merck, Germany). Nuclear Magnetic Resonance (NMR) spectra were recorded on Bruker (400/500 MHz, Germany) spectrometers. Infrared (IR) spectra were recorded on an IRAffinity-1S Model spectrometer (Shimadzu, Japan) or FTIR-Equinox 55 de Bruker (Germany). UV spectra were recorded on Spectrophotometer UVD-2970 (Labomed, US). Mass spectra were recorded on the MSQ Plus DAD Mass Spectrometer System (Vilber Lourmat, France).
Synthesis
The synthesis of chalcone derivatives was based on the SAR analysis and molecular docking results, which highlighted specific features for the simultaneous inhibition of α-glucosidase and α-amylase. Various chalcone synthesis pathways were available, allowing us to easily modify the scaffold. In terms of A ring substitution, it was observed that the attachment of polar substituents, particularly the amino functional group, enhanced the inhibitory activity of the chalcones. Therefore, we selected the N-benzylamino substituent, which could be readily attached to the A ring of the chalcones using substituted acetophenone. In terms of B ring modification, the attachment of functional groups such as hydroxyl, methoxyl, or halogen atoms (Cl or F) was found to further enhance the inhibitory potential. Considering the chemicals available in our laboratory, a series of N-benzylaminochalcone derivatives were synthesized for subsequent biological evaluation.
Chalcone derivatives were synthesized via Claisen–Schmidt condensation [47] (Fig. 2). Substituted acetophenone (5 mmol) and aryl-carboxaldehyde derivatives (5 mmol) were dissolved in methanol or ethanol (10 mL) with stirring. Potassium hydroxide (15 mmol) was added slowly in portions to give a dark yellow solution. The resulting solution was stirred at room temperature for 8–12 h, during which chalcone precipitated as the potassium salt. The solution or suspension was poured into cold 0.5 N HCl (10 mL), and further concentrated HCl was added until the solution was acidic. The resulting yellow solid was filtered, washed with water (2 × 20 mL), and recrystallized from a corresponding solvent or purified by flash column chromatography (silica gel, 30% ethyl acetate/hexane) to give the pure product.
α-Glucosidase inhibition assay
The in vitro performance of α-glucosidase inhibitory activity was based on the protocol used by Zhi-Wei Wang et al. [48] and Graciela Granados-Guzmán et al. [49]. Acarbose was used as the positive control. Accordingly, p-NPG was hydrolyzed by the α-glucosidase to form liberated p-nitrophenol (p-NP) absorbing 405–415 nm wavelengths. All test samples were dissolved in DMSO at different concentrations before being used in the assays. The sample solution (10 μL) was mixed with 10 μL of the α-glucosidase enzyme (0.5 U mL−1) and 150 μL of 50 mM phosphate buffer pH 6.8. The mixture was pre-incubated at 37 °C for 60 min. Then, 30 μL of 1 mM p–NPG solution was added, and the mixture was incubated at 37 °C for another 30 min. The optical absorbance was measured at 405 nm. Each experiment was done in triplicate. The percentage inhibition was calculated according to the formula (1).
where AbssampE is the absorbance of the sample solution with enzyme, AbssampNE is the absorbance of sample solution without enzyme (replaced by buffer), AbsctrE is the absorbance of the solution containing DMSO replacing the sample and enzyme, and AbsctrNE is the absorbance of the solution containing DMSO replacing the sample without enzyme (replaced by buffer). The IC50 value was obtained from the linear regression equation when plotting the logarithm of concentration with inhibitory activity (I%).
α-Amylase inhibition assay
The α-amylase inhibitory potential was performed in vitro by using the iodine starch method. The method was based on the studies of Rie Kusano et al. [50], Zhizhuang Xiao et al. [51], and Maryam Usman Ahmed et al. [52] with slight modification. Acarbose was used as the positive control. Sample or acarbose solutions were prepared by dissolving them in DMSO in various concentrations. The sample solution (25 μL) was mixed well with 25 μL of 1% soluble starch in distilled water. The mixtures were pre-incubated at 30 °C for 3 min. Then 25 μL of enzyme solution (2 U mL−1 in phosphate buffer pH 6.9) was added to each mixture. Subsequently, they were incubated at 30 °C for 15 min. The reaction was quenched by the addition of 50 μL of 1 M HCl, then 125 μL of 5 mM iodine solution (5 mM KI and 5 mM iodine). The mixtures were diluted four times and were measured the absorbance at 650 nm. The percentage inhibition was also calculated using the formula (1). The IC50 value was derived using the linear regression equation when the logarithm of concentration was plotted against the inhibitory activity (I%).
Molecular dynamics simulations
To further assess the binding stability of the small molecular inhibitors in the binding sites under a physiological-like environment, molecular dynamics simulations (MDS) were carried out using the GROMACS 2021 program [53]. Briefly, the protein backbone was treated using the built-in CHARMM-27 forcefield [54] and the small-molecule ligands were parameterized using the SwissParam server (https://www.swissparam.ch) [55]. The complexes were then put into virtual dodecahedron boxes, in which approximately 29,300 and 22,000 TIP3P solvent molecules were placed for α-glucosidase and α-amylase complexes, respectively. The systems were then neutralized by adding Na+ ions and underwent energy minimization using the steepest descent minimization algorithm with a maximum of 50,000 steps to avoid any geometrical incompatibilities. This is followed by two consecutive 100 ps equilibrium stages namely NVT and NPT equilibrium to stabilize the systems at physiological conditions of 1 bar pressure and 300 K temperature [56]. After achieving equilibrium, the MDS for each complex was produced in 100 ns with a timestep of 2 fs. The MD trajectories were recorded every 0.01 ns and visualized using the VMD [57] and PyMOL program [58]. The built-in commands of GROMACS 2021 were used to calculate the RMSD (root-mean-square deviation), RMSF (root-mean-square fluctuation), Rgyr (radius of gyration), and SASA (solvent-accessible surface area) values of the protein backbone or the heavy atoms of the ligands during the MDS. In this study, the hydrogen bonds were determined to occur if the bonding angle between the hydrogen donor (D) and acceptor (A) D-H⋯A larger than 120° with the distance between D and A not exceeding 3.5 Å [59].
ADMET evaluation
ADMET properties of the most potent dual inhibitors on the two enzymes were evaluated using the ADMETlab 2.0 platform (https://admetmesh.scbdd.com) [60] to assess their suitability for T2DM drug development. After the SMILES structures are submitted, the online tool will predict their characteristics including physicochemical properties, medicinal chemistry, pharmacokinetics, and toxicity.
