Abstract
Non-alcoholic fatty liver disease (NAFLD) is rapidly being recognized as the leading cause of chronic liver disease worldwide. Men1, encoding protein of menin, is a key causative gene of multiple endocrine neoplasia type 1 syndrome including pancreatic tumor. It is known that insulin that secretes by endocrine tissue pancreatic islets plays a critical role in hepatic metabolism. Mouse model of hemizygous deletion of Men1 was shown to have severe hepatic metabolism disorders. However, the molecular function of menin on lipid deposition in hepatocytes needs to be further studied. Transcriptome sequencing does show that expression suppression of Men1 in mouse hepatocytes widely affect signaling pathways involved in hepatic metabolism, such as fatty acid metabolism, insulin response, glucose metabolism and inflammation. Further molecular studies indicates that menin overexpression inhibits expressions of the fat synthesis genes Srebp-1c, Fas, and Acc1, the fat differentiation genes Pparγ1 and Pparγ2, and the fat transport gene Cd36, thereby inhibiting the fat accumulation in hepatocytes. The biological process of menin regulating hepatic lipid metabolism was accomplished by interacting with the transcription factor FoxO1, which is also found to be critical for lipid metabolism. Moreover, menin responds to insulin in hepatocytes and mediates its regulatory effect on hepatic metabolism. Our findings suggest that menin is a crucial mediation factor in regulating the hepatic fat deposition, suggesting it could be a potential important therapeutic target for NAFLD.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Non-alcoholic fatty liver disease (NAFLD), defined as excess fat accumulation in the liver, is rapidly becoming the most common cause for chronic liver disease worldwide. Nevertheless, the current treatment of NAFLD, there is no appropriate drug treatment, mainly through diet and lifestyle intervention [1,2,3]. Therefore, there is an urgent need to identify key factors that regulate liver fat metabolism to develop therapeutic targets for NAFLD.
Menin is a 610 amino acid protein encoded by Men1 gene on human 11q13 chromosome, which is a key causative agent of multiple endocrine neoplasia type 1 (MEN1) [4]. Menin has a tumor suppressing effect in the endocrine system [5,6,7]. Moreover, menin as a key scaffold protein interacts with a variety of transcription factors and participates in multiple biological processes, including metabolic processes [8,9,10,11]. Recent studies have shown that menin is involved in the glucose and lipid metabolism in liver. For example, liver-specific Men1 hemizygous deletion (Men1±) mutant mice show abnormal liver glucose tolerance and lipid metabolism after feeding on a high-fat diet [12]; Islet-specific Men1 knockout mouse obtain increased pancreatic cell proliferation and insulin secretion [13,14,15]; Menin affects fatty acid uptake in liver cells through targeting Cd36 gene transcription regulation[16]. Nonetheless, the mechanism by which menin regulates hepatic lipid metabolism is poorly understood.
It is well-known that the transcription factor forkhead box O1 (FoxO1) plays a crucial role in glucose and lipid metabolism [17]. Insulin enhances the interaction between menin and FoxO1 through Akt, therefore affecting the glucose metabolism in liver [18]. However, it is not clear whether menin binds to FoxO1 to regulate lipid metabolism. Therefore, the purpose of this study is to investigate the possible mechanism of menin regulates lipid metabolism, and whether it is interacting with FoxO1. Our study found that menin down-regulates lipid deposition in hepatocytes. This may be the consequent that menin interacts with FoxO1 targeting the transcription activities of Cd36 and Gk promoter and thereby regulating lipid metabolism in liver.
Materials and methods
Cell culture
Immortalized mouse normal (healthy) hepatocytes (NCTC-1469) used in this study were purchased from the National Cell Resource Center (Beijing, China). Cells were grown in Dulbecco’s modified Eagle’s medium-F12 (DMEM/F12; Gibco, Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Biological Industries, Israel), penicillin–streptomycin liquid (Sigma-Aldrich, St. Louis, MO, USA) at 37 °C in an atmosphere of 5% CO2.
Cell transfection
A recombinant expression plasmid of Men1 was constructed according to the Mus musculus Men1 sequence (NM_001168489). The plasmid was designated as pcDNA3.1 ( +)-mMen1, and pcDNA3.1( +)-vector was negative control plasmid. Men1- and FoxO1-specific short interfering (si) RNA (siG12315132207 and siB13415161059, designated as si-Men1 and si-FoxO1, respectively), as well as its negative control siRNA (siN05815122147, designated as NC), were designed and synthesized by Ribobio (Guangzhou, China). The cells were inoculated into 6-well plates and replaced with a fresh culture medium when there were approximately 1 × 106 cells in each well. Then, siRNA (50 nM) or plasmid (1.5 µg) was added and transferred by Lipofectamine® 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Total RNA and protein were isolated from the transfected cells 24 h post-transfection for further analysis.
Total RNA extraction and real-time PCR analysis
Total RNA was extracted using the RNASimple Total RNA Kit (Tiangen Biotech, Beijing, China) and reverse transcribed using PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa Bio, Shiga, Japan). The extracted RNA was reverse transcribed according to the instructions of the kit. In the 20 μL reverse transcription system, the maximum amount of total RNA used was 1000 ng. Real-time quantitative PCR was performed on the ABI 7500 instrument using the SYBR®Premix Ex TaqTM (Perfect Real-Time) kit (TaKaRa Bio) using the SYBR Green I dye method. The primer sequences are listed in Supplementary Table S1. The expression levels of the target mRNAs were normalized to the internal control β-actin mRNA. The 2−ΔΔCT method was used to calculate the relative abundance of mRNA. The results are representative of at least three independent experiments to determine the statistical significance.
