Abstract
The baculovirus Autographa californica multiple nucleopolyhedrovirus (AcMNPV) possesses a gene, ac-pcna or ac49, which encodes a protein with similarity to proliferating cell nuclear antigen (PCNA). Homologs of this gene code for DNA polymerase processivity factors and are essential in the DNA replication systems. But the function of ac-pcna still remains unclear. To define the function of Ac-pcna in AcMNPV and Sf-pcna in host Sf9 cells, Bac-to-Bac baculovirus expression system was used to generate two recombinant baculoviruses: AcMNPV-Ac-pcna-EGFP and AcMNPV-Sf-pcna-EGFP. Results indicated that AcMNPV-mediated overexpression of Ac-PCNA and Sf-PCNA could stimulate replication of AcMNPV genome in the host Sf9 cells. Meanwhile, either AcMNPV-Ac-pcna-EGFP or AcMNPV-Sf-pcna-EGFP had a significant stimulating effect on Sf9 genome replication during infection. We also found that Ac-PCNA and Sf-PCNA could promote the production of budded virus. Ac-PCNA could improve the transcription level of ie2 gene dramatically and further improved the transcription of late gene, for example 38 K and vp39, at 12 h p.i.. Moreover, insecticidal potency test showed that the larvae of Beet armyworm in the AcMNPV-Ac-pcna-EGFP and AcMNPV-Sf-pcna-EGFP groups had a higher mortality rate (83.33 and 91.67%), a lower pupation rate (16.67 and 8.33%), and a lower emergence rate (6.67 and 3.33%), compared with those in AcMNPV-EGFP group. The function of Ac-PCNA and Sf-PCNA was confirmed in this study, which provided the theoretical foundation for using and modifying AcMNPV.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Baculoviruses are large, enveloped and double-stranded DNA viruses. It can infect arthropods of the insect specifically orders Lepidoptera, Diptera, and Hymenoptera [1]. As a study model of baculovirus, Autographa californica multiple nucleopolyhedrovirus (AcMNPV) is the most studied and valuable virus in the known insect virus. Its genome approximately consists of a 134 kbp double-stranded DNA and contains 156 putative genes including those encoding DNA replication factors [2]. Baculovirus, as a new type of biological insecticide, is widely used in pest control.
Proliferating cell nuclear antigen (PCNA) expresses only in the nucleus of proliferating cells. PCNA is composed of three end-to-end homologous monomers. Each monomer contains two domains that have similar spatial structures. The two domains are connected by a interdomain connecting loop (IDCL). Therefore, the spatial structure of PCNA is a special hexagonal ring tertiary structure [3, 4]. PCNA is a helper protein for DNA polymerase δ, which can immobilize DNA polymerase δ on DNA strand while DNA is replicated [5]. PCNA forms a mobile platform on DNA, from which it interacts with several protein partners involved in Okazaki strand resolution, DNA modification, and cell-cycle control [6].
The structure of PCNA is highly conserved in different eukaryotic cells, although PCNA is not homologous in sequences [7]. According to bioinformatics analysis, Sf-pcna gene is 2392 bp in length and contains six exons. Its transcript is 1015 bp in length and its coding region is 783 bp. It is predicted to encode a protein containing 260 amino acids. In the course of evolution, baculovirus genomes have been subjected to high levels of gene loss and gene acquisition from their hosts [8]. A gene named AcOrf-49 is 771 bp in length, encodes a protein with sequence homology to PCNA, has been identified in AcMNPV, but its role in DNA replication has not been identified. The ORF of Ac-pcna encodes a 28.635 kDa protein with 256 amino acids residues. Its upstream has early promoter motifs, so it is a delayed early gene.
In our previous work, we identified 459 up-regulated and 112 down-regulated candidate genes with significantly differentially expression between the AcMNPV treatment and untreated Sf9 cells in transcriptome analysis [9]. We found that Sf-pcna gene, a down-regulated candidate gene, might be related to viral proliferation. Based on this work, the present study is to explore the function of Ac-PCNA and Sf-PCNA during AcMNPV infection process.
Materials and methods
Materials
AcMNPV and AcMNPV-EGFP virus, E. coli DH5α, E. coli DH10B, and pFastBacDual-EGFP (P10) were maintained in our laboratory. Sma I, Xho I, and T4 DNA ligase were purchased from Takara Biotechnology (Dalian) Co. Ltd. The anti-β-actin and anti-GFP antibody were purchased from Sangon Biotechnology (Shanghai, China) Co. Ltd. Cell lysis buffer and BCA Protein Assay kit were purchased from Beyotime Biotechnology Corp (Jiangsu, China). First-strand cDNA synthesis superMix and Tip green qPCR SuperMix were purchased from Transgen Biotechnology Corp (Beijing, China). pUCm-T Vector PCR products Cloning Kit was purchased from Sangon Biotechnology (Shanghai, China) Co. Ltd. Roxithromycin, vitamin C, and compound vitamin B tablets were purchased from Tianjin yabao pharmaceutical technology Co. Ltd (Tianjin, China). Beet armyworm (Spodoptera exigua Hubner) eggs were purchased from Jiyuan Baiyun Biotechnology Corp Ltd (Jiyuan, China).
Cell culture
Cell cultures were prepared and maintained according to standard cell culture procedures. The Sf9 cells were kindly provided by Prof. Jianzhen Zhang (Institute of Applied Biology, Shanxi University) and cultured at 27 °C in SIM SF serum-free insect cell culture medium (Sino Biological Inc., Beijing, China) [10].
