Abstract
Despite the current progress in cancer research and therapy, breast cancer remains the leading cause of mortality among half a million women worldwide. Migration and invasion of cancer cells are associated with prevalent tumor metastasis as well as high mortality. Extensive studies have powerfully established the role of prototypic second messenger cAMP and its two ubiquitously expressed intracellular cAMP receptors namely the classic protein kinaseA/cAMP-dependent protein kinase (PKA) and the more recently discovered exchange protein directly activated by cAMP/cAMP-regulated guanine nucleotide exchange factor (EPAC/cAMP-GEF) in cell migration, cell cycle regulation, and cell death. Herein, we performed the analysis of the Cancer Genome Atlas (TCGA) dataset to evaluate the essential role of cAMP molecular network in breast cancer. We report that EPAC1, PKA, and AKAP9 along with other molecular partners are amplified in breast cancer patients, indicating the importance of this signaling network. To evaluate the functional role of few of these proteins, we used pharmacological modulators and analyzed their effect on cell migration and cell death in breast cancer cells. Hence, we report that inhibition of EPAC1 activity using pharmacological modulators leads to inhibition of cell migration and induces cell death. Additionally, we also observed that the inhibition of EPAC1 resulted in disruption of its association with the microtubule cytoskeleton and delocalization of AKAP9 from the centrosome as analyzed by in vitro imaging. Finally, this study suggests for the first time the mechanistic insights of mode of action of a primary cAMP-dependent sensor, Exchange protein activated by cAMP 1 (EPAC1), via its interaction with A-kinase anchoring protein 9 (AKAP9). This study provides a new cell signaling cAMP–EPAC1–AKAP9 direction to the development of additional biotherapeutics for breast cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is a leading cause of death globally among women. Although a number of highly effective first-line therapies have prolonged survival, the recurrent behavior of cancer due to metastasis renders the disease incurable. Tumor metastasis is primarily mediated by detachment of cancer cells from primary tumor and their entry into the bloodstream or lymphatic vessels that are lined by the endothelial cells [1]. Several signaling pathways have been discovered till now which are involved in tumor progression. G-protein-coupled receptors (GPCRs) signaling is one of the signaling mechanism that activates adenylate cyclase (AC) and leads to the generation of second messenger molecule cAMP, involved in many known cell signaling pathways in cancer cells [2–4].
Earlier studies attributed all the effects of cAMP molecule to the activation of classical protein kinase A (PKA)-dependent pathway. More recently, exchange protein activated by cAMP (EPAC) has been shown to regulate several physiological functions by acting as guanine nucleotide exchange factors for small Ras-like GTPases (Rap-1 and Rap-2). EPAC has two isoforms, EPAC1 and EPAC2, with EPAC1 expressing predominantly in adipose tissue, blood vessels, kidney, and uterus, while the central nervous system, adrenal gland, and pancreas show the expression of EPAC2 [5]. EPAC proteins interact with several effectors like tubulin, ERM (ezrin–radixin–myosin) proteins, nucleoporin RanBP2, and A-kinase anchoring protein 9 (AKAP9) (commonly known as AKAP450/350/CG-NAP/PRKA9/Hyperion) [6–8] to exert pivotal role in regulating various cellular processes like cell adhesion, cell proliferation, cell survival and apoptosis, and vascular permeability [9, 10]. Altered expression of EPAC proteins has been reported in different carcinomas and it is different and cell specific in context of migration. In human prostate cancer, EPAC proteins have been shown to inhibit cell proliferation and migration [11], and interestingly accelerate migration in melanoma cells. The role of EPAC1 in integrin-mediated cell adhesion has been postulated in human ovarian carcinoma cells [12]. Moreover, EPAC1 over-expression is observed in pancreatic ductal adenocarcinoma [13]. The recent study showed increased uptake of sugar by EPAC/RAP1 in breast cancer cells prime to activation of oncogenic pathways [14].