Western blotting
After transfection of Men1-specific plasmid and/or siRNA, total protein was extracted from NCTC-1469 cells using RIPA lysis buffer (containing 1% phenylmethylsulfonyl fluoride) (Beyotime, Shanghai, China). The protein concentration was detected by the BCA kit (Beyotime). Protein (20 μg) were separated by 10% SDS-PAGE and transferred to the PVDF membrane (Millipore, Bedford, MA, USA). The membrane was incubated overnight with the primary antibodies menin (1:1000 dilution; Abcam, Cambridge, UK), FoxO1 (1:1000 dilution; Cell Signaling Technology, Beverly, MA, USA) or β-actin (1:1000 dilution; Beyotime). β-actin was used as a total protein loading control. It was incubated with horseradish peroxidase-conjugated secondary antibody (1:1000 dilution; Beyotime) for 4 h at 4 °C. Chemiluminescence detection was performed using BeyoECL Plus (Beyotime). The data shows the level of expression normalized to the corresponding negative control. The results are representative of three independent experiments that were used to determine the statistical significance.
Gene expression analysis by RNA-Seq
Three sets of RNAs with lower expression level of Men1 (31% ± 4.6%, n = 3) were sent for transcriptome sequencing (BaiMaiKe, Beijing, China). The RNA samples were quality guaranteed, followed by the construction of the library. The quality of the library was detected to meet the requirements before sequencing is done. The data were filtered to obtain Clean Data, and the data were compared with the specified reference genome. The resulting Mapped Data were used to evaluate the quality of the library, with attributes including the length of the inserted fragment, randomness of the fragment, discovery of new genes, and optimization of gene structure. Based on the expression level of all genes in different sample groups, the differences in the expression level of the genes were analyzed using Fold Change > 1.5 and false discovery rate (FDR) < 0.01 as the screening criteria. The functional differences of the differentially expressed genes (DEGs) were screened and the functional enrichment of different analytical methods was carried out.
Gene Ontology (GO) enrichment analysis
GO enrichment analysis of the DEGs was implemented by the GOseq R packages based Wallenius non-central hyper-geometric distribution [13, 19, 20], which can adjust for gene length bias in DEGs. The GO annotations contains three parts: the Biological Process, Molecular Function, and Cellular Component. The DEGs were analyzed by GO enrichment, and the GO annotations of each DEGs could be obtained. The Web Gene Ontology (WEG) Annotation Plot was used for GO function classifications [21].
Kyoto Encyclopedia of Genes and genomes (KEGG) pathway enrichment analysis
KEGG is a database resource for understanding high-level functions and utilities of biological systems [22], such as cells, organisms, and ecosystems, from molecular level information, especially large-scale molecular datasets generated by genome sequencing and other high-throughput experimental technologies. We used the KEGG Orthology Based Annotation System (KOBAS) software to test the statistical enrichment of differential expression genes in KEGG pathways [23].
Insulin treatment of NCTC-1469 cells
NCTC-1469 cells (8.0 × 105 cells/well) were inoculated in 6-well cell culture plates and incubated until the cells were 70% to 80% confluent. The blank control group was 10% FBS DMEM/F12 culture. Treated samples received insulin with concentration of 0.8, 2, 3.2, 4.8, 6 or 7.2 μg/mL. After continuous culture for 2, 4, 6, 8, 10, and/or 12 h, total intracellular RNA was extracted and RT-PCR was used to detect the changes in the expression level of Men1 in mouse normal (healthy) hepatocytes. The concentration and time point that affected the most significant changes in the expression level of Men1 mRNA was selected (Fig S1).
Oil Red O staining
The expression of menin was inhibited or enhanced by transfection of hepatocytes with siRNA (si-Men1) or plasmid (pcDNA3.1 ( +)-mMen1). The cells in the 6-well cell culture plate was added to 1 mL of 4% paraformaldehyde per well and the cells were fixed for at least 30 min. The cells were stained with the Oil Red O Stain Kit (Nanjing Jiancheng, Nanjing, China) according to the manufacturer's instructions. After washing with 1 × PBS, the images were observed and photographed under a microscope.
Isopropanol was used to extract oil red O and spectrophotometry was tested for the relative quantitative analysis of intracellular lipids [24]. In brief, 200 µl of 100% isopropanol was added to each well to extract oil red O and the mixtures were incubated for 10 min at room temperature. Then, 100 µl was transferred to a 96-well plate. OD value (absorbance) at 490 nm wavelength was measured by a microplate reader.
Cellular triglyceride (TG) assay
The level of TG in the cells was detected by ELISA. The cells were collected and disrupted by ultrasonication. Part of the cell lysates were detected protein concentration by BCA (Beyotime), and the other cell lysates were used for determination of TG using the Triglyceride assay kit (Nanjing Jiancheng). TG content was calculated (mmol/gprot) as (sample optical density [OD] value − blank OD value) / (calibration OD value − blank OD value) × calibrator concentration (mmol/L) / sample protein concentration (gprot/L). Samples comprised the transfected control group and the treatment group. The blank was 1 × PBS. The calibration product was provided as part of the kit.
Fatty hepatocyte model
Exposure of hepatocytes to oleic acid (OA) induces mRNA expression of enzymes involved in adipogenesis, accumulation of intracellular lipid droplets, and significantly increased triglyceride content, which has been widely used as a model of hepatocyte fatty liver [25, 26]. Therefore, hepatocytes were treated with oleic acid to establish the model of fatty hepatocytes. Cells were inoculated to a 96-well plate (1 × 104 cells/well). Cells were treated with different concentrations of OA (0, 0.1, 0.15, 0.2, 0.25, 0.3, 0.4, and 0.5 mM/L), and cell viability was detected after 24, 48, or 72 h. 50 μL MTT was added to each well, and incubation was continued for 4 h. The supernatant was discarded, 150 μL of dimethylsulfoxide was added to each well. The absorbance value of each well was measured by a microplate reader at a wavelength of 570 nm. The survival rate was determined by the formula: cell viability (T/C% expressed) = (test group average OD value / control group average OD value) × 100%. After 24, 48, and 72 h of treatment with the optimal concentration of OA, TG content was detected and Oil Red O staining was done. Finally, the fatty hepatocyte model was validated (Fig S2).