Results
Molecular docking models for α-glucosidase and α-amylase
From the structure of α-glucosidase co-crystallized with acarbose collected from the Protein Data Bank (PDB code: 2QMJ), an AutoDock grid box was determined from the location of the ligand as shown in Fig. 3a. The grid box had parameters including spacing = 1.0 Å; size_x, size_y, and size_z were 24, 24, and 26, respectively; center_x, center_y, and center_z were − 21.957, − 3.267, and − 7.522, respectively. The model was validated by both re-docking and ROC curves, the results of which are presented in Fig. 3b and c. Using the previously described grid box, acarbose was successfully re-docked into the active site of α-glucosidase and achieved the best docking energy of − 8.1 kcal mol−1 with an RMSD value between the heavy atoms of acarbose in the co-crystallization and re-docked conformation was 1.778 Å (Fig. 3b). Moreover, the docking model is also able to reproduce the main hydrophilic and hydrophobic interactions of acarbose in the binding site of α-glucosidase (Fig. 3d and e). Specifically, the OH groups of acarbose form many hydrogen bonds with the residues such as Thr205, Asn207, Arg526, Trp539, Asp542, and His600. In addition, the hydrophobic interactions between this drug with Tyr299, Trp406, and Phe575 were also re-established. The docking protocol using the above grid box parameter was performed and obtained an AUC-ROC value of 0.690 (> 0.5) and a \(\overline{\mathrm{TG} }\) value of 0.255 (> 0.25), indicating that the docking model could discriminate well between active and inactive compounds (Fig. 3c).
Molecular docking model for α-glucosidase based on the AutoDock package. a The 3D structure of α-glucosidase in the presence of a grid box was determined based on the position of the co-crystallized ligand acarbose. b Acarbose in co-crystallized conformation (with carbon atoms in pink) and re-docked conformation (with yellow carbons). c ROC curve obtained from docking process of active and decoy compounds into α-glucosidase. d Interactions of acarbose in co-crystallized conformation and e in re-docked conformation with the residues in the active site of the enzyme
Similarly, a suitable grid box for α-amylase was also identified based on the location of the co-crystallized ligand myricetin. As shown in Fig. 4a, this grid box has the following parameters: spacing = 1.0 Å; size_x, size y, and size_z were 22, 20, and 22, respectively; center_x, center_y, and center_z were 12.166, 19.236, and 42.288, respectively. Re-docking of myricetin was given the conformation illustrated in Fig. 4b with an RMSD value compared to the co-crystallization conformation of 1.6797 Å. A ROC curve was plotted from the docking results of the active and decoy datasets to the enzyme (Fig. 4c). The ΔGdock between myricetin and α-amylase was − 7.4 kcal mol−1. Myricetin in its re-docked conformation has re-established hydrogen bonds with the residues His101 and Asp197, and hydrophobic interactions with Tyr62 (Fig. 4d and e). The AUC-ROC and \(\overline{\mathrm{TG} }\) values obtained were 0.77 (> 0.5) and 0.39 (> 0.25), respectively. These validation results suggest that the docking models could be used for the next phase of virtual screening to discover novel inhibitors for α-glucosidase and α-amylase.
Molecular docking model for α-amylase constructed using the AutoDock Tools software. a The 3D structure of the enzyme in the presence of a grid box was identified based on the position of the co-crystallized ligand myricetin. b Myricetin in co-crystallized conformation (with carbon atoms in pink) and re-docked conformation (with carbon atoms in yellow). c ROC curve obtained from the docking of the true active compounds and corresponding decoy dataset into α-amylase. d Interactions of myricetin in co-crystallized conformation and e in re-docked conformation with the residues in the active site of α-amylase
SAR analysis of known inhibitors of α-glucosidase and α-amylase
SAR of flavonoids inhibiting α-glucosidase
In this SAR analysis, we consider the standardized IC50 values and Autodock Vina docking energies of 51 known α-glucosidase inhibitors. The detailed data are demonstrated in Table S2. The compounds in this list may have a chalcone, flavone, or flavanone scaffold. The enzyme inhibitory activity of flavonoids depends on their substituents as well as the bulkiness of the molecule. Because of the shallow and narrow active site of α-glucosidase, most flavonoids bind to the gatekeeper residues of the cavity rather than to the catalytic residues located inside the active site (Fig. 5a). By analyzing Protein–Ligand Interaction Fingerprints (PLIF) using MOE software, we found that most of the investigated flavonoids bind to α-glucosidase by hydrogen bonding with Asp542 and Asp327 by possessing OH groups. In addition, due to the presence of aromatic rings in the flavonoid scaffold, they could also form arene interactions with the aromatic amino acids such as Tyr299 and Trp406 (Fig. 5b). These are four of the key residues in the catalytic cavity of the enzyme. Notably, several inhibitors have given good IC50 values and strong docking energies due to the deep penetration into the active site. They bound well to the enzyme and formed hydrogen bonds with the most important residues, such as Asp203, Asp443, Asp542, and Arg526 in the catalytic cavity.
The chalcone derivatives registered the IC50 values of 9.8 to 409 μM and ΔGdock of − 8.3 to − 7.4 kcal mol−1. To increase the binding capacity to the active site of α-glucosidase, most studies have focused on developing substituents in the A ring (Fig. 5c). Particularly, the N-substituents toluene sulfonamides result in strong inhibition capacity, especially when the substituent is at the R2 position (with IC50 < 10 μM), while the introduction of − NH2, − OH, − Cl, − F, − NO2, and − Br substituents results in moderate inhibitors. On the B ring, the − OH substituent in the para and/or meta positions exhibits good inhibitory activity as it could form extra hydrogen bonds with important nucleophilic catalysis residues such as Asp327, Asp443, and Asp542.
On the other hand, the flavone and flavanone derivatives recorded the ΔGdock value varying from − 9.4 to −7.2 kcal mol−1. Similar to chalcones, flavanones and flavones have also been focused on expanding the structure in the A ring (Fig. 5d). Adding a sulfonamide group or a chalcone fragment on the A ring gives very strong α-glucosidase inhibitors (IC50 < 2 μM) and good docking energies, while adding a sugar fragment or a 7–hydroxy–2H-chromene–2–one moiety on the A ring at different positions also establishes good inhibitors compared with acarbose. However, when attaching bulky alkoxy substituents to either the A or B ring, the enzyme inhibitory activity decreased markedly (IC50 > 2500 μM) and the docking energies were also poor. On the B ring, most compounds demonstrate that the − OH substituent leads to stronger inhibitors than other substituents.
SAR of flavonoids inhibiting α-amylase
A list of 69 published α-amylase inhibitors with their IC50 values and Autodock Vina docking energies are presented in Table S3. Most known α-amylase inhibitors are chalcones. Contrary to the shallow active site of α-glucosidase, the active site of α-amylase forms a “V” shape, therefore the ligands could penetrate deeply and fit into the cavity (Fig. 6a). As a result, they could form hydrogen bonds with the three important residues in the catalytic site, including Asp197 and Glu233, hence enhancing the inhibitory activity. Simultaneously, hydrophobic interactions (π–π) were also formed between the aromatic nucleus of the investigated ligand and the hydrophobic residues Tyr59 and Tyr62 (Fig. 6b). The aromatic B ring plays an important role in the inhibition of α-amylase activity. However, the activity is strong or weak depending on the different substituents attached to this aromatic ring.