Tumor metastasis involves microtubule-meditated movement of tumor from primary to secondary site and could be a very good target against breast cancer [15, 16]. Microtubule movement towards periphery of cells is controlled by microtubule-organizing center (MTOC) [17, 18]. Specific proteins like AKAPs regulate MTOC activity by acting as regulatory scaffolds for proteins involved during cell cycle [19, 20]. AKAP is a family of scaffold proteins which are functionally related and bind to PKA. Binding of PKA to these AKAP proteins confines the activity of PKA to specific sets of substrates. One of these AKAP proteins, AKAP9, is confined intra-cellularly to the centrosome and is also present in Golgi apparatus, modulating microtubule dynamics in human endothelial cells [21]. The mutations in the centrosome matrix protein, AKAP9, are demonstrated statistically to be associated with the risk of breast cancer [22, 23]. However, the role of AKAP9 in breast cancer is not extremely clear. In the present study, we hypothesized that EPAC proteins might be involved in modulating microtubule architecture in breast cancer via its effector AKAP9. The role of EPAC proteins in pathogenesis of various diseases including cancer can be elucidated due to the availability of EPAC-Specific Inhibitors (ESIs) [24, 25]. ESI-09, an inhibitor of EPAC1/2, blocks Rap1 activation, Akt phosphorylation, and EPAC-mediated insulin secretion in pancreatic B-cells. Although the involvement of EPAC1 is shown in pancreatic [26] and prostrate carcinomas [11], its role in breast cancer is not fully explored. The data obtained from The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov) analysis revealed that the gene set consisting of EPAC1, AKAP9, and PKA is altered in a number of invasive breast carcinoma cases.
Here, we present the first evidence to ascertain the role of EPAC1 in cell migration and cell death in breast cancer via AKAP9 destabilization as demonstrated by in silico TCGA Analysis. Although cAMP has been shown to regulate endothelial cells permeability, the molecular basis of this signaling in breast cancer has not yet been established. Herein, we report that cAMP-dependent EPAC1 affects cell migration as well as regulates the process of apoptosis in breast cancer cells via AKAP9 destabilization.
Materials and methods
Reagents and antibodies
3-[5-(tert-butyl) isoxazol-3-yl]-2-[2-(3-chlorophenyl) hydrazono]-3-oxopropanenitrile (ESI-09) obtained from BioLog Life Science Institute (Bremen, Germany) is a competitive, membrane-permeable inhibitor of both exchange proteins EPAC1 and EPAC2. PKA-AKAP Inhibitor peptide (St-Ht31) and cAMP-PKA inhibitory peptide (PKI) were purchased from Promega (USA). KH7, Tubulin, Triton X-100 solution, and Formaldehyde solution were purchased from Sigma-Aldrich, (USA), anti-rabbit AKAP9 (BD Biosciences, USA); Alexa 488 and 594 anti-mouse and rabbit, and DAPI-Antifade (Life technologies-Invitrogen, USA) were used in experiments.
Cell culture
MCF-7 cells were maintained as monolayer in 25 cm2 culture flasks (T-25) in DMEM medium (GIBCO, St. Louis, USA) supplemented with high glucose, l-glutamine, pyridoxine hydrochloride, 110 mg/l sodium pyruvate, 3.7 g/l NaHCO3, 10% heat inactivated FBS (Fetal Bovine Serum, Gibco USA), and antibiotics (Pen-Strep 10,000 units/ml, Gibco USA) in humidified CO2 incubator at 37 °C. The medium was changed 10–12 h prior to experiment. MCF-7 cells were regularly sub-cultured thrice a week in a seeding density of 40,000–50,000 cells/cm2 area. The cells used for each experiment were of less than ten passage number.
TCGA analysis
Data from TCGA dataset were extracted and analyzed for cAMP pathway mediators for amplification, deletion, mutation, and alteration. The data were analyzed statistically, and P-value was calculated. The survival rate was also calculated with respect to the cluster containing these genes and represented as graph with P value.
Wound healing assay
MCF-7 cells were grown in 12-well plates at a density of ∼2 × 105 cells per well. After 24 h of seeding, the cells were treated with 10 µM concentrations of all compounds (ESI-05, ESI-09, KH7, GGTI, and St-Ht31) for 30 min. The medium was removed, and the cells were scratched by 200-μl pipette micro tip through the center of the plate and imaged at 0, 6, 12, 18, and 24 h. Healing rate was calculated by counting five to seven fields per image according to the following equation: % wound closure = (initial wound width − wound width 24 h post-treatment with all compounds/control)/initial wound width × 100. The results presented as the average of three independent experiments performed over multiple days. Similar wound healing assay was also performed with MDA-MB-231 cells using ESI-09 at a concentration of 10 µM for 24 h, and cell migration was measured for 0, 12, and 24 h.