Chromatin immunoprecipitation (ChIP)
NCTC-1469 cells (1–2 × 107 cells) were cross-linked with 1% formaldehyde and lysed in 1 mL SDS lysis buffer. Then, the chromatin was sheared into 200–1000 bp fragments using a Bioruptor ultrasonic apparatus. For each immunoassay, ultrasound-treated cell lysates were filtered by agarosaccharide beads and 1% supernatant was taken as input. The remainder was divided into two tubes, and 1.6 μg anti-rabbit menin (Abcam) and anti-rabbit IgG antibodies (Cell Signaling Technology) were added for ChIP analysis. Precipitated DNA was used as a template for quantitative real-time PCR using sybr-premix Ex Taq II (TaKaRa Bio). The primer pair to screen the 1200 bp promoter region of Gk and Cd36 was designed and synthesized. Details are provided in the supplemental data (Table S2). For quantitative real-time PCR, Ct values for each ChIP were obtained from triplicate reactions. The results represent three separate ChIP experiments, in which the amount of chromatin obtained from immunoprecipitation was quantified relative to the amount in the input sample, and the input percentage of chromatin obtained from immunoprecipitation was quantified relative to the amount in the input sample.
Statistical analysis
The results are expressed as mean ± SD. The difference was statistically analyzed using SAS 8.2 statistical software (SAS Institute, Cary, NC) for single factor analysis of variance (ANOVA). Significant difference was denoted P < 0.05 (*) and P < 0.01 (**).
Results
Inhibition of expression of menin in hepatocytes affects signaling pathways involved in glycolipid metabolism and inflammatory responses
To explore the role of menin in the liver, NCTC-1469 mouse normal hepatocytes were used as an in vitro hepatocyte model. Menin expression was knocked down by transfection of the cells with Men1-specific siRNA, After transfection for 24 h, the mRNA expression level of Men1 in the si-Men1 group was 31% ± 4.6% (n = 3), compared to that in the negative control group (Fig. 1A). Triplicates of the menin expression inhibited cells, as well as their negative control cells, were submitted for transcriptome sequencing analysis. A total of 157 differentially expressed genes (DEGs) were identified, among which 51 genes were upregulated and 106 genes were down-regulated. Gene expression validation results of randomly selected genes (four down-regulated and eight upregulated genes) showed that the correlation coefficient (R2) of qRT-PCR data with sequencing data was 0.916 (Fig. 1B), suggesting the accurate and reliable identification of DEGs upon lower expression of Men1.
Knockdown of Men1 in hepatocytes causes changes in signaling pathways involved in glycolipid metabolism and inflammatory responses. A Mouse normal hepatocytes NCTC-1469 cells were transfected with Men1-specific siRNA (si-Men1) or control (NC) for 24 h, n = 3. The expression level of Men1 mRNA were reduced to 30.2% ± 0.58% via qRT-PCR. B Three sets of the Men1-knockdown RNA, as well as their control, were performed RNA-seq. The high correlation coefficient (R2 = 0.92) of fold changes for representative differentially expressed genes between RNA-seq and qRT-PCR data indicated the present analysis was stable and repeatable. C KEGG analysis showed that the differentially expressed genes after Men1-knockdown enriched into signaling pathways such as glucose metabolism, lipid metabolism and inflammatory responses. Data are means ± SEM of 3 independent experiments per each group. ***P < 0.001
KEGG enrichment analysis of DEGs identified pathways including the adipocytokine signaling pathway related to fat metabolism, tumor necrosis factor signaling pathway related to inflammation and glycolipid metabolism, and a variety of inflammatory pathways (Fig. 1C). GO analysis revealed the involvement of the DEGs in the insulin response, glucose metabolism, fatty acid metabolism, and inflammatory response (Table 1). These results indicated that modification of menin expression in mouse hepatocytes affects inflammation and glycolipid metabolism pathways, which are closely related to the occurrence of fatty liver disease in vivo [27].
Inhibition of menin expression promotes fat accumulation in hepatocytes
To further investigate whether menin has an effect on fat deposition in hepatocytes, Men1 expression was inhibited by transfection of specific siRNAs (si-Men1) into mouse hepatocytes (NCTC-1469 cells, Fig. 2A, B). The inhibition of Men1 caused significantly decreased expression of Gk gene, which was involved in glycolytic pathway (Fig. 2C). However, the expression of the fatty acid synthesis genes Srebp-1c, Acc1, Fas and the fatty acid transporter gene Cd36 were all significantly increased, as well as the expression of adipose differentiation related gene Pparγ1 and Pparγ2 [28, 29].
The knockdown of Men1 promotes fat accumulation in hepatocytes. NCTC-1469 cells were transfected with Men1-specific siRNA (si-Men1) or control (NC) for 24 h. A, B The expression levels of Men1 mRNA (A) and menin protein (B) were detected via qRT-PCR and western blotting. C After knockdown of Men1, the expression of genes related to lipid synthesis, differentiation and fat transport was changed via qRT-PCR. D After knockdown of Men1, the cellular triglyceride (TG) were significantly increased by ELISA. E The lipid drops were increased in Men1-knockdown hepatocytes via absorbance value of the extracted oil red O dye at 490 nm. Data are means ± SEM of 3 independent experiments per each group. *P < 0.05, **P < 0.01, ***P < 0.001
To further validate the effect of menin on lipid metabolism in mouse hepatocytes, we examined the triglyceride (TG) content after knockdown of menin. The TG content was significantly increased upon menin expression inhibition compared to the negative control (Fig. 2D). Moreover, to detect whether the content of lipid droplets changed, we stained the transfected cells with Oil Red O and extracted with absolute ethanol to determine the absorbance value. The menin knockdown cells showed significant increase absorbance value, indicating significant increase in lipid droplet content (Fig. 2E). These results suggested that the decreased expression of menin enhances fat synthesis, transport and deposition in mouse hepatocytes.