Chemicals
All chemicals were reagent grade and purchased from Sangon Biotechnology (Shanghai, China) Co. Ltd. PBS buffer: 6.5 mM Na2HPO4, 2.9 mM NaH2PO4, 141 mM NaCl; 5 × SDS-PAGE loading buffer: 250 mM Tris/HCI (pH 6.8), 10% SDS, 0.5% bromophenol blue, 10% (v/v) glycerol, and 5% (v/v) 2-mercaptoethanol; 1 × Tris/glycine buffer: 25 mM Tris base, 190 mM glycine, 0.1% SDS, pH 8.3; Western transfer buffer: 192 mM Glycine, 25 mM Tris, 20% methanol; PBST: 6.5 mM Na2HPO4, 2.9 mM NaH2PO4, 141 mM NaCl, 0.1% Tween 20; Blocking buffer: 6.5 mM Na2HPO4, 2.9 mM NaH2PO4, 141 mM NaCl, 0.1% Tween 20 and 10% (w/v) powdered milk.
Prediction of three-dimensional (3D) structure of Ac/Sf-PCNA
The amino acid sequence of Ac-PCNA and Sf-PCNA was aligned using DNAMAN (Fig. 1a). We obtained domains of Ac/Sf-PCNA in the GenBank database and SMART (Simple Modular Architecture Research Tool) database. Then, these sequences were submitted to the Swiss-Model server, which was a fully automated protein structure homology-modeling server, to predict the three-dimensional structure of Ac/Sf-PCNA (Fig. 1b, c).
Amino acid sequence alignment and prediction of three-dimensional structure of Ac/Sf-PCNA. a Sequence alignment analysis of Ac-PCNA and Sf-PCNA. The amino acid sequence of Ac-PCNA and Sf-PCNA was aligned using DNAMAN. Conserved residues were marked with black notes. b 3D simulation structure of Ac-PCNA. Swiss-Model server was used to predict the three-dimensional structure of Ac-PCNA. Distribution of N-terminal domain in Ac-PCNA was marked with yellow notes. And distribution of C-terminal domain in Ac-PCNA was marked with blue notes. c 3D simulation structure of Sf-PCNA. (Color figure online)
Construction of AcMNPV-Ac-pcna-EGFP and AcMNPV-Sf-pcna-EGFP
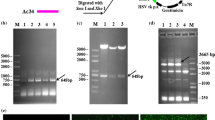
For generation of AcMNPV-Ac-pcna-EGFP and AcMNPV-Sf-pcna-EGFP, the transcription of Ac-pcna gene and Sf-pcna gene was driven by P10 promoter. As shown in Fig. 2a and b, PCR amplification of 771 bp Ac-pcna gene was performed by AcMNPV genome, forward primer (5′-TACCCGGGATGTTCGAAGCGGAA-3′) and reverse primer (5′-GCTCGAGGAAAAATTTCCTCGTCGTT-3′). As shown in Fig. 3a and c, 783 bp Sf-pcna gene was also amplified by PCR using Sf9 cDNA, forward primer (5′-TACCCGGGTATGTTCGAAGCGCGT-3′) and reverse primer (5′-GCCTCGAGGACTCTCTTCTTCCTC-3′). Two PCR products were cloned into the pFastBacDual-EGFP vector, respectively, to generate the transfer vector pFastBacDual-Ac-pcna-EGFP and pFastBacDual-Sf-pcna-EGFP. These plasmids, pFastBacDual-Ac-pcna-EGFP and pFastBacDual-Sf-pcna-EGFP, were transformed into the Escherichia coli strain DH10 Bac and transposed into the AcMNPV genome, followed by PCR verification with primer M13-F (5′-CGCCAGGGTTTTCCCAGTCACGAC-3′) and M13-R (5′-CACACAGGAAACA GCTATGAC-3′). On the other hand, the existence of gentamicin- and kanamycin-resistant genes in the recombinant bacmid could act as selection markers. The transformed DH10B cells were incubated at 37 °C for 4 h in SOC (0.5% yeast extract, 2% tryptone, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, 20 mM glucose) medium with gentle shaking before being spread onto an LB agar plate containing kanamycin and gentamicin. Positive clones with both gentamicin and kanamycin resistance were selected after 48-h incubation at 37 °C. For transfection, ~ 2 × 105 Sf9 cells were cultured in 24-well plate and then transfected with 1 µg recombinant Bacmid-Ac-pcna-EGFP bacmid DNA and Bacmid-Sf-pcna-EGFP bacmid DNA, respectively, using 3 µL TransFast reagent solution (Promega, USA) according to the manufacturer’s specification. At 120-h post-transfection, cells were examined for monitoring enhanced green fluorescent protein (EGFP) expression using fluorescence microscopy and supernatant from the transfection was harvested to infect a new batch of Sf9 cells to, respectively, generate AcMNPV-Ac-pcna-EGFP and AcMNPV-Sf-pcna-EGFP.