Regarding the chalcone structures, the recorded IC50 values ranged from 21 to 40 μM while the docking energy varied between − 8.5 and − 7.1 kcal mol−1, depending on the variation of substituents attached to both aromatic rings. On the B ring and at the para position (R′3), the enzyme inhibitory activity decreased gradually according to the substituents in the following order: –SCH3 > –Cl > –OCH3 > –Br > –C6H5 (Fig. 6c). The − SCH3 and − OCH3 groups play an important role in α-amylase inhibition as the sulfur or oxygen atom helped direct the binding conformation and formed multiple interactions with residues in the active site. Meanwhile, the phenyl or benzyloxy substituents on this position resulted in a bulkier structure and, therefore, was predicted to cover a larger surface in the binding site with a better ΔGdock < − 8 kcal mol−1, yet the actual IC50 values of these chalcones only indicate moderate inhibition (i.e., compound BC_2018_12 with IC50 = 34.6 μM and ΔGdock = − 8.5 kcal mol−1; compound BC_2018_5 with IC50 = 34.6 μM and ΔGdock = −8.3 kcal mol−1). In addition, the halogen or alkyl halogen substituents also demonstrated good ΔGdock due to the formation of a hydrophobic interaction (π-alkyl) between the halogen atoms (–Cl, –F, –Br) and the hydrophobic residues Trp59, Trp406, and Phe575 in the active site. Concerning the modification of the R1 site, the phenyl ring results in the basic chalcone structure while the replacement of it with another heterocyclic could weaken the inhibitory potency. Particularly, the 2-pyridyl-containing chalcones demonstrate better activity than the 3-pyridyl ones, as the ΔGdock of these chalcones ranged from − 7.5 to − 6.8 kcal mol−1. Within these heterocyclic-containing chalcones, the introduction of –OCH3, –F, –Cl, –NO2, or –Br substituents on the B ring significantly decreases the inhibitory potency. To be specific, the addition of the –Br substituent at any position on this ring negatively impacts the activity, while the addition of the –NO2 substituent at the meta position (compound BC_2021_16 with IC50 = 27.57 μM, Table S3) demonstrates stronger activity than the para position (compound BC_2021_18 with IC50 = 51.94 μM, Table S3). Especially, the replacement of the phenyl ring with [(4-methylphenyl)sulfonyl]-1H-benzimidazol-2-yl significantly enhances the inhibitory activity against α-amylase (i.e., compound IJC_2016_3I with IC50 = 24.86 and compound IJC_2016_3G with IC50 = 37.29 μM). Typically, their ΔGdock ranged from − 8.4 to − 7.4 kcal mol−1. This is because this substituent helps form extra hydrogen bonds with the three important residues Asp197, Glu233, Asp300 and hydrophobic interactions with Trp59 and Tyr62, revealed by molecular modeling results.
For the flavone scaffold, the introduction of substituents caused the IC50 values to range from 18.25 to 871 μM and the ΔGdock to range from − 8.5 to − 7.0 kcal mol−1. Regarding the variation on ring A, the addition of the –OH groups simultaneously at three positions 1, 2, and 3, or at 1 and 3 enhances the inhibitory activity, as long as the ring B is kept clean. Replacing the –OH group at position 2 with the − OCH3 group could lead to a decrease in enzyme inhibitory activity (from 22 to 871 μM). On the B ring, the introduction of the − OH group at the R′1 position demonstrates the best activity, while that at either R′2 or R′3 decreases the activity sharply by 20 times. The activity of flavone derivatives decreased with the addition of − Cl, − F, − NO2, or − Br substituents on the B ring (Fig. 6d).
Structural features required for flavonoid derivatives to have dual inhibitory effects on the two enzymes
By separate SAR analysis on flavonoid compounds with α-glucosidase or α-amylase inhibitory activity, several common structural properties were identified. First, the majority of reported enzyme inhibitors belong to the chalcone sub-class of flavonoids. On the A ring, aromatic substituents containing additional polar functional groups are found in both strong inhibitors of α-glucosidase and α-amylase. The presence of these substituents enables the inhibitors to form hydrogen bonds and hydrophobic interactions with residues in the catalytic site of the enzymes. On the B ring, − OH, − OCH3, or − SCH3 substitutions have introduced strong inhibitors, but the substitution of the halogen groups (especially the − Cl group) also gave potential inhibitors.
Virtual screening using molecular docking and identification of the top in silico hit compounds
All 378 flavonoid derivatives in our in-house database were successfully docked into the active site of α-glucosidase and α-amylase. The compound ID, coupled with their ∆Gdock-glucosidase and ∆Gdock-amylase, are detailed in Table S4. Regarding the α-glucosidase docking result, our compounds could bind to α-glucosidase with Autodock Vina ΔGdock range from − 9.7 to − 5.6 kcal mol−1 as shown in the diagram in Fig. 7a. Most ligands had docking energy of − 8.0 to − 6.0 kcal mol−1. Of these, 33 investigated compounds have better docking energy than acarbose (ΔGdock-glucosidase = − 8.1 kcal mol−1) as shown in Fig. 7b.
Results of virtual screening using molecular docking to identify dual inhibitory flavonoid derivatives for α-glucosidase and α-amylase. a Histogram and b density plot of Autodock Vina docking energies of 378 flavonoid compounds in the in-house database on α-glucosidase. c Histogram and d density plot docking energies of 378 compounds in the database on α-amylase. e The docking energies of the 36 most potential compounds capable of simultaneously binding to the two enzymes with a threshold of less than − 8.0 kcal mol−1
At the same time, the flavonoid derivatives in our database could also bind to α-amylase with a ΔGdock ranging from − 9.2 to − 6.2 kcal mol−1 as shown in the histogram in Fig. 7c. In particular, we have found 215 flavonoids with better docking energies than both myricetin and acarbose (with respective ΔGdock-amylase being − 7.4 and − 6.7 kcal mol−1) as illustrated in Fig. 7d. Based on the results of virtual screening using molecular docking, 36 compounds were selected for further analysis because they had docking energies on both enzymes less than or equal to − 8.0 kcal mol−1 (Fig. 7e). Their chemical structures, docking energies, and interactions with residues in the active sites of α-glucosidase and α-amylase are detailed in Table S5.
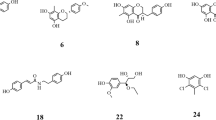
Of these 36 compounds, together with the previously described SAR analysis, results in the accumulation of 12 compounds which concurrently satisfy our proposed SAR and have a docking affinity lower than − 8.0 kcal mol−1. These flavonoid derivatives were submitted to an in vitro biological test to evaluate the inhibitory activity against α-glucosidase and α-amylase. Their structure, along with their ID and their binding pattern with α-glucosidase and α-amylase, is available in Table 1. As can be seen, all the selected structures compose an N-benzylaminochalcone structure. According to the SAR analysis, the introduction of a substituent containing both aromatic rings and polar functional groups (such as 2- or 4-hydroxybenzylamine) to the para position of the A ring could help the flavonoids engage deeper in the binding pocket of α-glucosidase by forming both hydrogen bonds and hydrophobic contacts. Especially for α-amylase binding, chalcones with large but still flexible substituents such as the hydroxybenzylamine fragment on the A ring will fit into the V-shaped active site of the enzyme. Meanwhile, the B ring was kept clear or attached substituents such as − OCH3, − Cl, or − F to enhance the activity against α-amylase. The presence of the − OH and − NH groups in the selected flavonoid structures is expected to form multiple hydrogen bonds with the catalytic residues inside the binding cavity of the two enzymes.