Overexpression of Men1 inhibits fat accumulation in hepatocytes
To further verify the effect of menin on lipid metabolism, hepatocytes were transfected with the Men1-overexpressed plasmid pcDNA3.1( +)-mMen1 to examine its metabolic effects. After transfection, Men1 mRNA and menin protein levels were significantly elevated compared to vector controls (Fig. 3A, B). Enhanced expression of menin caused significantly increased expression of Gk. Moreover, the expression of Fas and Srebp-1c were decreased (p = 0.081 and p = 0.052, respectively), and the expression of Acc1, Cd36, Pparγ1 and Pparγ2 were obviously decreased (p < 0.05) (Fig. 3C). These results were opposite to those observed with that of menin inhibition, suggesting that an increased expression of menin has an inhibitory effect on fat synthesis. This was further verified by two different assays. Firstly, the amount of TG was observably reduced upon menin overexpression compared to negative controls (Fig. 3D). Furthermore, to detect whether menin could rescue the fat deposition in fatty hepatocytes induced by Oleic acid (OA), Oil Red O staining was used to assess the content of lipid droplets. The measurement of the absorbance due to the stain showed that the overexpression of menin reduced the fat deposition in OA-induced hepatocytes (p > 0.05), indicating that menin suppresses the fat deposition (Fig. 3E).
The overexpression of Men1 inhibits fat accumulation in hepatocytes. NCTC-1469 cells were transfected with pcDNA3.1( +)-mMen1 (mMen1) or control (Vector) for 24 h to overexpress Men1. A, B The expression levels of Men1 mRNA (A) and menin protein (B) were detected via qRT-PCR and western blotting. C After overexpression of Men1, the expression of genes related to lipid synthesis, differentiation and fat transport was changed via qRT-PCR. D After overexpression of Men1, the cellular triglyceride (TG) were significantly decreased by ELISA. E NCTC-1469 cells were divided into 4 groups: normal (healthy) cells were transfected with Vector (Ctrl + Vector); normal cells were transfected with mMen1 (Ctrl + mMen1); Fatty liver model cells were transfected with Vector (OA + Vector); Fatty liver model cells were transfected with mMen1 (OA + mMen1). The cellular lipid drops were decreased after overexpression of Men1 via absorbance value of the extracted Oil Red O dye at 490 nm. Data are means ± SEM of 3 independent experiments per each group. *P < 0.05, **P < 0.01
Menin mediates the effect of insulin on lipid metabolism by responding to insulin surrounding the hepatocytes
Insulin is important in regulating cell metabolism of different tissues, including the liver. To test the effect of insulin on Men1 expression in hepatocytes, NCTC-1469 cells were treated with insulin. The results showed that Men1 expression was gradually suppressed dependent on the insulin concentration (Fig S1), with the lowest Men1 expression level observed at 6 μg/mL insulin at 8 h post treatment (Fig. 4A). The expression levels of Pparγ1, Pparγ2, Cd36, Srebp-1c, Fas and Acc1 were significantly elevated (Fig. 4B). These results suggest that menin may be involved in the regulation of insulin on hepatocyte metabolism.
Insulin-treated hepatocytes inhibits Men1/menin expression and promotes fat accumulation. NCTC-1469 cells were treated with 6 μg/mL insulin or negative control PBS (Ctrl) for 8 h. A The expression level of Men1 mRNA and it protein menin were both detected to be inhibited compared with control, via qRT-PCR and western blotting. B After insulin treatment, the expression of genes related to lipid synthesis, differentiation and fat transport was up-regulated. Data are means ± SEM of 3 independent experiments per each group. *p < 0.05
Menin associates with the promoters of Cd36 and Gk gene in hepatocytes
Based on the above results, we found no matter menin expression was enhanced or inhibited, the expression of Cd36 and Gk would be affected (Figs. 2C and 3C). Thus, we hypothesized that menin might recruit transcription factors and associate with promoter regions of target genes, modulating their transcriptions. Thus, a ChIP assay was performed in hepatocytes. We initially designed four amplicons (P1, P2, P3, and P4) to determine whether menin binds to the promoter regions of Cd36 and Gk. (Fig. 5A, C). It was found that menin bound to the regions detectable by P1 (P = 0.18), P2 (P = 0.19), P3 (P = 0.18) and P4 (P = 0.014) for Cd36 gene (Fig. 5B). Whereas, for Gk gene, menin bound to the regions detectable by P1 (P = 0.28), P2 (P = 0.22), P3 (P < 0.01) and P4 (P < 0.01). These results suggested that menin may regulate lipid metabolism and glycolysis in hepatocytes by binding to the Cd36 and Gk promoters.
Cd36 and Gk are transcriptional targets of menin. A and B Four amplicons (P1, P2, P3, and P4) were designed to detect the indicated regions (upstream 1000 bp) of the Cd36 (A) or Gk (C) by menin-ChIP-PCR assays in NCTC-1469 cells. B and D ChIP-qPCR analysis of menin binding to Cd36 (B) or Gk (D) promoter. The results showed as the percentage of input by quantifying the amount of chromatin obtained from immunoprecipitation relative to the amount in the input samples. Data are means ± SEM of 3 independent experiments per each group. **P < 0.01, *P < 0.05
Menin and FoxO1 interact and co-regulate lipid metabolism in hepatocytes
FoxO1 has been shown to play a crucial role in glucose and lipid metabolism in the liver [17]. To investigate the effect of FoxO1 on metabolism-related genes in hepatocytes, the expression of FoxO1 was inhibited by transfection of si-Foxo1 and followed by detection of target gene expression. The results showed that inhibition of FoxO1 expression (39.3% ± 3.6% of control cells, n = 3) decreased the expression of Srebp-1c, Acc1, Fas, Cd36, Pparγ1, Pparγ2 and Mtp, but increased the expression of Gk (Fig. 6A). Moreover, the knockdown of FoxO1 significantly reduced lipid deposition in both healthy and fatty liver cells (Fig. 6B). Thus, Both menin and FoxO1 can regulate the same target gene expression, but in an opposite regulatory direction (Figs. 2 and 3), indicating a coordinating acting model of menin and FoxO1 in controlling the lipid metabolism in mouse hepatocytes. This model was confirmed by co-IP assays in both menin overexpressed and knockdown NCTC-1469 cells. The results showed that the amount of FoxO1 binding to menin was significantly increased upon menin knockdown, while, significantly decreased upon menin overexpression (Fig. 6C, D).