Construction of recombinant AcMNPV-Ac-pcna-EGFP. a Schematic representation of pFB-D-Ac-pcna-EGFP. b PCR amplification of Ac-pcna gene. Lane 1: pFastBacDual-EGFP (P10) plasmid. Lanes 2–3: PCR product of Ac-pcna gene from AcMNPV genome. The arrow showed the Ac-pcna gene fragment. c Identification of recombination plasmid pFastBauDual-Ac-pcna-EGFP. Lanes 1–2: pFB-D-Ac-pcna-EGFP plasmid. Lane 3: negative control. Lanes 4–5: PCR product of Ac-pcna gene from pFB-D-Ac-pcna-EGFP. Lanes 6–7: Digestion product of pFB-D-Ac-pcna-EGFP by Sma I and Xho I. The arrow showed the Ac-pcna gene fragment. d Identification of recombination Bacmid-Ac-pcna-EGFP. Lanes 1–4: PCR product of Ac-pcna-EGFP-Gentamicin fused gene from Bacmid-Ac-pcna-EGFP templet. The arrow showed Ac-pcna-EGFP-Gentamicin fused gene fragment. e Fluorescence microscopic observation of three generations of recombinant AcMNPV-Ac-pcna-EGFP. f Western blot analysis of expression level of Ac-PCNA during the infection process in Sf9 cells. g Photograph of the AcMNPV-Ac-pcna-EGFP-infected Sf9 cells sample obtained with a fluorescence microscope. The fluorescence intensity of EGFP reflected the amount of progeny virus production. The second generation virus was obtained from the Sf9 cells infected with the first generation one, and so on. Scale bar: 100 µm
Construction of the recombinant AcMNPV-Sf-pcna-EGFP. a Schematic representation of pFB-D-Sf-pcna-EGFP. b Electrophoresis analysis of total RNA of Sf9 cells (Lanes 1–2). c PCR amplification of Sf-pcna gene. Lanes 1–2: PCR product of Sf-pcna gene from the cDNA of Sf9 cells. The arrow showed the Sf-pcna gene fragment. d Identification of recombination plasmid pFastBauDual-Sf-pcna-EGFP. Lanes 1–4: Digestion product of pFB-D-Sf-pcna-EGFP by Sma I and Xho I. The arrow showed the Sf-pcna gene fragment. e Identification of recombination bacmid Bacmid-Sf-pcna-EGFP. Lanes 1–4: PCR product of Sf-pcna-EGFP-Gentamicin fused gene from Bacmid-Sf-pcna-EGFP templet. The arrow showed Sf-pcna-EGFP-Gentamicin fused gene fragment. f Fluorescence microscopic observation of three generations of the recombinant baculovirus AcMNPV-Sf-pcna-EGFP. g Western blot analysis of expression level of Sf-PCNA during the infection process in Sf9 cells. h Photograph of the AcMNPV-Sc-pcna-EGFP-infected Sf9 cells sample obtained with a fluorescence microscope. Scale bar: 100 µm
Plaque assay
Sf9 insect cells were seeded in 6-well plate at 50% confluence, and 1 h later the medium was replaced with 1 mL serum-free insect cell culture medium. After 30-min incubation at room temperature, 500 µL serially diluted AcMNPV-Ac-pcna-EGFP and AcMNPV-Sf-pcna-EGFP was respectively added to each well and incubated for 1 h at room temperature. The cells were washed once with fresh serum-free insect cell culture medium before 1.5% (w/v) nutrient agarose overlay was added. Plaques were counted 7 days later [11].
Western blot analysis
Monolayer cultures of 6 × 106 Sf9 cells in 6-well microtiter plates were infected with AcMNPV-Ac/Sf-pcna-EGFP at a multiplicity of infection (MOI) of 5, respectively. The total protein and nuclear protein samples were, respectively, boiled in 5 × SDS-PAGE loading buffer at 95 °C for 10 min, and indicated amounts of proteins were separated using 10% (w/v) SDS-PAGE and transferred to PVDF membranes for western blot analysis. The membranes were blocked for 1 h in blocking buffer with slow rotation on an orbital shaker. PVDF membranes incubated with primary anti-β-actin (diluted at 1:2000) and anti-GFP antibody (diluted at 1:2000) overnight at 4 °C, washed three times with PBST for 15 min. Blots were incubated with species-specific secondary (horseradish peroxidase-conjugated goat anti-rabbit IgG, diluted at 1:20,000) for 2 h with slow rotation on an orbital shaker, washed three times with PBST for 15 min and then developed using Odyssey infrared imaging system (LI-COR, USA).
Absolute quantification PCR analysis of genome DNA replication of AcMNPV and Sf9 cells
In order to draw the standard curve, the upstream sequences derived from the AcMNPV hr3 region and the unique section derived from the Sf21 hsp90 gene were cloned into the vector pUCm-T to form the standard: pUCm-T-hr3 and pUCm-T-hsp90, which was used to detect the copy number of the viral genome and the cell genome, respectively. After the standard was gradient diluted (10−1–10−7), the copy number corresponding to different concentrations of plasmid was calculated according to the formula: plasmid copy number (copies/μL) = 6.022 × 1023 (copies/mol) × plasmid concentrations (ng/μL)/plasmid molecular weight (ng/mol). And then according to the different copy number standard corresponding to the Ct value, the standard curve was drawn and it could reflect the linear relationship of Ct value and the copy number. When the sample was measured, the templates of the standard and the sample were amplified simultaneously. And the initial copy number of the sample was obtained according to the Ct value of the unknown sample and the standard curve.
An equivalent number of Sf9 cells was added into a 12-well plate, followed by adding the medium to give 1000 µL. Sf9 cells were infected with AcMNPV, AcMNPV-Ac-pcna-EGFP and AcMNPV-Sf-pcna-EGFP up to 72 h, at a multiplicity of infection (MOI) of 5, respectively. The supernatant and cell precipitation were respectively collected to extract total genome DNA. The copy number of Sf9 cells and virus genome DNA were, respectively, quantitated by absolute quantification PCR analysis. The sequence of primers used to amplify Ac-pcna, Sf-pcna, hr3, and hsp90 were listed in Supplement Table 1. Then, amplification of hr3 was used to detect copy number of virus genome DNA, and amplification of hsp90 was used to detect copy number of Sf9 cells genome DNA. Promotion rate was calculated according to the following equation: promotion rate (%) = (T − C)/C × 100%. T and C represented the copy number of genome of the treated sample and the control, respectively.