Chemical synthesis
All flavonoid derivatives demonstrated the potential activities against both α-glucosidase and α-amylase by in silico screening are chalcone-structural compounds, including F336, F337, F341, F342, F347, F352, F353, F364, F368, F369, F370, and F378. They were synthesized with high purity via Claisen–Schmidt condensation (over 96%, performed by HPLC). This process afforded the desired chalcones with an average yield of 45–85%. The 1H-NMR spectra of synthesized chalcones displayed two doublets at δ 6.2–8.0 ppm with characteristic coupling constant (J) of 15–16 Hz, which confirms the formation of chalcones (possessing an α,β-unsaturated carbonyl ketone). This higher coupling constant value indicates all synthetic compounds were geometrically pure and were exclusively trans (E) isomers [61]. The information from 1H-NMR, 13C-NMR, HPLC, and MS of all synthesized chalcones was attached to the Supplementary Information.
Biological assays
A preliminary assessment of the inhibitory activity of the screened compounds was conducted using a uniform concentration of 1 mM. Repeating previous studies, acarbose exhibits dual inhibition of α-glucosidase and α-amylase. As can be seen in Fig. 8a, regarding the α-glucosidase inhibition, eight over twelve compounds were able to inhibit the enzyme all over 60% and more strongly than acarbose (56,52%). The inhibitory activity of F336, F347, F364, and F370 could not be determined because the reaction mixtures were cloudy. At lower concentrations, the IC50 values of all 12 compounds against α-glucosidase were determined and detailed in Table 2.
For the α-amylase inhibition assay, the inhibitory activities of the flavonoid compounds and acarbose at 1 mM concentration vary significantly, yet stand far below 50% (Fig. 8b). Therefore, a higher concentration of substances was used in this experiment. The detailed inhibitory activity, regarding the inhibitory percentage and IC50 value of the tested compounds, is shown in Table 2. Based on the ratio of I% of the investigated flavonoids to acarbose, Fig. 8c shows that F337, F352, F353, F368, and F369 have a better dual inhibitory effect on the two enzymes than acarbose.
α-Glucosidase inhibitory activity
In Table 2, for ease of comparison, we present the percentage of α-glucosidase inhibition at 0.5 mM because the twelve compounds were all soluble at this concentration. In this assay, the obtained IC50 value of acarbose was 609.67 ± 13.61 µM, which was comparable to the findings of Nasli Esfahani et al. [62] or Zawawi et al. [63]. Surprisingly, all the 12 tested compounds were able to inhibit α-glucosidase stronger than acarbose. Most compounds have higher I% values than the positive control compound. Additionally, their IC50 values range from 34.28 to at most 554.25 µM, compared to only 609.67 µM for acarbose. These results indicate that the combination of SAR analysis and molecular modeling was able to produce outstanding results.
α-Amylase inhibitory activity
In terms of α-amylase inhibition, the reaction mixtures of the majority of the tested compounds were turbid when performed at concentrations greater than 20 mM. Therefore, there were only five compounds with retrievable IC50 values. However, the results are also very promising as all five compounds (F341, F347, F369, F370, and F378) could inhibit the α-amylase at much lower IC50 values than acarbose, with the most potent compound being F369 (IC50 = 2.55 ± 0.14 mM).
In summary, our computational approaches, together with the SAR analysis, were able to retrieve outstanding candidates for the treatment of T2DM. From the present data, we have identified 5 novel flavonoid derivatives with dual inhibitory effects against α-glucosidase and α-amylase with determined IC50 values, including the compound F337, F347, F369, F370, and F378. These hit compounds inhibit α-glucosidase approximately 2 times more potently than acarbose and α-amylase approximately 2–14 times more potently than acarbose. They were subjected to MDS to simulate the interaction between their small molecular structure and the enzymes thereby comprehending the mechanism of inhibition of these compounds.
Molecular dynamics simulations (MDS)
The semi-flexible docking algorithm of the Autodock Vina algorithm only treats ligands as flexible, while the protein sidechains are kept rigid. Although a fully flexible algorithm has also been developed in this docking program, its flexibility still limits to certain residues surrounding the ligands. Therefore, a series of 100 ns (ns) MDS was conducted to gain insights into the inhibitory mechanism of our investigated compounds.
Stability of protein–ligand complexes
The stability of the complexes was evaluated using the RMSD, SASA, and Rgyr values of the protein backbone during the MDS period. The mean values of these values during the MDS trajectories are shown in Table 3.
For comparison purposes, the binding states of acarbose inside the binding cavities of the two enzymes of interest were also extracted. The initial stage of acarbose inside the binding pocket of α-amylase was created using the previously described docking protocol with the acarbose structure extracted from the α-glucosidase co-crystallized complex used in our study. The initial stage was assessed to be comparable to the crystal structure of α-amylase-acarbose available on the PDB (PDB ID: 1XCX) [12] before being undergone a 100 ns MDS. The binding conformations of the acarbose in complex with α-glucosidase and α-amylase are available in Fig. S1.
Regarding the α-glucosidase complexes, as can be seen in Table 3, there was no significant difference between the flavonoid-protein complexes and the acarbose-protein complexes. The detailed RMSD value along the 100 ns duration (Fig. 9 and Fig. S2) also shows that all the complexes could reach equilibrium within the first 1 ns, indicating the high stability of the protein–ligand complexes during the MD simulations. Other interpreting values, such as SASA and Rgyr, also aligned well with this observation.
Analysis of molecular dynamics simulations using 100 ns trajectories. a RMSD value of carbon backbone of α-glucosidase, b RMSF value of Cα of α-glucosidase and c the number of hydrogen bonds with α-glucosidase during the MDS with different ligands; d RMSD value of carbon backbone of α-amylase, b RMSF value of Cα of α-amylase and c the number of hydrogen bonds with α-amylase during the MDS with different ligands
In terms of the α-amylase complexes, although there was an observable variance between the RMSD value of the acarbose–protein complex and that of the compound F347, F369, F370, and F378, the detailed value along the MD trajectories, nevertheless, shows the stability of the protein structure, as most of them still could reach equilibrium within 40 ns of MDS (Fig. S2). However, this is not the case for compound F378, although have reaching equilibrium in the first 50 ns, the RMSD value skyrocketed to 0.20 nm, before stabilizing back to 0.13 nm in the last 20 ns. This could be explained by the high fluctuation of the loop consisting of Asn350, which is far away from the targeted binding cavity and could be omitted (Fig. 9).
As for hydrogen bonding, it is noteworthy that the number of hydrogen bonds formed between the chalcones and the enzymes remained relatively low compared to the interactions between acarbose and the enzymes. Throughout the simulations, the chalcones were able to form a limited number of hydrogen bonds, ranging from one to six, whereas acarbose exhibited a more substantial number of hydrogen bonds, ranging from five to a maximum of eighteen. This difference can be attributed to the presence of hydroxyl substituents in the acarbose structure.