Menin regulates the expression of genes involved in lipid metabolism by interacting with FoxO1. A NCTC-1469 cells were transfected with si-FoxO1 or negative control (NC) for 24 h to suppress FoxO1. Then, the expression of genes related to lipid synthesis, differentiation and fat transport was changed via qRT-PCR. B NCTC-1469 cells were divided into 4 group: normal (healthy) cells were transfected with NC (Ctrl + NC); normal cells were transfected with si-Foxo1 (Ctrl + si-Foxo1); Fatty liver model cells were transfected with NC (OA + NC); Fatty liver model cells were transfected with si-Foxo1 (OA + si-Foxo1). The cellular lipid drops were decreased after overexpression of Men1 via absorbance value of the extracted Oil Red O dye at 490 nm. C Menin-immunoprecipitation (IP) assay were conducted in Men1-knockdown (si-Men1) and Men1-overexpressed (mMen1) NCTC-1469 cells, as well as their negative control cells (NC and Vector), together with IgG-IP control experiments. Foxo1-immunoblot assay results indicated that Menin could bind to FoxO1, not only in Men1-knockdown (si-Men1) and/or Men1-overexpressed (mMen1) cells but also in their negative control cells. (D) The quantitative results showed that the combination of menin and Foxo1 was significantly increased upon Men1 expression inhibition (si-Men1) compared with negative control (NC). However, the combination of menin and Foxo1 was significantly decreased upon Men1-overexpressed (mMen1). Data are means ± SEM of 3 independent experiments per each group. **P < 0.01, *P < 0.05
Discussion
Non-alcoholic fatty liver disease (NAFLD) is the most common chronic liver disease, accounting for approximately 20% ~ 30% of the global prevalence [30]. NAFLD is associated with a variety of metabolic disorder, mainly fat deposition in liver, but the critical molecular determinants of its pathogenesis remain largely unexplored. Although menin is a focus of tumor-associated research, it is seldom studied in lipid and glucose metabolism in liver. Our collective results indicate that menin plays an important role in regulating glucose metabolism and lipid metabolism in the liver. Altered menin expression in mouse hepatocytes can cause abnormalities including glucose metabolism, lipid metabolism and inflammatory response (Table 1, Fig. 1). Furthermore, overespression of menin inhibits triglyceride deposition in hepatocytes by regulating the expression of genes involved in fatty acid differentiation, synthesis and transport in hepatocytes (Figs. 2 and 3), indicating menin’s important role in maintaining normal liver function. The function of menin in lipid metabolism is consistent with previous reports, and fat deposition were increased in hepatocytes after menin knockdown [12, 16].
Menin regulates the lipid metabolism by coordinating with FoxO1 and targeting the same gene expression that are closely associated with fat deposition
We found that overexpression of menin suppressed the expression of genes related to fat differentiation (Pparγ1 and Pparγ2), transport (Cd36) and synthesis (Srebp-1c, Acc1, and Fas) in hepatocytes, while increased the expression of the key enzyme gene for glycogen synthesis genes (Gk) (Fig. 3). Meanwhile, menin overexpression reduced the TG content in hepatocytes, and attenuated the fat deposition in fatty hepatocytes (Fig. 3). And, the inhibition of menin had the opposite effect in hepatocytes (Fig. 2). Furthermore, we found that menin can bind to promoter regions of Cd36 and Gk gene in hepatocyte (Fig. 5). Many studies have shown that inhibiting the expression of genes related to glucose production, including Gk, is essential for adipogenesis [31]. Interestingly, previous study also found that menin can deacetylate histone H3 by binding to the promoter region of Cd36 via recruiting NAD-dependent deacetylase sirtuin 1 (SIRT1), thereby inhibiting the expression of Cd36 and reducing the uptake of fatty acids by hepatocytes [16]. These results demonstrate that menin can modulate the lipid metabolism in hepatocytes by binding to the promoter region of lipid metabolism-related genes, such as Cd36, Gk.
Menin usually plays its regulatory role through acting as a key scaffold protein that interacts with a variety of proteins, such as transcription factors [8,9,10,11]. It is well-known that the transcription factor FoxO1 is involved in lipid metabolism and plays a critical role in the development of NAFLD [17, 32]. Moreover, overexpression of FoxO1 in mice promotes liver fat deposition [33, 34]. Intriguingly, our results found inhibition of FoxO1 significantly reduced the fat deposition in both healthy and fatty hepatocytes (Fig. 6B), which was exactly consistent with the effect of menin overexpression (Fig. 2D, E) by targeting the same gene expression (Figs. 2C, 3C and 6A). Studies have shown that FoxO1 binds to the Gk and/or Cd36 promoter region and regulates its expression [35, 36]. Here we also found that menin binds to the Gk and/or Cd36 promoter region thereby regulating their expression, through interacting with FoxO1 in mouse hepatocytes (Fig. 6B). Thus, it suggests that menin regulates the lipid metabolism in hepatocytes by interacting with transcription factor FoxO1 and targeting the expression of lipid metabolism-related genes, including Cd36, Gk.