Cell adhesion assay
Adhesion assay was performed using breast cancer cell line MDA-MB-231 as described [27]. Briefly, the cells were grown in 6-well plates to a density of ∼4 × 105 cells per well. After 48 h of seeding, the cells were trypsinized and re-plated in another plate alone and with ESI-09 10 µM for 24 h. The cells were imaged at 0, 12, and 24 h, and numbers of attached cells were counted as average of ten different fields in a single well. The results presented here are the average of three independent experiments performed over multiple days.
Immunofluorescence assay
MCF-7 cells were plated on glass cover slips in FBS-supplemented media with 1% antibiotic. The cover slips were fixed in 4% formaldehyde in phosphate buffer saline (PBS) for 10–15 min and then permeabilized with 0.1% Triton X-100 in PBS for 10 min. Cells were blocked in 1% FBS in PBS and then immune-stained with primary antibody mouse anti-tubulin and rabbit anti-AKAP9 for 2 h, followed by Alexa 488 anti-mouse secondary antibody and Alexa 594 anti-rabbit secondary antibody at room temperature for 2 h. The cover slips were then mounted with vector shield mounting reagent with DAPI and sealed. The counting of AKAP9 at the MTOC was performed in three independent experiments and results shown are the average of them.
Cell cycle and morphology analysis
MCF-7 cells were seeded in 6-well plates at a density of ~3.6 × 105 cells per well in 2 ml of complete DMEM, and treated with EPAC inhibitor, ESI-09 (10 and 50 μM), for 24 h. The cells without any treatment were taken as negative control cells, and etoposide-treated cells (10 μM) were included as positive control for the experiments. The morphological changes were observed and the images were captured under an inverted light microscope (Olympus, PA, USA) at 24 and 48 h.
For cell cycle analysis, MCF-7 cells were seeded in 6-well plates at a density of ~3.6 × 105 cells in 2 ml of complete growth culture media and treated with ESI-09 (10 and 50 μM) for 24 and 48 h. Following incubation, the adherent cells were detached by trypsinization and pelleted at 1000×g for 5 min. The floating cells in each well were collected and pelleted in the same way. Cells were washed twice with PBS and re-suspended in PBS-chilled methanol (1:9) at 4 °C overnight. The cells were washed once with PBS, and treated with 20 μg/ml RNase (Sigma-Aldrich, St. Louis, MO, USA), for 30 min at 37 °C. Before analysis, the cells were washed with PBS, and stained with 10 μg/ml propidium iodide (stock-1 mg/ml, Sigma-Aldrich, St. Louis, MO, USA) and incubated at room temperature in dark for 20 min. Finally, the cells were re-suspended in 200 μl of PBS for acquisition. DNA content of 25,000 cells was analyzed by flow cytometer (Becton Dickinson, CA, USA), and the population of cells in each cell cycle phase was determined by using the Cell Quest™ software.
Determination of apoptosis by annexin/PI assay
To determine apoptosis in MCF-7 cells treated with ESI-09, the Annexin/PI assay was performed as described below. Briefly, the cells were seeded in 6-well plates at 3.6 × 105 cells in 2 ml of complete culture media and treated with ESI-09 (50 μM). Following incubation, the cells were trypsinized and pelleted at 1000×g after 6 and 48 h. The cells were rinsed with chilled PBS and re-suspended in 1× binding buffer (1.5 M NaCl, 0.1 M HEPES, 25 mM CaCl2). 5 μl of Annexin V-fluorescein isothiocyanate (FITC) (BD Pharmingen) and 10 μg/ml propidium iodide (PI) were added to cells and incubated for 20 min at room temperature in dark. Finally, the cells were transferred to the FACS tubes for sorting. About 10,000 cells were analyzed by FACS Calibur™ flow cytometer (Becton Dickinson, CA, USA). The desired population was first gated based on size and granularity. This gated population was then assessed and gated for fluorescence in FL1 and FL2 channels. Stained cells were also mounted on glass slides for imaging using confocal microscope (Nikon).