Budded virus (BV) production in Sf9 cells
To determine the effect of AcMNPV-Ac-pcna-EGFP and AcMNPV-Sf-pcna-EGFP on BV virus production in Sf9 cells, monolayer culture of 3 × 106 Sf9 cells in 6-well plate were infected with AcMNPV-EGFP, AcMNPV-Ac-pcna-EGFP, and AcMNPV-Sf-pcna-EGFP for 1-4 d post-infection (d p.i.) at a MOI of 5, respectively. Followed by incubation for 2 h, the virus-containing culture medium was removed and fresh medium was added. Culture medium and cells were harvested at different detection time points. Cells were resuspended in PBS buffer and released the viruses by repeated freeze–thaw cycles. BV production was determined by the plaque assay.
Quantitative RT-PCR analysis
Total RNA was isolated from infected Sf9 cells using Trizol. Single-strand cDNA was synthesized from total RNAs using an EasyScript First Strand cDNA Synthesis SuperMix for RT-PCR. The qPCR was conducted with TransStart Tip Green qPCR SuperMix and the StepOne™ Real-Time PCR System (Applied Biosystem). The β-actin gene was used as a housekeeping reference gene. The sequence of primers used to amplify 38 K, vp39, ie2, and β-actin was listed in Supplement Table 2. The cycling profile used for qPCR was as follows: a preheating step for enzyme activation at 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, and 60 °C for 30 s. The relative transcription level was calculated by using the 2−ΔΔCT method.
Anti-insect activity analysis
As the permissive host for AcMNPV, the larvae of Beet armyworm were used to compare the virulence of AcMNPV-EGFP, AcMNPV-Ac-pcna-EGFP, and AcMNPV-Sf-pcna-EGFP. The larvae were reared in 24-well microtiter plates with artificial diet, under the condition of 28 ± 2 °C, 70–80% humidity, and natural photoperiod. The biological activity against second instar larvae of the recombinant virus AcMNPV-Ac-pcna-EGFP or AcMNPV-Sf-pcna-EGFP was evaluated. The mortality, pupation, and emergence rate of larvae in AcMNPV-Ac/Sf-pcna-EGFP and AcMNPV-EGFP treatment groups were determined by feeding of 2rd instar larvae of Beet armyworm on a piece of artificial diet (27 mm3) with 20 μL, 1 × 107 pfu mL−1 of each viruses daily for five days, respectively. There were 60 larvae in each treatment group. The average weight, mortality, pupation, and emergence rate of larvae were recorded daily.
Statistical analysis
For each experiment, three independent experiments were performed. Statistical significance was assessed using t test by using excel. Results were considered significant for P < 0.05 and very significant for P < 0.01 or P < 0.001.
Results
Amino acid sequence alignment and prediction of 3D structure of Ac/Sf-PCNA
Homologous sequence alignment showed that Ac-PCNA had 43.73% amino acid sequence homology with Sf-PCNA, but the C-terminal and N-terminal regions in these proteins were highly conservative (Fig. 1a). Swiss-Model server and SwissPdbViewer 3.7 software were used to predict the three-dimensional structure of Ac/Sf-PCNA (Fig. 1b, c). Ac-PCNA and Sf-PCNA had the high homology with 4cs5.1.A template in PDB database, so their structures were constructed by using 4cs5.1.A model as the template. Distribution of N-terminal domain in Ac/Sf-PCNA was marked with yellow notes. And distribution of C-terminal domain in Ac/Sf-PCNA was marked with blue notes. For Ac-PCNA, the N-terminal domain corresponded to 1–122 amino acids residues, and the C-terminal domain corresponded to 125–247 amino acids residues. For Sf-PCNA, the N-terminal domain corresponded to 1–124 amino acids residues, and the C-terminal domain corresponded to 127–254 amino acids residues. The template 4cs5.1.A is the Pacific white leg shrimp Litopenaeus vannamei Proliferating Cell Nuclear Antigen (LvPCNA). This protein is a member of the sliding clamp family of proteins, that binds DNA replication and DNA repair proteins through a motif called PIP-box (PCNA-Interacting Protein) [12].
Construction of recombinant virus AcMNPV-Ac-pcna-EGFP and AcMNPV-Sf-pcna-EGFP
As shown in Fig. 2c, enzyme digestion identification and DNA sequencing confirmed pFastBauDual-Ac-pcna-EGFP was constructed successfully. PCR product of Ac-pcna-EGFP-Gentamicin fused gene (3788 bp) from Bacmid-Ac-pcna-EGFP templet was shown in Fig. 2d. The Bacmid-Ac-pcna-EGFP was transfected into Sf9 cells. At 120 h p.t., cells were examined for EGFP expression by fluorescence microscopy and the supernatant was harvested to infect a new batch of Sf9 cells to generate AcMNPV-Ac-pcna-EGFP (Fig. 2e). Western blot analysis and green fluorescence detection showed that the expression of Ac-PCNA-EGFP was detectable at 12 h p.i. in Sf9 cells, and the expression of Ac-PCNA-EGFP were significantly enhanced with the infection time (Fig. 2f, c). Similar to the construction process of AcMNPV-Ac-pcna-EGFP, the plasmid pFastBauDual-Sf-pcna-EGFP, Bacmid-Sf-pcna-EGFP, and AcMNPV-Sf-pcna-EGFP were constructed and sequencing confirmed (Fig. 3a–f). And the expression level of Sf-PCNA-EGFP was significantly enhanced with the infection time (Fig. 3g, h).