Menin mediates the hepatic lipid deposition by responding to insulin in hepatocytes
Results in this study also showed that insulin can significantly reduced the Men1 expression and up-regulated the expression of lipid metabolism-associated genes (Fig. 4) that were also targeted by menin (Fig. 2C). The mechanism of insulin targeting the expression of menin in hepatocytes was also found present in mammary epithelial cells [37]. Furthermore, it was also demonstrated that insulin could regulated the glucose metabolism of liver through FoxO1 [38]. Meanwhile, Wuescher et al. found that insulin regulated the localization of menin in the cytoplasm and the interaction ability with FoxO1 in hepatocytes by Akt [18]. Interestingly, menin can suppress the Akt activity and regulate its cellular localization [38]. In addition, it was found the expression of FoxO1 was controlled by menin in hepatocytes, with promoted transcription upon menin inhibition and inhibited transcription upon menin overexpression (Fig S3). We presumed that there was a negative feedback loop in hepatocytes between menin and FoxO1, which had been found present between menin and miR-24-3p in mammary epithelial cells, too [11]. For example, overexpression of menin inhibited the transcription activity of FoxO1, causing attenuated binding amount of FoxO1 recruited by the adaptor protein menin, and thus inhibited fat deposition. Taken together, these results suggest that menin acts an important mediator in hepatocytes that responds to insulin, modulating its recruitment ability of binding to FoxO1, therefore regulating the activity of genes related to lipid metabolism, such as Cd36 and Gk, and ultimately affects fat deposition in the liver (Fig. 7). A rescue experiment was performed and confirmed this model (Fig S4). The increased expression of Cd36 in insulin treated cells tends to be balanced to normal level (negative control) upon menin over-expression (Ins + mMen1). However, it is worthy to mention that menin might not be the only mediator that responds to insulin in hepatocytes. In another word, insulin might control the glucolipid metabolism in hepatocytes through different ways, since we observed that the expression levels of genes changed more dramatically upon insulin treatment than that happened upon menin inhibition (Figs. 4B and 2C).
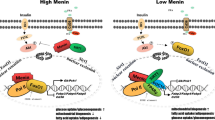
Menin mediates hepatic metabolism through interacting with transcription factor FoxO1. Menin acts an important mediator in hepatocytes that responds to insulin, modulating its recruitment ability of binding to FoxO1, therefore regulating the activity of genes related to lipid metabolism, such as Cd36 and Gk, and ultimately affects fat deposition in the liver. Please see the text for further details. IR insulin receptor, IRS IR substrate, Akt also known as protein kinase B, Pol II RNA polymerase II, Cd36 a fatty acid transporter, Gk Glucokinase. The solid line represents fluxes; The dotted line represents possible effects or fluxes
Although our research revealed part of the mechanism by which menin regulates lipid metabolism in hepatocytes, this study has only been explored in vitro and needs to be studied in mouse models in the future. Although the commonly used NASH mouse model (diet-induced, genetic or a combination of more than one intervention) causes lipid accumulation in the liver and histological appearance is similar to that of NASH patients, the proteomics, lipidomic and transcription Omics is not similar to NASH patients [39, 40]. Studies have shown that the inbred isogenic offspring of a C57BL/6J mouse crossed with a s129/SvlmJ mouse showed obesity, insulin resistance, hypoadiponectinemia, adipose tissue inflammation, dyslipidemia, cell signaling, transcriptomic and lipidomic NASH patients [39, 41]. This NASH mouse model may be widely used in the future.
In summary, our study is the first to show that menin inhibits lipid deposition in hepatocytes by binding to Cd36 and Gk promoter regions associated with lipid metabolism. Moreover, menin interacts with the transcription factor FoxO1 and may participate in insulin-mediated hepatic lipid metabolism by regulating the expression of target genes related to lipid metabolism. Our findings provide new insights into the mechanism of menin liver lipid metabolism and may provide a theoretical basis for the clinical treatment of NAFLD patients.
Data availability
The data used to support the findings of this study are available from the corresponding author upon request.
Abbreviations
- Acc1:
-
Acetyl CoA carboxylase
- Akt, also named as PKB:
-
Protein kinase B
- Cd36:
-
A fatty acid transporter
- FoxO1:
-
Forkhead box O1
- ChIP:
-
Chromatin immunoprecipitation
- DEGs:
-
Differentially expressed genes
- Fas:
-
Fatty acid synthase
- Gk:
-
Glucokinase
- GO:
-
Gene ontology
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- MEN1:
-
Multiple endocrine neoplasia type 1
- NAFLD:
-
Nonalcoholic fatty liver disease
- OA:
-
Oleic acid
- Pparγ:
-
Peroxisome proliferator-activated receptor gamma
- qRT-PCR:
-
Quantitative reverse transcription polymerase chain reaction
- SIRT1:
-
Sirtuin 1
- Srebp-1c:
-
Sterol regulatory element-binding protein 1C
- T2DM:
-
Type 2 diabetes mellitus
- TG:
-
Triglyceride
References
Younossi ZM (2019) Non-alcoholic fatty liver disease - A global public health perspective. J Hepatol 70:531–544. https://doi.org/10.1016/j.jhep.2018.10.033