Despite the lower number of hydrogen bonds, our chalcones demonstrated remarkable inhibitory activity. These findings align with the results obtained from the structure–activity relationship (SAR) analysis and molecular docking, highlighting the significance of hydrophobic interactions between the chalcones and the digestive enzymes. The subsequent section provides a more in-depth analysis of the interactions of flavonoids with the enzymes, shedding further light on their inhibitory mechanisms.
Stability of investigated flavonoid compounds during the MDS
Regarding the α-glucosidase simulation, as previously described, because of the shallow nature of the binding cavity of the enzyme, most flavonoids could not penetrate deeply to contact with the catalytic residues. This is also clearly demonstrated in our MDS results (Fig. 10). Apart from the compound F369, most of the chalcones underwent transformations during the MD simulations to fit in the binding pocket but could only interact with the gatekeeper residues of the cavity, except for the F378 ligand. Particularly, this small molecule could not stabilize in the docked binding cavity, as it rotated noticeably around the binding site before freely moving in the solvation box. This could be explained by the shortage of hydrogen bond donor and acceptor atoms in the structure of the F378 ligand. Although not being stabled in the anticipated binding pocket, most of our small molecules could outperform acarbose, in terms of their IC50 value against α-glucosidase. The instability of the ligands during the MDS suggests that the inhibitory mechanism of our investigated ligands toward α-glucosidase could be via uncompetitive, non-competitive, or a mixed manner. Further discussion is available in the Discussion part.
On the other hand, our chalcones were able to bind consistently to the active site of human pancreatic α-amylase, revealed by the MDS results. As can be seen in Fig. 11, although the chalcones also needed to undergo some transformations, they managed to stabilize inside the anticipated binding pocket. The binding capability of the chalcones could be explained by the deeper cavity of the enzyme, therefore increasing the possibility of the chalcones interacting with the catalytic residues inside the pocket. The results from our MDS suggest that our chalcones could competitively inhibit α-amylase. However, further experimental evaluations need to be conducted to confirm the hypothesis.
Predicted ADMET properties of the top hit compounds
Table S6 contains detailed information on the medicinal chemistry properties, ADME, and toxicity of the five potential dual inhibitors of α-glucosidase and α-amylase. Their physicochemical properties are also illustrated in Fig. S3. According to the mechanism of inhibiting the hydrolysis and digestion of polysaccharide chains to treat T2DM, we consider a shortlist of the ADMET properties presented in Table 4. The results predicted by the ADMETlab 2.0 show that these 5 chalcones are accepted by the Lipinski and Golden Triangle rules. A more favorable ADMET profile may be seen in compounds that adhere to the Golden Triangle guideline. All compounds had good oral bioavailability, as indicated by the Human Intestinal Absorption (HIA) probability. They were not P-glycoprotein substrates but may be inhibitors of this transport protein (except for the compound F337). The probability of blood–brain barrier penetration from these compounds was also not high (except for the compound F370). In addition, these chalcones were predicted to have no acute toxicity. In general, they have no serious cardiovascular toxicity (as predicted by the hERG blockers classification). Four of the five compounds were predicted to be associated with varying degrees of carcinogenicity, but none reached the highest predicted level (+ + +). This property needs to be considered for optimization in the next stages of drug development. Finally, for the liver, these substances have low predicted human hepatotoxicity and drug-induced liver injury.
Discussion
α-Glucosidase and α-amylase are two promising targets for the treatment of T2DM. However, currently available therapeutic agents such as acarbose, miglitol, and voglibose are not widely used because of adverse drug reactions. This raises the need for a more effective agent that can simultaneously inhibit α-glucosidase and α-amylase for a better sugar blood-maintaining outcome with minimal side effects. Flavonoids, a ubiquitous naturally occurring class, have long been considered to be active in many systems and have multiple benefits to human health [64,65,66,67], therefore they could be a safe and well-tolerated approach for the treatment of T2DM. In this study, by combining both the computational and experimental approaches, we retrieved twelve N-benzylaminochalcones that were able to inhibit α-glucosidase stronger than the commercially available drug acarbose, five of which were able to inhibit simultaneously the α-amylase enzyme, also with a stronger activity than acarbose.
Regarding the α-glucosidase inhibition, although being able to achieve outstanding in vitro results, our MDs results show that most of the compounds were not able to bind stably in the desired binding site. This indicates that our investigated compounds may not inhibit this enzyme via the competitive mechanism. This is not unusual as multiple research in literature have reported that chalcones may inhibit α-glucosidase via either competitive [41], non-competitive [36], or mixed-type inhibition [68]. Within the same study, even the introduction of the − OH group could transit the inhibitory mechanism from uncompetitive to competitive [69]. Therefore, a case-by-case inhibitory mechanism evaluation should be conducted to confirm the hypothesis.
Concerning α-amylase inhibition, the stability of the ligands inside the binding cavity of α-amylase suggests that these compounds could inhibit enzymes competitively. This hypothesis aligns well with the co-crystallized structure of α-amylase with myricetin (also a flavonoid structure) [23] and the majority of studies in the literature [41, 42, 69].
The discovered chalcones exhibit significant potential as more effective and safer therapeutic agents for the treatment of diabetes mellitus. Their ability to act as multi-target inhibitors, targeting both α-glucosidase and α-amylase enzymes simultaneously, provides distinct advantages in managing blood glucose levels. Moreover, some chalcones have shown inhibition of other putative targets for diabetes treatment, such as protein tyrosine phosphatase 1B, aldose reductase, and dipeptidyl peptidase-4 [20]. This multi-target characteristic makes flavonoids strong anti-diabetic agents, even at low concentrations, thus reducing the risk of adverse effects. Additionally, their natural origin and predicted safe ADMET profiles make them attractive alternatives to synthetic drugs. The antioxidant and anti-inflammatory properties of flavonoids further contribute to their potential benefits in diabetes management by reducing oxidative stress and inflammation, both implicated in the pathogenesis of the disease [70]. These findings highlight the unique advantages of the discovered flavonoids in dual-target inhibition and their potential as safer therapeutic agents for the management of diabetes mellitus. Further research and clinical investigations are warranted to fully explore their efficacy, pharmacokinetic properties, and long-term safety in diabetic patients.
It is important to acknowledge the limitations of our current study. The small size of the library, comprising a limited range of flavonoid scaffolds, might explain why all the selected compounds share the same N-benzylaminochalcone structure. However, to the best of our knowledge, these twelve chalcones are being investigated for their anti-α-glucosidase and α-amylase dual-target inhibition for the first time, making this research a valuable starting point for rational design and synthesis in future studies. Another limitation is that, although several approaches exist to determine the inhibitory kinetics of small molecules toward the two digestive enzymes such as the Lineweaver–Burk plot or NMR spectroscopy, we were unable to conduct these experiments due to limited resources in our laboratory. As an alternative approach, we employed molecular dynamics simulations to gain insights into the molecular mechanisms of these chalcones toward the two enzymes. Nevertheless, follow-up experimental results are needed to confirm the computational findings. Furthermore, although our primary objective was to investigate safer anti-α-glucosidase inhibitors, our study did not assess whether the compounds have fewer adverse effects than acarbose or not. Therefore, further in vivo research using animal models will be conducted to evaluate the safety of the discovered compounds.