AcMNPV-mediated overexpression of Ac-PCNA and Sf-PCNA improved virus DNA replication in Sf9 cells
To analyze the effect of AcMNPV-Ac-pcna-EGFP and AcMNPV-Sf-pcna-EGFP on virus DNA replication, we examined the copy number of virus genome extracted from Sf9 cells precipitation and supernatant infected with AcMNPV, AcMNPV-Ac-pcna-EGFP and AcMNPV-Sf-pcna-EGFP, respectively. As shown in Fig. 4a, the copy number of the virus genome increased gradually after infection with the recombinant virus AcMNPV-Ac-pcna-EGFP, AcMNPV-Sf-pcna-EGFP during 36–72 h p.i., and reached the maximum value at 72 h p.i. However, the copy number of the intracellular virus genome was essentially constant and maintained at a low level after infection with the wild-type virus AcMNPV. Moreover, the copy number of the viral genomic DNA in the AcMNPV-Ac-pcna-EGFP treatment group was 110.86, 560.48, and 731.29 times higher than that in wild-type virus treatment group at 36, 48, and 72 h p.i., respectively. And the copy number of the viral genomic DNA in the AcMNPV-Sf-pcna-EGFP treatment group was 45.08, 37.50, and 909.89 times higher than that in wild-type virus treatment group, respectively. These results suggested that Ac-PCNA and Sf-PCNA could promote the replication of virus genome DNA.
Ac-PCNA and Sf-PCNA improved DNA replication of virus and Sf cells. a Absolute quantification PCR analysis of copy number of AcMNPV genome in infected Sf9 cells. b Absolute quantification PCR analysis of copy number of AcMNPV genome in medium. Sf9 cells were infected with AcMNPV and AcMNPV-Ac/Sf-pcna-EGFP at 36 h–72 h p.i., respectively. Absolute quantification PCR was performed to determine AcMNPV genome DNA replication. In order to draw the standard curve, pUCm-T-hr3 was used to amplify hr3. c Absolute quantification PCR analysis of copy number of Sf9 genome in infected Sf9 cells. Sf9 cells were infected with AcMNPV and AcMNPV-Ac/Sf-pcna-EGFP at 36 h–72 h p.i, respectively. Absolute quantification PCR was performed to determine Sf9 genome DNA replication. In order to draw the standard curve, pUCm-T-hsp90 was used to amplify hsp90. Data shown are mean values of triplicate assays ± SD. The error bars represent standard deviations. Student’s t tests were performed on the two selected important data. **Means the difference is significant at the 0.01 level. d The promotion rate analysis of AcMNPV and Sf9 genome in the AcMNPV-Ac-pcna-EGFP and AcMNPV-Sf-pcna-EGFP treatment groups
In the meantime, Fig. 4b shows virus genome copies in supernatant of cells in three treatment groups. The copy number of the viral genome in the extracellular supernatant increased slowly and reached the maximum at 72 h p.i., when the cells were infected with the recombinant virus AcMNPV-Ac-pcna-EGFP or AcMNPV-Sf-pcna-EGFP. And the copy number of the viral genome in the extracellular supernatant increased sharply at 72 h p.i. after infection with wild-type virus AcMNPV. So, the number of viral genome copies in AcMNPV-Ac-pcna-EGFP and AcMNPV-Sf-pcna-EGFP treatment groups was most significantly increased compared with that in AcMNPV-treated group during the infection process. The copy number of the viral genomic DNA in the AcMNPV-Ac-pcna-EGFP treatment group was 1919.63, 2100.42, and 399.78 times higher than that in wild-type virus treatment group and the copy number of the viral genomic DNA in the AcMNPV-Sf-pcna-EGFP treatment group was 4328.29, 4190.15, and 792.69 times higher than that in wild-type virus treatment group at three time points. It also confirmed that Ac-PCNA and Sf-PCNA could promote the replication of virus genome DNA.
AcMNPV-Ac-pcna-EGFP and AcMNPV-Sf-pcna-EGFP could improve DNA replication of host Sf9 cells
To analyze the effect of AcMNPV-Ac-pcna-EGFP and AcMNPV-Sf-pcna-EGFP on Sf9 genome DNA replication, we examined the copy number of Sf9 cells genome extracted from Sf9 cells precipitation infected with AcMNPV, AcMNPV-Ac-pcna-EGFP, and AcMNPV-Sf-pcna-EGFP, respectively. As shown in Fig. 4c, the copy number of the host genome was detected and there were significantly differences among these treatment groups during the virus infection process. The copy number of Sf9 genome increased gradually after infection with the AcMNPV, AcMNPV-Ac-pcna-EGFP, AcMNPV-Sf-pcna-EGFP during 36-72 h p.i. and reached the maximum value at 72 h p.i. However, the copy number of Sf9 genome was increased slowly after infection with the wild-type virus. Furthermore, compared with that in black treatment group, the copy number of host genomic DNA was low in general in wild-type virus treatment group at all times. Nevertheless, the copy number of the host genomic DNA in recombinant virus treatment groups was higher than that in black treatment group at 72 h p.i. The copy number of Sf9 genomic DNA in the AcMNPV-Ac-pcna-EGFP treatment group was 9.83 and 270.27 times higher than that in wild-type virus treatment group at 36 and 72 h p.i., respectively. And the copy number of the Sf9 genomic DNA in the AcMNPV-Sf-pcna-EGFP treatment group was 147.39, 29.30, and 31.58 times higher than that in wild-type virus treatment group at 36, 48, and 72 h p.i., which suggested that AcMNPV-Ac-pcna-EGFP and AcMNPV-Sf-pcna-EGFP had influence on Sf9 genome DNA replication, and Ac-PCNA and Sf-PCNA could promote the replication of Sf9 genome DNA. In order to continue the virus replication in the host cell, both wild-type virus and recombinant virus would inhibit apoptosis of the host cells and promoted replication of the host cells genome, although they had a negative impact on the cells in the process of infection.