Younossi ZM, Loomba R, Rinella ME, Bugianesi E, Marchesini G, Neuschwander-Tetri BA, Serfaty L, Negro F, Caldwell SH, Ratziu V, Corey KE, Friedman SL, Abdelmalek MF, Harrison SA, Sanyal AJ, Lavine JE, Mathurin P, Charlton MR, Chalasani NP, Anstee QM, Kowdley KV, George J, Goodman ZD, Lindor K (2018) Current and future therapeutic regimens for nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Hepatology 68:361–371. https://doi.org/10.1002/hep.29724
Rotman Y, Sanyal AJ (2017) Current and upcoming pharmacotherapy for non-alcoholic fatty liver disease. Gut 66:180–190. https://doi.org/10.1136/gutjnl-2016-312431
Chandrasekharappa SC, Guru SC, Manickam P, Olufemi SE, Collins FS, Emmert-Buck MR, Debelenko LV, Zhuang Z, Lubensky IA, Liotta LA, Crabtree JS, Wang Y, Roe BA, Weisemann J, Boguski MS, Agarwal SK, Kester MB, Kim YS, Heppner C, Dong Q, Spiegel AM, Burns AL, Marx SJ (1997) Positional cloning of the gene for multiple endocrine neoplasia-type 1. Science 276:404–407. https://doi.org/10.1126/science.276.5311.404
Song TY, Lim J, Kim B, Han JW, Youn HD, Cho EJ (2014) The role of tumor suppressor menin in IL-6 regulation in mouse islet tumor cells. Biochem Biophys Res Commun 451:308–313. https://doi.org/10.1016/j.bbrc.2014.07.113
Iyer S, Agarwal SK (2018) Epigenetic regulation in the tumorigenesis of MEN1-associated endocrine cell types. J Mol Endocrinol 61:R13-r24. https://doi.org/10.1530/jme-18-0050
Gao SB, Hua X, Jin GH (2008) Menin regulates endocrine diseases by controlling histone modification and gene transcription. Ann Endocrinol (Paris) 69:426–432. https://doi.org/10.1016/j.ando.2008.06.001
Matkar S, Thiel A, Hua X (2013) Menin: a scaffold protein that controls gene expression and cell signaling. Trends Biochem Sci 38:394–402. https://doi.org/10.1016/j.tibs.2013.05.005
Ehrlich L, Hall C, Meng F, Lairmore T, Alpini G, Glaser S (2017) A review of the scaffold protein menin and its role in hepatobiliary pathology. Gene Expr 17:251–263. https://doi.org/10.3727/105221617x695744
Shi K, Liu X, Li H, Lin X, Yan Z, Cao Q, Zhao M, Xu Z, Wang Z (2017) Menin modulates mammary epithelial cell numbers in bovine mammary glands through cyclin D1. J Mammary Gland Biol Neoplasia 22:221–233. https://doi.org/10.1007/s10911-017-9385-8
Qiaoqiao C, Li H, Liu X, Yan Z, Zhao M, Xu Z, Wang Z, Shi K (2019) MiR-24-3p regulates cell proliferation and milk protein synthesis of mammary epithelial cells through menin in dairy cows. J Cell Physiol 234:1522–1533. https://doi.org/10.1002/jcp.27017
Wuescher L, Angevine K, Patel PR, Mensah-Osman E (2012) Menin liver-specific hemizygous mice challenged with high fat diet show increased weight gain and markers of metabolic impairment. Nutr Diabetes 2:e34. https://doi.org/10.1038/nutd.2012.7
Yang Y, Gurung B, Wu T, Wang H, Stoffers DA, Hua X (2010) Reversal of preexisting hyperglycemia in diabetic mice by acute deletion of the Men1 gene. Proc Natl Acad Sci U S A 107:20358–20363. https://doi.org/10.1073/pnas.1012257107
Schnepp RW, Chen YX, Wang H, Cash T, Silva A, Diehl JA, Brown E, Hua X (2006) Mutation of tumor suppressor gene Men1 acutely enhances proliferation of pancreatic islet cells. Cancer Res 66:5707–5715. https://doi.org/10.1158/0008-5472.can-05-4518
Barbu A, Lejonklou MH, Skogseid B (2016) Progranulin stimulates proliferation of mouse pancreatic islet cells and is overexpressed in the endocrine pancreatic tissue of an MEN1 mouse model. Pancreas 45:533–540. https://doi.org/10.1097/mpa.0000000000000509
Cao Y, Xue Y, Xue L, Jiang X, Wang X, Zhang Z, Yang J, Lu J, Zhang C, Wang W, Ning G (2013) Hepatic menin recruits SIRT1 to control liver steatosis through histone deacetylation. J Hepatol 59:1299–1306. https://doi.org/10.1016/j.jhep.2013.07.011
Wu Y, Pan Q, Yan H, Zhang K, Guo X, Xu Z, Yang W, Qi Y, Guo CA, Hornsby C, Zhang L, Zhou A, Li L, Chen Y, Zhang W, Sun Y, Zheng H, Wondisford F, He L, Guo S (2018) Novel mechanism of Foxo1 phosphorylation in glucagon signaling in control of glucose homeostasis. Diabetes 67:2167–2182. https://doi.org/10.2337/db18-0674
Wuescher L, Angevine K, Hinds T, Ramakrishnan S, Najjar SM, Mensah-Osman EJ (2011) Insulin regulates menin expression, cytoplasmic localization, and interaction with FOXO1. Am J Physiol Endocrinol Metab 301:E474–E483. https://doi.org/10.1152/ajpendo.00022.2011
Yue SJ, Zhao YQ, Gu XR, Yin B, Jiang YL, Wang ZH, Shi KR (2017) A genome-wide association study suggests new candidate genes for milk production traits in Chinese Holstein cattle. Anim Genet 48:677–681. https://doi.org/10.1111/age.12593
Yan Z, Wang Z, Zhang Q, Yue S, Yin B, Jiang Y, Shi K (2020) Identification of whole-genome significant single nucleotide polymorphisms in candidate genes associated with body conformation traits in Chinese Holstein cattle. Anim Genet 51:141–146. https://doi.org/10.1111/age.12865
Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Bolund L, Wang J (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34:W293–W297. https://doi.org/10.1093/nar/gkl031
Hashimoto K, Goto S, Kawano S, Aoki-Kinoshita KF, Ueda N, Hamajima M, Kawasaki T, Kanehisa M (2006) KEGG as a glycome informatics resource. Glycobiology 16:63r–70r. https://doi.org/10.1093/glycob/cwj010
Mao X, Cai T, Olyarchuk JG, Wei L (2005) Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21:3787–3793. https://doi.org/10.1093/bioinformatics/bti430
Zhao NQ, Li XY, Wang L, Feng ZL, Li XF, Wen YF, Han JX (2017) Palmitate induces fat accumulation by activating C/EBPβ-mediated G0S2 expression in HepG2 cells. World J Gastroenterol 23:7705–7715. https://doi.org/10.3748/wjg.v23.i43.7705
Yan D, Dou QL, Wang Z, Wei YY (2015) Establishment of a hepatocyte steatosis model using Chang liver cells. Genet Mol Res 14:15224–15232. https://doi.org/10.4238/2015.November.25.10