TUNEL assay
The TUNEL assay was performed using APO-BrdU TUNEL Assay Kit (Molecular Probes, Invitrogen) as per manufacturer’s protocol. Briefly, ESI-09-treated cells were trypsinized and pelleted at 1000×g after 24 and 48 h. The cells were fixed with 1% paraformaldehyde in PBS for 15 min on ice. The cells were then washed twice with PBS and re-suspended in ice-cold 70% (v/v) methanol for minimum of 30 min at −20 °C. The fixed cells were incubated in 50 µl of DNA labeling solution containing bromodeoxyuridine triphosphate (Br-dUTP) and a terminal deoxynucleotidyl transferase (TdT), for 60 min at 37 °C. Following incubation, bound Br-dUTP was detected using Alexa Fluor 488 dye-labeled anti-BrdU monoclonal antibody and the DNA was counterstained with propidium iodide (PI)/RNase solution for flow analysis. The negative and positive control cells provided in the kit were processed in similar way as described above. A minimum of 10,000 events was counted for each sample. The analysis was performed using the Cell Quest software. Etoposide was also used as an external positive control apart from the kit.
Results
Alteration of the gene set in breast cancer patients
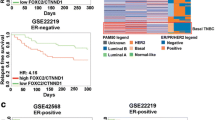
With the objective to identify the role of cAMP pathway in breast cancer progression, the effector genes related to this pathway were analyzed in TCGA breast cancer dataset. Analysis of TCGA dataset showed that gene set consisting of EPAC1, AKAP9, and PKA is altered and amplified in a number of invasive breast carcinoma cases. Additionally, the amplification of the gene set is associated with poor survival. Identifying the contribution of AKAP9, a member of the AKAP family of proteins that are known to temporally and spatially compartmentalize cAMP-induced signals, to specific EPAC functions may help to explain how EPAC fulfills its diverse roles within a single cell. In silico data provide a probable hint towards the involvement of the novel PKA, EPAC1, and AKAP9 complex in breast cancer (Fig. 1a, b).
cAMP signaling pathway molecules such as RAPGEF3, AKAP9, PRKAR1A, PRKAR1B, PRKAR2A, PRKAR2B, PRKACA, PRKACB might play a role in breast cancer. a RAPGEF3/EPAC- and AKAP9-associated genes are amplified in breast cancer. b Survival curve indicates that this gene set is extremely important for survival
Epac1 inhibition leads to reduction in breast cancer cell migration
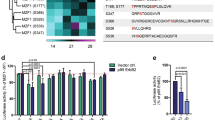
MCF-7 cells were treated with cAMP modulators and analyzed for their involvement in cell migration. Control cells completely migrated after 24 h. We used upstream KH7 and downstream GGTI regulators of cAMP signaling, but ESI-09 exhibits more pronounced effect compared to all other molecules tested signifying its efficacy for inhibition of migration in breast cancer cells. However, ESI-05, a specific inhibitor of EPAC2, did not have any significant effect on cell migration suggesting EPAC1 plays an important role in breast cancer cell migration. Quantitative analysis of cell migration inhibition is also depicted in (Fig. 2a). Migration analysis was also done for MDA-MB-231 cells for 24 h. ESI-09-treated cells showed less migration as compared with the control cells (Fig. 2b). Cell adhesion and migration is a crucial phenomenon for spread of cancer cells via metastasis. While our results have shown that ESI-09 is inhibiting cell migration in both breast cancer cell lines, we also analyzed its effect on cells adherence by treating the cells with ESI-09 for 24 h. Reduction in number of cells attached on to the surface after 24 h confirmed the effect of ESI-09 on cell adhesion in MDA-MB-231 cells. Quantitative analysis is also shown by bar graph (Fig. 2c).
cAMP modulators regulate breast cancer cell migration and adhesion. a The cell migration was analyzed using the conventional wound healing method. The cells treated with specific cAMP modulators were compared with control. St-Ht31 failed to inhibit cell migration as compared with the control, while ESI-09 exhibits efficient cell migration reduction. The quantitative analysis of cell migration inhibition is shown by bar graph. b Treatment of ESI-09 showed reduced cell migration in MDA-MB-231 cells. c Cell adhesion assay with MDA-MB-231 treated with ESI-09 inhibitor for 24 h. The treated cells showed reduced attachment on to the surface as compared to control
Pharmacological modulation of cAMP-regulated Epac1 affects microtubule redistribution via AKAP9 delocalization in breast cancer cells
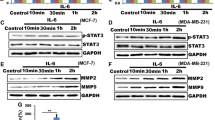
Cell movement occurs due to the involvement of cytoskeletal rearrangement. We performed immunofluorescence assays to investigate the effect of cAMP signaling modulators on cell cytoskeleton and discovered that microtubule distribution in ESI-09 treated cells was affected. We also observed AKAP9 mislocalization after the specific treatment with ESI-09. Immunostaining of AKAP9 in all cAMP modulator-treated cells is depicted in Fig. 3. It provides an indication that the disturbance in microtubules and its associated effector AKAP9 could be the possible mechanism of action.