Conclusion
In this research, a mini-SAR analysis from the literature has been conducted, the results from which were combined with molecular modeling techniques to yield twelve highly potent α-glucosidase inhibitors. Most notably, five out of these compounds were able to inhibit simultaneously the α-amylase enzyme with outstanding IC50 value, compared to acarbose. In addition, MD simulations were conducted to estimate the inhibitory mechanism of the dual-target inhibitors. The results suggest that these compounds could inhibit α-glucosidase non-competitively while inhibiting α-amylase competitively. Further evaluation using X-ray crystallization or Lineweaver Burk plot should be conducted to confirm the hypothesis.
References
Sapra A, Vaqar S, Bhandari P (2019) Diabetes Mellitus. In: StatPearls [Internet] (ed). StatPearls Publishing, Treasure Island (FL), pp 1–12
IDF Diabetes Atlas. Available at https://www.diabetesatlas.org. Accessed 30 Dec 2022
Deshpande AD, Harris-Hayes M, Schootman M (2008) Epidemiology of Diabetes and diabetes-related complications. Phys Ther 88:1254–1264. https://doi.org/10.2522/ptj.20080020
ElSayed NA, Aleppo G, Aroda VR, Bannuru RR, Brown FM, Bruemmer D, Collins BS, Hilliard ME, Isaacs D, Johnson EL, Kahan S, Khunti K, Leon J, Lyons SK, Perry ML, Prahalad P, Pratley RE, Seley JJ, Stanton RC, Gabbay RA, and on behalf of the American Diabetes A (2022) Classification and diagnosis of diabetes: standards of care in diabetes—2023. Diabetes Care 46:S19–S40. https://doi.org/10.2337/dc23-S002
ElSayed NA, Aleppo G, Aroda VR, Bannuru RR, Brown FM, Bruemmer D, Collins BS, Hilliard ME, Isaacs D, Johnson EL, Kahan S, Khunti K, Leon J, Lyons SK, Perry ML, Prahalad P, Pratley RE, Seley JJ, Stanton RC, Gabbay RA, Association obotAD, (2022) 9. Pharmacologic approaches to glycemic treatment: standards of care in diabetes—2023. Diabetes Care 46:S140–S157. https://doi.org/10.2337/dc23-S009
Ghani U (2020) Chapter one - Introduction, rationale and the current clinical status of oral α-glucosidase inhibitors. In: Ghani U (ed) Alpha-glucosidase inhibitors. Elsevier, Amsterdam, pp 1–15
Gong L, Feng D, Wang T, Ren Y, Liu Y, Wang J (2020) Inhibitors of α-amylase and α-glucosidase: potential linkage for whole cereal foods on prevention of hyperglycemia. Food Sci Nutr 8:6320–6337. https://doi.org/10.1002/fsn3.1987
Poovitha S, Parani M (2016) In vitro and in vivo α-amylase and α-glucosidase inhibiting activities of the protein extracts from two varieties of bitter gourd (Momordica charantia L.). BMC Complement Altern Med 16:185. https://doi.org/10.1186/s12906-016-1085-1
Chiasson J-L, Josse RG, Leiter LA, Mihic M, Nathan DM, Palmason C, Cohen RM, Wolever TM (1996) The effect of acarbose on insulin sensitivity in subjects with impaired glucose tolerance. Diabetes Care 19:1190–1193. https://doi.org/10.2337/diacare.19.11.1190
Brayer GD, Luo Y, Withers SG (1995) The structure of human pancreatic α-amylase at 1.8 Å resolution and comparisons with related enzymes. Protein Sci 4:1730–1742. https://doi.org/10.1002/pro.5560040908
Proença C, Ribeiro D, Freitas M, Fernandes E (2022) Flavonoids as potential agents in the management of type 2 diabetes through the modulation of α-amylase and α-glucosidase activity: a review. Crit Rev Food Sci Nutr 62:3137–3207. https://doi.org/10.1080/10408398.2020.1862755
Li C, Begum A, Numao S, Park KH, Withers SG, Brayer GD (2005) Acarbose rearrangement mechanism implied by the kinetic and structural analysis of human pancreatic α-amylase in complex with analogues and their elongated counterparts. Biochemistry 44:3347–3357. https://doi.org/10.1021/bi048334e
Sim L, Willemsma C, Mohan S, Naim HY, Pinto BM, Rose DR (2010) Structural basis for substrate selectivity in human maltase-glucoamylase and sucrase-isomaltase N-terminal domains. J Biol Chem 285:17763–17770. https://doi.org/10.1074/jbc.M109.078980
Sim L, Quezada-Calvillo R, Sterchi EE, Nichols BL, Rose DR (2008) Human intestinal maltase–glucoamylase: crystal structure of the N-terminal catalytic subunit and basis of inhibition and substrate specificity. J Mol Biol 375:782–792. https://doi.org/10.1016/j.jmb.2007.10.069
Ren L, Qin X, Cao X, Wang L, Bai F, Bai G, Shen Y (2011) Structural insight into substrate specificity of human intestinal maltase-glucoamylase. Protein Cell 2:827–836. https://doi.org/10.1007/s13238-011-1105-3
Le M-T, Trinh D-TT, Ngo T-D, Tran-Nguyen V-K, Nguyen D-N, Hoang T, Nguyen H-M, Do T-G-S, Mai TT, Tran T-D, Thai K-M (2022) Chalcone derivatives as potential inhibitors of P-glycoprotein and NorA: an in silico and in vitro study. BioMed Res Int 2022:9982453. https://doi.org/10.1155/2022/9982453
Ashraf J, Mughal EU, Sadiq A, Naeem N, Muhammad SA, Qousain T, Zafar MN, Khan BA, Anees M (2020) Design and synthesis of new flavonols as dual ɑ-amylase and ɑ-glucosidase inhibitors: structure-activity relationship, drug-likeness, in vitro and in silico studies. J Mol Struct 1218:128458. https://doi.org/10.1016/j.molstruc.2020.128458
Ullah A, Munir S, Badshah SL, Khan N, Ghani L, Poulson BG, Emwas A-H, Jaremko M (2020) Important flavonoids and their role as a therapeutic agent. Molecules 25:5243. https://doi.org/10.3390/molecules25225243
Zhu J, Chen C, Zhang B, Huang Q (2020) The inhibitory effects of flavonoids on α-amylase and α-glucosidase. Crit Rev Food Sci Nutr 60:695–708. https://doi.org/10.1080/10408398.2018.1548428
Mahapatra DK, Asati V, Bharti SK (2015) Chalcones and their therapeutic targets for the management of diabetes: structural and pharmacological perspectives. Eur J Med Chem 92:839–865. https://doi.org/10.1016/j.ejmech.2015.01.051
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28:235–242. https://doi.org/10.1093/nar/28.1.235
Khoo CM (2017) Diabetes Mellitus treatment. In: Quah SR (ed) International encyclopedia of public health, 2nd edn. Academic Press, Oxford, pp 288–293
Williams LK, Li C, Withers SG, Brayer GD (2012) Order and disorder: differential structural impacts of myricetin and ethyl caffeate on human amylase, an antidiabetic target. J Med Chem 55:10177–10186. https://doi.org/10.1021/jm301273u
Lasko TA, Bhagwat JG, Zou KH, Ohno-Machado L (2005) The use of receiver operating characteristic curves in biomedical informatics. J Biomed Inform 38:404–415. https://doi.org/10.1016/j.jbi.2005.02.008
Erickson JA, Jalaie M, Robertson DH, Lewis RA, Vieth M (2004) Lessons in molecular recognition: the effects of ligand and protein flexibility on molecular docking accuracy. J Med Chem 47:45–55. https://doi.org/10.1021/jm030209y
Mysinger MM, Carchia M, Irwin JJ, Shoichet BK (2012) Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. J Med Chem 55:6582–6594. https://doi.org/10.1021/jm300687e