Rafiei H, Omidian K, Bandy B (2019) Dietary polyphenols protect against oleic acid-induced steatosis in an in vitro model of NAFLD by modulating lipid metabolism and improving mitochondrial function. Nutrients. https://doi.org/10.3390/nu11030541
Engin A (2017) Non-alcoholic fatty liver disease. Adv Exp Med Biol 960:443–467. https://doi.org/10.1007/978-3-319-48382-5_19
Higuchi N, Kato M, Shundo Y, Tajiri H, Tanaka M, Yamashita N, Kohjima M, Kotoh K, Nakamuta M, Takayanagi R, Enjoji M (2008) Liver X receptor in cooperation with SREBP-1c is a major lipid synthesis regulator in nonalcoholic fatty liver disease. Hepatol Res 38:1122–1129. https://doi.org/10.1111/j.1872-034X.2008.00382.x
Luo H, Zhou Y, Hu X, Peng X, Wei H, Peng J, Jiang S (2015) Activation of PPARgamma2 by PPARgamma1 through a functional PPRE in transdifferentiation of myoblasts to adipocytes induced by EPA. Cell Cycle 14:1830–1841. https://doi.org/10.1080/15384101.2015.1033594
Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M (2016) Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 64:73–84. https://doi.org/10.1002/hep.28431
Titchenell PM, Quinn WJ, Lu M, Chu Q, Lu W, Li C, Chen H, Monks BR, Chen J, Rabinowitz JD, Birnbaum MJ (2016) Direct hepatocyte insulin signaling is required for lipogenesis but is dispensable for the suppression of glucose production. Cell Metab 23:1154–1166. https://doi.org/10.1016/j.cmet.2016.04.022
Lai S, Zhou H, Xiong W, Gilbert M, Huang Z, Yu J, Yin W, Wang L, Chen Q, Li Y, Mu D, Zeng L, Ren X, Geng M, Zhang Z, Cui B, Li T, Wang D, Li Z, Wardrop NA, Tatem AJ, Yu H (2017) Changing epidemiology of human brucellosis, China, 1955–2014. Emerg Infect Dis 23:184–194. https://doi.org/10.3201/eid2302.151710
Altomonte J, Cong L, Harbaran S, Richter A, Xu J, Meseck M, Dong HH (2004) Foxo1 mediates insulin action on apoC-III and triglyceride metabolism. J Clin Invest 114:1493–1503. https://doi.org/10.1172/jci19992
Qu S, Altomonte J, Perdomo G, He J, Fan Y, Kamagate A, Meseck M, Dong HH (2006) Aberrant Forkhead box O1 function is associated with impaired hepatic metabolism. Endocrinology 147:5641–5652. https://doi.org/10.1210/en.2006-0541
Zhang W, Patil S, Chauhan B, Guo S, Powell DR, Le J, Klotsas A, Matika R, Xiao X, Franks R, Heidenreich KA, Sajan MP, Farese RV, Stolz DB, Tso P, Koo SH, Montminy M, Unterman TG (2006) FoxO1 regulates multiple metabolic pathways in the liver: effects on gluconeogenic, glycolytic, and lipogenic gene expression. J Biol Chem 281:10105–10117. https://doi.org/10.1074/jbc.M600272200
Ren B, Best B, Ramakrishnan DP, Walcott BP, Storz P, Silverstein RL (2016) LPA/PKD-1-FoxO1 signaling axis mediates endothelial cell CD36 transcriptional repression and proangiogenic and proarteriogenic reprogramming. Arterioscler Thromb Vasc Biol 36:1197–1208. https://doi.org/10.1161/atvbaha.116.307421
Li H, Liu X, Wang Z, Lin X, Yan Z, Cao Q, Zhao M, Shi K (2017) MEN1/Menin regulates milk protein synthesis through mTOR signaling in mammary epithelial cells. Sci Rep 7:5479. https://doi.org/10.1038/s41598-017-06054-w
Os I, Zhang W, Wasserman DH, Liew CW, Liu J, Paik J, DePinho RA, Stolz DB, Kahn CR, Schwartz MW, Unterman TG (2015) FoxO1 integrates direct and indirect effects of insulin on hepatic glucose production and glucose utilization. Nat Commun 6:7079. https://doi.org/10.1038/ncomms8079
Friedman SL, Neuschwander-Tetri BA, Rinella M, Sanyal AJ (2018) Mechanisms of NAFLD development and therapeutic strategies. Nat Med 24:908–922. https://doi.org/10.1038/s41591-018-0104-9
Eng JM, Estall JL (2021) Diet-induced models of non-alcoholic fatty liver disease: food for thought on sugar, fat, and cholesterol. Cells. https://doi.org/10.3390/cells10071805
Asgharpour A, Cazanave SC, Pacana T, Seneshaw M, Vincent R, Banini BA, Kumar DP, Daita K, Min HK, Mirshahi F, Bedossa P, Sun X, Hoshida Y, Koduru SV, Contaifer D Jr, Warncke UO, Wijesinghe DS, Sanyal AJ (2016) A diet-induced animal model of non-alcoholic fatty liver disease and hepatocellular cancer. J Hepatol 65:579–588. https://doi.org/10.1016/j.jhep.2016.05.005
Acknowledgements
We are grateful to Prof. Shaorong Gao in Tongji University for his helpful discussions and excellent suggestions for the project and manuscript. We also thank all present and past members of our laboratory who have contributed comments and ideas.
Funding
This work was financially supported by the Natural Science Foundation of Shandong (ZR2020MC166; ZR2013CM013), the National Key Research and Development Program of China (2021YFD1200900), the National Natural Science Foundation of China (31402054), the Key Project of Agricultural Fine Breeding of Shandong Province (2019LZGC011, 2016LZGC030).
Author information
Authors and Affiliations
Contributions
Shi KR, Wang SX; Liu TJ and Du HX participated in the execution of all experiments, analyzed the data, and drafted the manuscript; Liu TJ, Sun LL, Xu ZJ, and Li RR participated in construction of the expression plasmids, data analysis and discussion; Yu Y and Mao YJ participated in experimental design, coordinated the experiments. All authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest concerning this article.
Ethical approval
All process was completed on the basis of the Guidelines for Care and Use of Laboratory Animals of Shandong Agricultural University and approved by the Animal Ethics Committee of Shandong Agricultural University.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, S., Liu, T., Sun, L. et al. Menin regulates lipid deposition in mouse hepatocytes via interacting with transcription factor FoxO1. Mol Cell Biochem 477, 1555–1568 (2022). https://doi.org/10.1007/s11010-022-04392-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-022-04392-6