cAMP modulators regulate AKAP9 delocalization and microtubule redistribution. Effect of cAMP modulators on AKAP9 delocalization and tubulin distribution in MCF-7 cells. ESI-09 results in a dramatic mislocalization of AKAP9 and tubulin structure collapse indicating a functional interaction between EPAC and AKAP9 proteins in breast cancer cells. The quantitative analysis of delocalization of AKAP9 from MTOC in MCF-7 cells treated with different cAMP modulators (Control, KH7, ESI05, ESI-09) is shown by bar graph
ESI 09 increased sub G1 population in cell cycle
Treatment with various cAMP modulators revealed that these compounds show different effects on cell cycle in MCF-7 cells. Treatment with EPAC inhibitor (ESI-09) at 10 μM significantly increased the population of cells at sub G1 phase with a concomitant decrease in the G2/M cells as compared to the control. At higher concentrations of 50 μM, a significant increase in the number of cells at sub G1 phase was observed for ESI-09 at 48 h (Fig. 4a). However, St-Ht31 and cAMP-PKA inhibitor peptide (PKI) showed no changes in cell cycle distribution at 10 μM, even after treatment for 48 h (Fig. 4b). The effect of drugs is compared with our positive control drug etoposide at 10 μM, which is known to cause apoptosis in cancer cells. Our results indicate that among all cAMP modulators used in this study, ESI-09 is more effective in causing cell death.
ESI-09 modulates cell cycle in MCF-7 cells. MCF-7 cells were treated with different cAMP modulators, and changes in cell cycle stages were analyzed by flow cytometry. a EPAC inhibitor (ESI-09) at 10 and 50 µM concentrations for 24 and 48 h. Etoposide (10 µM), used as a positive control, showed highest number of apoptotic cells. b PKA–AKAP9 interaction inhibitory peptide (St Ht31) and cAMP–PKA inhibitor peptide (PKI) at 10 and 50 µM concentrations for 24 and 48 h. Histograms represent the amount of DNA on X-axis and number of cells on Y-axis. M1, M2, M3, and M4 represent the stages sub G 1 , G 0 /G 1 , S, and G 2 /M stage of cell cycle measured by fluorescence intensity at 617 nm (FL2).The treatment with ESI-09 showed increased sub G1 population as compared to control
ESI-09 induced morphological changes and inhibited growth in breast cancer cells
To determine whether ESI-09 can cause apoptosis in MCF-7 breast cancer cells, we first looked for characteristics of apoptosis like cellular shrinkage, chromatin condensation, membrane blabbing, and formation of apoptotic bodies in treated cells. Morphological study showed that ESI-09 induced growth inhibition and apoptosis in MCF-7 cells. The cells demonstrated cellular shrinkage at 24 h, and this phenomenon became more obvious after 48 h of treatment. At higher concentration of ESI-09 (50 μM), the growth inhibition was more pronounced and the cells detached from the substratum (Fig. 5a) at 24 h. Majority of the cells treated with ESI-09 at 50 μM detached from the substratum and cellular shrinkage was observed at 48 h.