In order to compare the promotion rate of the host and viral genomic DNA replication after the infection of AcMNPV-Ac-pcna-EGFP and AcMNPV-Sf-pcna-EGFP, the histogram was plotted. As shown in Fig. 4d, in the two recombinant virus treatment groups, the promotion rate of the virus genome was much higher than that of the host genome. The promotion rate of the virus genome of AcMNPV-Sf-pcna-EGFP treatment group was 1.24 fold of that in AcMNPV-Ac-pcna-EGFP treatment group at 72 h p.i., and the promotion rate of the Sf9 genome was higher in AcMNPV-Sf-pcna-EGFP treatment group at 36 and 48 h p.i. It indicated that the promoting effect of Ac-PCNA and Sf-PCNA on the genome of the virus was significantly stronger than these on the genome of the host. Compared with the Ac-PCNA, Sf-PCNA was more capable of promoting viral and host genome replication.
Ac-PCNA and Sf-PCNA promoted budded virus (BV) production in Sf9 cells
In order to determine when Ac-PCNA and Sf-PCNA entered the nucleus and played function as nuclear proteins, western blot analysis of EGFP, Ac-PCNA-EGFP, and Sf-PCNA-EGFP expression in the nucleus was performed at 12–144 h p.i., respectively. As shown in Fig. 5a, the expression of Ac-PCNA-EGFP was at the beginning of 12 h p.i., and continued to increase until 72 h p.i., and gradually decreased at 72–144 h p.i. The expression of Sf-PCNA-EGFP was at the beginning of 24 h p.i., and continued to increase until 96 h p.i., and gradually decreased at 96–144 h p.i. The nuclear location of EGFP was not detected until 144 h p.i., which was caused by rupture of the nuclear membrane in the late infection process. Western blot for nucleoprotein indicated that expression of Ac-PCNA-EGFP could be detected at 12 h p.i., which was 12 h ahead of expression of Sf-PCNA-EGFP. Combining Figs. 2f and 3g, we found that the initiation time of expression of Ac-PCNA and Sf-PCNA was consistent with the time when they entered into the nucleus, respectively. In summary, Ac-PCNA and Sf-PCNA played their roles as soon as they transferred into the nucleus.
Effects of Ac-PCNA and Sf-PCNA on the BV production. a Western blot analysis of nuclear accumulation of Ac-PCNA-EGFP, Sf-PCNA-EGFP, and EGFP in AcMNPV-EGFP- and AcMNPV-Ac/Sf-pcna-EGFP-infected Sf9 cells. Sf9 cells were infected with AcMNPV-EGFP, AcMNPV-Ac/Sf-pcna-EGFP for 12–144 h p.i. at a MOI of 5. b Viral titer analysis of BV in AcMNPV-EGFP and AcMNPVAc/Sf-pcna-EGFP treatment groups. Error bars represent the standard deviation. Double asterisk means the difference is significant at the 0.01 level
It is common knowledge that in order to expedite amplification, baculovirus could inhibit host cells apoptosis which was induced by virus DNA replication. So, we examined BV production in virus-infected cells. As shown in Fig. 5b, there had more BV production in the AcMNPV-Ac-pcna-EGFP- and AcMNPV-Sf-pcna-EGFP- infected cells compared with that in AcMNPV treatment groups during the virus infection process. BV production basically maintained at the highest level at 72–96 h p.i. in three treatment groups. BV production in AcMNPV-Ac-pcna-EGFP- and AcMNPV-Sf-pcna-EGFP-infected cells, respectively, was 14.53 and 19.83 fold of that in AcMNPV-infected cells at 72 h p.i. In addition, BV production in AcMNPV-Sf-pcna-EGFP-infected cells was higher than that in AcMNPV-Ac-pcna-EGFP-infected cells at 36-96 h p.i., which was 2.74, 1.45, 1.37, and 1.42 fold of that in AcMNPV-Ac-pcna-EGFP-infected cells at 36–96 h p.i., respectively. In total, Ac-PCNA and Sf-PCNA promoted budded virus (BV) production in Sf9 cells and Sf-PCNA had better promoting effect on the production of BV.
Ac-PCNA was more favorable to improve the transcription level of 38 K, vp39, and ie2 gene
The process of viral DNA replication is considered essential for late gene expression derived BV formation. Since the expression of late genes in baculovirus is coupled to viral DNA replication. Transcription of late genes is dependent upon DNA replication. Then, quantitative RT-PCR was performed to detect the transcription level of late genes, 38 K and vp39. As shown in Fig. 6a and b, the transcription level of 38 K and vp39 genes reached maximum value at 12 h p.i. in the AcMNPV-Ac-pcna-EGFP treatment group and it was 53.87 and 38.47 times of that in AcMNPV-EGFP treatment group at 12 h p.i., respectively. Compared with that in AcMNPV-EGFP treatment group, the transcription level of 38 K and vp39 genes was lower integrally at 24–48 h p.i. in the AcMNPV-Ac-pcna-EGFP treatment group. But it increased gradually in the AcMNPV-Ac-pcna-EGFP treatment group and was similar to that in AcMNPV-EGFP treatment group.