Empereur-Mot C, Zagury J-F, Montes M (2016) Screening explorer–an interactive tool for the analysis of screening results. J Chem Inf Model 56:2281–2286. https://doi.org/10.1021/acs.jcim.6b00283
Empereur-mot C, Guillemain H, Latouche A, Zagury J-F, Viallon V, Montes M (2015) Predictiveness curves in virtual screening. J Cheminform 7:52. https://doi.org/10.1186/s13321-015-0100-8
Molecular Operating Environment (MOE) (2022) Version 2022.10. Chemical Computing Group Inc., Montreal
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) Open babel: an open chemical toolbox. J Cheminform 3:33. https://doi.org/10.1186/1758-2946-3-33
Eberhardt J, Santos-Martins D, Tillack AF, Forli S (2021) AutoDock Vina 1.2.0: new docking methods, expanded force field, and python bindings. J Chem Inf Model 61:3891–3898. https://doi.org/10.1021/acs.jcim.1c00203
Adasme MF, Linnemann KL, Bolz SN, Kaiser F, Salentin S, Haupt VJ, Schroeder M (2021) PLIP 2021: expanding the scope of the protein–ligand interaction profiler to DNA and RNA. Nucleic Acids Res 49:W530–W534. https://doi.org/10.1093/nar/gkab294
Liu M, Yin H, Liu G, Dong J, Qian Z, Miao J (2014) Xanthohumol, a prenylated chalcone from beer hops, acts as an α-glucosidase inhibitor in vitro. J Agric Food Chem 62:5548–5554. https://doi.org/10.1021/jf500426z
Proença C, Freitas M, Ribeiro D, Oliveira EFT, Sousa JLC, Tomé SM, Ramos MJ, Silva AMS, Fernandes PA, Fernandes E (2017) α-Glucosidase inhibition by flavonoids: an in vitro and in silico structure–activity relationship study. J Enzyme Inhib Med Chem 32:1216–1228. https://doi.org/10.1080/14756366.2017.1368503
Sun H, Song X, Tao Y, Li M, Yang K, Zheng H, Jin Z, Dodd RH, Pan G, Lu K, Yu P (2018) Synthesis & α-glucosidase inhibitory & glucose consumption-promoting activities of flavonoid–coumarin hybrids. Future Med Chem 10:1055–1066. https://doi.org/10.4155/fmc-2017-0293
Seo WD, Kim JH, Kang JE, Ryu HW, Curtis-Long MJ, Lee HS, Yang MS, Park KH (2005) Sulfonamide chalcone as a new class of α-glucosidase inhibitors. Bioorg Med Chem Lett 15:5514–5516. https://doi.org/10.1016/j.bmcl.2005.08.087
Chatsumpun N, Sritularak B, Likhitwitayawuid K (2017) New biflavonoids with α-glucosidase and pancreatic lipase inhibitory activities from boesenbergia rotunda. Molecules 22:1862. https://doi.org/10.3390/molecules22111862
Cheng N, Yi W-B, Wang Q-Q, Peng S-M, Zou X-Q (2014) Synthesis and α-glucosidase inhibitory activity of chrysin, diosmetin, apigenin, and luteolin derivatives. Chin Chem Lett 25:1094–1098. https://doi.org/10.1016/j.cclet.2014.05.021
Chen Y-G, Li P, Li P, Yan R, Zhang X-Q, Wang Y, Zhang X-T, Ye W-C, Zhang Q-W (2013) α-Glucosidase inhibitory effect and simultaneous quantification of three major flavonoid glycosides in microctis folium. Molecules 18:4221–4232. https://doi.org/10.3390/molecules18044221
Tajudeen Bale A, Mohammed Khan K, Salar U, Chigurupati S, Fasina T, Ali F, Kanwal WA, Taha M, Sekhar Nanda S, Ghufran M, Perveen S (2018) Chalcones and bis-chalcones: as potential α-amylase inhibitors; synthesis, in vitro screening, and molecular modelling studies. Bioorg Chem 79:179–189. https://doi.org/10.1016/j.bioorg.2018.05.003
Saleem F, Kanwal KKM, Chigurupati S, Solangi M, Nemala AR, Mushtaq M, Ul-Haq Z, Taha M, Perveen S (2021) Synthesis of azachalcones, their α-amylase, α-glucosidase inhibitory activities, kinetics, and molecular docking studies. Bioorg Chem 106:104489. https://doi.org/10.1016/j.bioorg.2020.104489
Sahnoun M, Trabelsi S, Bejar S (2017) Citrus flavonoids collectively dominate the α-amylase and α-glucosidase inhibitions. Biologia 72:764–773. https://doi.org/10.1515/biolog-2017-0091
Li K, Yao F, Xue Q, Fan H, Yang L, Li X, Sun L, Liu Y (2018) Inhibitory effects against α-glucosidase and α-amylase of the flavonoids-rich extract from Scutellaria baicalensis shoots and interpretation of structure–activity relationship of its eight flavonoids by a refined assign-score method. Chem Cent J 12:82. https://doi.org/10.1186/s13065-018-0445-y
Meshram G, Vala V, Wagh P, Deshpande SS (2016) Ultrasound accelerated synthesis of novel benzimidazole derived chalcones as glucosidases inhibitor and antimicrobial agents. Indian J Chem Sect B 55:613–623
Kiruthiga N, Prabha T, Selvinthanuja C, Srinivasan K, Sivakumar T (2018) Design, synthesis and evaluation of recent flavones as anti-diabetics. Int J Chem Pharm Anal 5:1–12
Lo Piparo E, Scheib H, Frei N, Williamson G, Grigorov M, Chou CJ (2008) Flavonoids for Controlling starch digestion: structural requirements for inhibiting human α-amylase. J Med Chem 51:3555–3561. https://doi.org/10.1021/jm800115x
Tran T-D, Nguyen T-C-V, Nguyen N-S, Nguyen D-M, Nguyen T-T-H, Le M-T, Thai K-M (2016) Synthesis of novel chalcones as acetylcholinesterase inhibitors. Appl Sci 6:198. https://doi.org/10.3390/app6070198
Wang Z-W, Wang J-S, Luo J, Kong L-Y (2013) α-Glucosidase inhibitory triterpenoids from the stem barks of Uncaria laevigata. Fitoterapia 90:30–37. https://doi.org/10.1016/j.fitote.2013.07.005
Granados-Guzmán G, Castro-Ríos R, Waksman de Torres N, Salazar-Aranda R (2018) Optimization and validation of a microscale in vitro method to assess α-glucosidase inhibition activity. Curr Anal Chem 14:458–464
Kusano R, Ogawa S, Matsuo Y, Tanaka T, Yazaki Y, Kouno I (2011) α-Amylase and lipase inhibitory activity and structural characterization of acacia bark proanthocyanidins. J Nat Prod 74:119–128. https://doi.org/10.1021/np100372t
Xiao Z, Storms R, Tsang A (2006) A quantitative starch-iodine method for measuring alpha-amylase and glucoamylase activities. Anal Biochem 351:146–148. https://doi.org/10.1016/j.ab.2006.01.036
Ahmed MU, Ibrahim A, Dahiru NJ, Mohammed HS (2020) Alpha amylase inhibitory potential and mode of inhibition of oils from Allium sativum (Garlic) and Allium cepa (Onion). Clin Med Insights Endocrinol Diabetes 13:1179551420963106. https://doi.org/10.1177/1179551420963106
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2:19–25. https://doi.org/10.1016/j.softx.2015.06.001
Mackerell AD Jr, Feig M, Brooks Iii CL (2004) Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem 25:1400–1415. https://doi.org/10.1002/jcc.20065
Zoete V, Cuendet MA, Grosdidier A, Michielin O (2011) SwissParam: A fast force field generation tool for small organic molecules. J Comput Chem 32:2359–2368. https://doi.org/10.1002/jcc.21816
Bhardwaj VK, Singh R, Sharma J, Rajendran V, Purohit R, Kumar S (2021) Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J Biomol Struct Dyn 39:3449–3458. https://doi.org/10.1080/07391102.2020.1766572
Humphrey W, Dalke A, Schulten K (1996) VMD: Visual molecular dynamics. J Mol Graph 14:33–38. https://doi.org/10.1016/0263-7855(96)00018-5
Schrödinger LLC., The PyMOL Molecular Graphics System. 2020. 2.0.