ESI-09 induces morphological changes and apoptosis in MCF-7 cells. a Cells treated with ESI-09 for 24 and 48 h showed significant changes in their morphology. Cellular shrinkage characteristic of apoptosis was seen after treatment with ESI-09 (white arrow heads). b (i) The percentage of viable and apoptotic cells was determined by flow cytometry using Annexin V/PI assay after treatment with ESI-09 (50 µM). Lower left quadrant (Annexin V−/PI−) represents viable cells, whereas lower right (Annexin V+/PI−) and upper (PI+) quadrants represent the percentage of early apoptotic and late apoptotic/secondary necrotic cells, respectively. (ii) Confocal images showed surface staining by Annexin V-FITC (green) in early apoptotic cells (6 h) after treatment with ESI-09. On the other hand, after 48 h with the increase in permeabilization of treated cells, the staining of propidium iodide increases (red, shown by arrow). Control untreated cells showed no staining with Annexin V-FITC or PI. c Flow cytometry analysis of DNA fragmentation during apoptosis in MCF-7 cells by TUNEL assay. DNA histograms represent effect of ESI-09 on breast cancer cells at different concentrations. Etoposide is used as positive control for the experiment. The quantitative analysis of TUNEL profile is shown by bar graph (black negative control, yellow fixed cells tunel positive control, green ESI-09 10 μM, pink ESI-09 50 μM, blue etoposide 10 μM). (Color figure online)
ESI-09 induced apoptosis in breast cancer cells
It has been previously shown that during apoptosis, the cells expose phosphatidylserine (PS) on the outer surface of plasma membrane. This exposed PS functions as a tag for specific recognition by macrophages leading to phagocytosis of the dying cells [28]. Exposed PS has high affinity for Annexin V, a phospholipid-binding protein [29]. To further investigate that ESI-09 induced apoptosis in MCF-7 cells, the exposure of PS was studied by flow cytometry and imaging using Annexin V-FITC. Treatment with ESI-09 significantly increased the percentage of early apoptotic cells (Annexin V+/PI−) (24.6%) as compared to the control (9.2%) (Fig. 5b(i)). After 48 h of treatment, the percentage of dead cells or late apoptotic cells (Annexin V+/PI+) was 19.7% as compared to the control 4.23%. Confocal microscopy was also performed which also supports our FACS analysis. Green and red show Annexin V and PI staining, respectively (Fig. 5b(ii)).
ESI-09 induced DNA fragmentation in breast cancer cells
DNA strand breaks are a landmark phenomenon observed in apoptotic cells. Here, we detected DNA fragmentation in MCF-7 cells after treatment with ESI-09 by flow cytometry. Etoposide (10 μM) was used as positive control for the experiments. Additionally, fixed human lymphoma cell line provided in the kit as control was also analyzed as a positive control. The frequency of DNA strand breaks (TUNEL positive cells) was found to be more at higher concentrations of ESI-09 (50 µM) as evident by clear shift in peaks (Fig. 5c). After 48 h of treatment with ESI-09, even low concentration (10 µM) of ESI-09 induced apoptosis.
Discussion
Deciphering the regulatory mechanism of adhesion and migration in breast cancer cells is highly important in order to explain the players promoting metastasis in the very deadly form of cancer. In silico, we conducted a comprehensive analysis of genes involved in breast cancer by TCGA and found that effectors and mediators of cAMP signaling pathway like EPAC1, PKA, and AKAP9 are amplified in breast cancer (Fig. 1). It concludes that pathways involving AKAP9 and EPAC1 offer potential therapeutic targets in the treatment of breast cancer. Furthermore, we explored the role of these cAMP-dependent mediators in regulation of cell migration and observed that EPAC1 is involved in breast cancer progression via interaction with AKAP9. Previous studies have shown the role of these EPAC proteins in endothelial cell permeability and cell–cell adhesion [30, 31]. In the present study, we examined the effects of different types of cAMP modulators, which affect cAMP pathway at different levels in breast cancer cells (Fig. 6). These inhibitors work as upstream or downstream regulators in cAMP signaling [3, 32, 33].
ESI-09 is a known potent inhibitor of EPAC1/EPAC2. Studies have been performed to elucidate the binding efficiency of ESI-09 towards EPAC1/2. EPAC1 is a known primary sensor of cAMP pathway, and ESI-09 worked as a competitive inhibitor of cAMP signaling through PKA-independent pathway [25, 34]. ESI-09 opened a new window towards the development of new therapeutics by inhibiting EPACs. ESI-09 as compared to other cAMP modulators is more efficient, and substituting some moieties on it can lead to increase the potency of action [25].
To determine the role of EPAC1 in cell migration, breast cancer cell lines, luminal-like MCF-7 cells, and basal-like MDA-MB-231 cells [35] were treated with ESI-09 inhibitor. Migration inhibition shown by ESI-09 in both of the breast cancer cell lines suggests significance of PKA-independent EPAC1-mediated pathway in cancer progression. Migration of tumor cells relies on to attachment of cells to the substratum; we also analyzed the adhesion of these cells with the treatment of ESI-09 and found reduced cells attachment on to the surface as compared to control cells.