Transcription level analysis of 38 K, vp39, and ie2 gene in AcMNPV-Ac-pcna-EGFP-infected Sf9 cells. a–c Transcription level analysis of 38 K, vp39, and ie2 gene was brought forward in AcMNPV-Ac-pcna-EGFP treatment group. Data shown are mean values of triplicate assays ± SD. Error bars represent the standard deviation. Student’s t tests were performed on the two selected important data. **Means the difference is significant at the 0.01 level
IE2 is the main transactivator in baculovirus and is involved in the activation of late expression factor genes. To look further into the relationship among Ac-pcna, ie2, 38 K, and vp39, we examined the role of Ac-pcna in regulating the expression of ie2 gene. As shown in Fig. 6c, compared with that in AcMNPV-EGFP treatment group, the transcription level of ie2 gene was increased significantly during the early infection stage (before 24 h p.i.) and dropped slowly during the late infection stage (after 24 h p.i.) in the AcMNPV-Ac-pcna-EGFP treatment group. The transcription level of ie2 gene in the AcMNPV-Ac-pcna-EGFP treatment group, respectively, was 10.80, 13.13, and 3.82 fold of that in AcMNPV-EGFP treatment group at 12, 24, and 36 h p.i., respectively. These results confirmed that Ac-PCNA was more favorable to improve the transcription level of 38 K, vp39, and ie2 gene.
AcMNPV-Sf-pcna-EGFP had higher anti-insect activity than AcMNPV-Ac-pcna-EGFP
In this section, the anti-insect activity analysis was performed to detect insecticidal rate of AcMNPV-Sf-pcna-EGFP and AcMNPV-Ac-pcna-EGFP. As shown in Fig. 7 and Supplement Tables 3–6, compared with control and AcMNPV-EGFP treatment groups, the larvae in the recombinant virus treatment groups were growing slowly, and had higher mortality rate, lower pupation rate and emergence rate. Moreover, the results in AcMNPV-Ac-pcna-EGFP and AcMNPV-Sf-pcna-EGFP treatment groups were compared with each other. As shown in Fig. 7a, the average growth rate of larvae in the AcMNPV-Sf-pcna-EGFP treatment group was faster than that in the AcMNPV-Ac-pcna-EGFP treatment group at the middle infection stage (9–12 d p.i.); compared with AcMNPV-Ac-pcna-EGFP treatment group, the larvae in AcMNPV-Sf-pcna-EGFP treatment group were slowly growing at the late infection stage (12–13 d p.i.). As shown in Fig. 7b, the average mortality rate of larvae in the AcMNPV-Ac-pcna-EGFP and AcMNPV-Sf-pcna-EGFP treatment groups were 83.33 and 91.67%, respectively. Pupation rate was 16.67 and 8.33% in the AcMNPV-Ac-pcna-EGFP and AcMNPV-Sf-pcna-EGFP treatment groups, respectively (Fig. 7c). Emergence rate is also one of the important indices for pesticide activity; it was 6.67 and 3.33% in the AcMNPV-Ac-pcna-EGFP and AcMNPV-Sf-pcna-EGFP treatment groups, respectively (Fig. 7d). And the half-lethal time of the AcMNPV-Sf-pcna-EGFP treatment group was shortest in three virus-treated groups (Supplement Table 4). From the above results, we concluded that recombinant AcMNPV-Sf-pcna-EGFP had higher anti-insect activity than AcMNPV-Ac-pcna-EGFP.
Anti-insect activity analysis of AcMNPV-Ac/Sf-pcna-EGFP. a Weight analysis of Beet armyworm larvae infected by the virus. Second instar larvae of Beet armyworm were fed with 20 µL, 1 × 107 pfu/mL AcMNPV-EGFP, AcMNPV-Ac-pcna-EGFP, and AcMNPV-Sf-pcna-EGFP by adding to the surface of the small piece of diet (27 mm3) daily for five days, respectively. Each treatment group had 60 larvae. Control treatment consisted of uninfected larvae. The regression analysis (by SigmaPlot software) showed the relationship between the weight of larvae and treatment time (d p.i.). b–d Mortality rate, pupation rate, and emergence rate analysis, respectively
Discussion
The study of baculovirus has become a hot spot due to their specificity in killing pest since chemical pesticides has made human health in danger. In the baculoviral development cycles, DNA replication is the central link. The proliferating cell nuclear antigen-like protein gene (pcna) is associated with baculoviral genome DNA replication. In transcriptome analysis, we identified Sf-pcna gene, a down-regulated candidate gene, with significantly differentially expression in between the AcMNPV treatment and untreated Sf9 cells [9]. In this study, the function of Ac-PCNA and Sf-PCNA was confirmed in vitro and in vivo. Ac-PCNA and Sf-PCNA were confirmed to stimulate virus DNA replication and induce Sf9 cell DNA replication. Both Ac-pcna and Sf-pcna were necessary for budded virus (BV) production by stimulating replication of virus genome DNA. Crawford et al. reported that cells infected with a recombinant virus where the C terminus of the vPCNA gene was deleted, exhibited a slight delay in late gene expression [13]. However, in 1999, it was reported that the viral homolog of pcna did not influence late gene expression in transient transfection assay [14]. In this study, Ac-PCNA could stimulate replication of virus genome DNA and further was benefit to budded virus (BV) production, which was achieved by promoting the expression of late genes.