Guerra F, Siemers M, Mielack C, Bondar A-N (2018) Dynamics of long-distance hydrogen-bond networks in photosystem II. J Phys Chem B 122:4625–4641. https://doi.org/10.1021/acs.jpcb.8b00649
Dong J, Wang N-N, Yao Z-J, Zhang L, Cheng Y, Ouyang D, Lu A-P, Cao D-S (2018) ADMETlab: a platform for systematic ADMET evaluation based on a comprehensively collected ADMET database. J Cheminform 10:29. https://doi.org/10.1186/s13321-018-0283-x
Yadav P, Lal K, Kumar A, Guru SK, Jaglan S, Bhushan S (2017) Green synthesis and anticancer potential of chalcone linked-1,2,3-triazoles. Eur J Med Chem 126:944–953. https://doi.org/10.1016/j.ejmech.2016.11.030
Nasli Esfahani A, Iraji A, Alamir A, Moradi S, Asgari MS, Hosseini S, Mojtabavi S, Nasli-Esfahani E, Faramarzi MA, Bandarian F, Larijani B, Hamedifar H, Hajimiri MH, Mahdavi M (2022) Design and synthesis of phenoxymethybenzoimidazole incorporating different aryl thiazole-triazole acetamide derivatives as α-glycosidase inhibitors. Mol Divers 26:1995–2009. https://doi.org/10.1007/s11030-021-10310-7
Zawawi NKNA, Taha M, Ahmat N, Ismail NH, Wadood A, Rahim F (2017) Synthesis, molecular docking studies of hybrid benzimidazole as α-glucosidase inhibitor. Bioorg Chem 70:184–191. https://doi.org/10.1016/j.bioorg.2016.12.009
Halliwell B, Rafter J, Jenner A (2005) Health promotion by flavonoids, tocopherols, tocotrienols, and other phenols: direct or indirect effects? Antioxidant or not? Am J Clin Nutr 81:268S-276S. https://doi.org/10.1093/ajcn/81.1.268S
Fan X, Fan Z, Yang Z, Huang T, Tong Y, Yang D, Mao X, Yang M (2022) Flavonoids-natural gifts to promote health and longevity. Int J Mol Sci 23:2176. https://doi.org/10.3390/ijms23042176
Xu SL, Zhu KY, Bi CW, Yan L, Men SW, Dong TT, Tsim KW (2013) Flavonoids, derived from traditional Chinese medicines, show roles in the differentiation of neurons: possible targets in developing health food products. Birth Defects Res C Embryo Today 99:292–299. https://doi.org/10.1002/bdrc.21054
Lam TP, Tran NVN, Pham LHD, Lai NVT, Dang BTN, Truong NLN, Nguyen-Vo SK, Mai TT, Tran TD (2023) Flavonoids as dual-target inhibitors against α-glucosidase and α-amylase: a systematic review of in vitro studies. J Chem. https://doi.org/10.26434/chemrxiv-2023-cdlf8-v3
Ryu HW, Lee BW, Curtis-Long MJ, Jung S, Ryu YB, Lee WS, Park KH (2010) Polyphenols from broussonetia papyrifera displaying potent α-glucosidase inhibition. J Agric Food Chem 58:202–208. https://doi.org/10.1021/jf903068k
Rocha S, Sousa A, Ribeiro D, Correia CM, Silva VLM, Santos CMM, Silva AMS, Araújo AN, Fernandes E, Freitas M (2019) A study towards drug discovery for the management of type 2 diabetes mellitus through inhibition of the carbohydrate-hydrolyzing enzymes α-amylase and α-glucosidase by chalcone derivatives. Food Funct 10:5510–5520. https://doi.org/10.1039/C9FO01298B
Basu S, Debroy R, Kumar H, Singh H, Ramaiah S, Anbarasu A (2023) Bioactive phytocompounds against specific target proteins of Borrelia recurrentis responsible for louse-borne relapsing fever: genomics and structural bioinformatics evidence. Med Vet Entomol 37:213–218. https://doi.org/10.1111/mve.12623
Funding
This work was supported by the University of Medicine and Pharmacy at Ho Chi Minh City for Thanh-Dao Tran under Grant number 162/2019/HĐ-ĐHYD.
Author information
Authors and Affiliations
Contributions
TTM and T-DT contributed to conceptualization; TTM, M-HP, T-TN, and T-V-PN contributed to methodology; TTM, M-HP, T-TT, T-TN, T-PL, and NV-TL contributed to formal analysis and investigation; TTM, T-TT, and M-HP contributed to writing - original draft preparation;TTM, T-PL, and C-VTV contributed to writing - review and editing; T-DT, K-MT, and T-V-PN contributed to funding acquisition; T-DT, K-MT, and C-VTV contributed to resources; T-DT and K-MT contributed to supervision. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mai, T.T., Phan, MH., Thai, T.T. et al. Discovery of novel flavonoid derivatives as potential dual inhibitors against α-glucosidase and α-amylase: virtual screening, synthesis, and biological evaluation. Mol Divers 28, 1629–1650 (2024). https://doi.org/10.1007/s11030-023-10680-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-023-10680-0