Our experiments also demonstrated that Ac-pcna promoted the expression of late genes 38 K and vp39 by promoting the transcription of ie2. Most of the late gene-encoded proteins associated with viral structures. The advanced expression of late genes is favorable for the assembly of mature viruses. The expression of the immediate early genes is entirely dependent on the expression product of the host cell. And increased copy number of genomic DNA could promote the expression of immediate early genes, ie1 and ie2, the major transactivators in baculovirus. Both of them are involved in the activation of late expression factor genes, which are necessary for virus propagation [15]. Meantime, it is known that the 39 K gene requires both IE1 and IE2 activators for efficient activation in insect cells [16].
Finally, these recombinant baculoviruses, AcMNPV-Ac-pcna-EGFP and AcMNPV-Sf-pcna-EGFP, had effective insecticidal activity, which was due to the large proliferation of progeny virus caused by the promotion of genome DNA replication by Sf-PCNA and Ac-PCNA.
In conclusion, this study disclosed the function and mechanism of Sf-PCNA and Ac-PCNA during the AcMNPV infection process.
References
Peng K, Wu M, Deng F, Song J, Dong C, Wang H, Hu Z (2010) Identification of protein-protein interactions of the occlusion-derived virus-associated proteins of Helicoverpa armigera nucleopolyhedrovirus. J Gen Virol 91:659–670
Ayres MD, Howard SC, Kuzio J, Lopez-Ferber M, Possee RD (1994) The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202:586–605
Takahashi T, Caviness VS Jr (1993) PCNA-binding to DNA at the G1/S transition in proliferating cells of the developing cerebral wall. J Neurocytol 22:1096–1102
Maga G, Hubscher U (2003) Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J Cell Sci 116:3051–3060
Xing G, Kirouac K, Shin YJ, Bell SD, Ling H (2009) Structural insight into recruitment of translesion DNA polymerase Dpo4 to sliding clamp PCNA. Mol Microbiol 71:678–691
De Biasio A, Blanco FJ (2013) Proliferating cell nuclear antigen structure and interactions: too many partners for one dancer? Adv Protein Chem Struct Biol 91:1–36
Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J (1994) Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 79:1233–1243
Herniou EA, Olszewski JA, Cory JS, O’Reilly DR (2003) The genome sequence and evolution of baculoviruses. Annu Rev Entomol 48:211–234
Wei L, Cao L, Miao Y, Wu S, Xu S, Wang R, Du J, Liang A, Fu Y (2017) Transcriptome analysis of Spodoptera frugiperda 9 (Sf9) cells infected with baculovirus, AcMNPV or AcMNPV-BmK IT. Biotechnol Lett 39:1129–1139
Mehrabadi M, Hussain M, Matindoost L, Asgari S (2015) The baculovirus antiapoptotic p35 protein functions as an inhibitor of the host RNA interference antiviral response. J Virol 89:8182–8192
Wu C, Wang S (2012) A pH-sensitive heparin-binding sequence from Baculovirus gp64 protein is important for binding to mammalian cells but not to Sf9 insect cells. J Virol 86:484–491
Carrasco-Miranda JS, Lopez-Zavala AA, Arvizu-Flores AA, Garcia-Orozco KD, Stojanoff V, Rudiño-Piñera E, Brieba LG, Sotelo-Mundo RR (2014) Crystal structure of the shrimp proliferating cell nuclear antigen: structural complementarity with WSSV DNA polymerase PIP-box. PLoS ONE 9:e94369
Crawford AM, Miller LK (1988) Characterization of an early gene accelerating expression of late genes of the baculovirus Autographa californica nuclear polyhedrosis virus. Virol. 62:2773–2781
Li L, Harwood SH, Rohrmann GF (1999) Identification of additional genes that influence baculovirus late gene expression. Virology 255:9–19
Lu A, Miller LK (1995) The roles of eighteen baculovirus late expression factor genes in transcription and DNA replication. J Virol 69:975–982
Carson DD, Guarino LA, Summers MD (1988) Functional mapping of an AcNPV immediately early gene which augments expression of the IE-1 trans-activated 39 K gene. Virology 162:444–451
Acknowledgements
This project was supported by grants from ‘National Natural Science Foundation of China (No.31272100 and 31372199)’, ‘Natural Science Foundation of Shanxi Province (No.2014011038-1)’.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors state that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1
Sequence of primers used to amplify Ac-pcna, Sf-pcna, hr3, hsp90 (DOC 26 kb)
Supplementary Table 2
Sequence of primers used to amplify β-actin, 38K, vp39, ie2 in qPCR (DOC 27 kb)
Supplementary Table 3
Average weight of Beet armyworm larvae (g) (DOC 35 kb)
Supplementary Table 4
Mortality rate analysis of Beet armyworm larvae (DOC 45 kb)
Supplementary Table 5
Pupation rate analysis of Beet armyworm larvae (%) (DOC 33 kb)
Supplementary Table 6
Emergence rate analysis of Beet armyworm larvae (%) (DOC 33 kb)
Rights and permissions
About this article
Cite this article
Fu, Y., Wang, R. & Liang, A. Function analysis of Ac-PCNA and Sf-PCNA during the Autographa californica multiple nucleopolyhedrovirus infection process. Mol Cell Biochem 443, 57–68 (2018). https://doi.org/10.1007/s11010-017-3210-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-017-3210-y