Thereafter, we studied the effect of ESI-09 on cell cycle and cell morphology in MCF-7 cells. Our results showed that ESI-09-treated MCF-7 cells exhibited cellular shrinkage, indicating induction of apoptosis in these cells. Furthermore, the cell cycle analysis revealed arrest of cells at sub G1 phase at 10 µM concentration of ESI-09 between 24 and 48 h as confirmed by flow cytometric analysis (Fig. 4). The effect of drug was more pronounced at 50 µM concentration of ESI-09. Since in our study we did not see significant effect of PKA and cAMP modulators on cell cycle, we propose a possible role of PKA-independent EPAC-mediated pathway in regulating cell cycle in breast cancer cells.
In tumor microenvironment, a number of things such as increase in vascular permeability, angiogenesis, and immune suppression may lead to tumor progression [36]. EPAC1 has been known to play an important role in vascular permeability [21] and also known to participate in Treg-mediated suppression [37]. The two major cAMP modulated pathways, PKA and EPAC1, regulate T cell functions in diverse ways. This preliminary study will help in basic understanding of involvement of cAMP effectors in breast cancer as well as proposing therapeutic strategies to combat the disease. Therefore, studying the mechanism of action of Epac1 pathway is extremely essential to elucidate the intricate signaling networks that regulate Treg cells, breast cancer cells, etc.
Conclusion
In the present study, we have shown that the blocking of EPAC1 by its specific inhibitor ESI-09 can lead to inhibition of migration and apoptosis in breast cancer cells. Further, we confirmed that inhibition of EPAC1 leads to mislocalization of AKAP9. Our study opens up new avenues for development of therapeutics against breast cancer by targeting effector molecules of cAMP signaling. EPAC1 signaling can emerge as an essential target for development of potential therapeutics for breast cancer treatment. Deciphering the role of EPAC1 on vascular permeability will help in designing novel therapeutics for breast cancer. The use of cAMP elevating agents and RNAi-mediated knockdown of EPAC1 will help in further dissecting the role of EPAC1 in breast cancer.
Abbreviations
- AKAP:
-
A-kinase anchoring protein
- PKA:
-
Protein kinase A
- EPAC:
-
Exchange protein activated by cAMP
- TCGA:
-
The Cancer Genome Atlas
- ESI:
-
EPAC-specific inhibitor
References
Guan X (2015) Cancer metastases: challenges and opportunities. Acta Pharm Sin B 5:402–418. doi:10.1016/j.apsb.2015.07.005
Dorsam RT, Gutkind JS (2007) G-protein-coupled receptors and cancer. Nat Rev Cancer 7:79–94. doi:10.1038/nrc2069
Gidon A, Feinstein TN, Xiao K, Vilardaga JP (2016) Studying the regulation of endosomal cAMP production in GPCR signaling. Methods Cell Biol 132:109–126. doi:10.1016/bs.mcb.2015.10.007
Zhou C, Dai X, Chen Y, Shen Y, Lei S, Xiao T, Bartfai T, Ding J, Wang MW (2016) G protein-coupled receptor GPR160 is associated with apoptosis and cell cycle arrest of prostate cancer cells. Oncotarget 7:12823–12839. doi:10.18632/oncotarget.7313
Gloerich M, Bos JL (2010) Epac: defining a new mechanism for cAMP action. Annu Rev Pharmacol Toxicol 50:355–375. doi:10.1146/annurev.pharmtox.010909.105714
Gloerich M, Ponsioen B, Vliem MJ, Zhang Z, Zhao J, Kooistra MR, Price LS, Ritsma L, Zwartkruis FJ, Rehmann H, Jalink K, Bos JL (2010) Spatial regulation of cyclic AMP-Epac1 signaling in cell adhesion by ERM proteins. Mol Cell Biol 30:5421–5431. doi:10.1128/MCB.00463-10
Liu C, Takahashi M, Li Y, Dillon TJ, Kaech S, Stork PJ (2010) The interaction of Epac1 and Ran promotes Rap1 activation at the nuclear envelope. Mol Cell Biol 30:3956–3969. doi:10.1128/MCB.00242-10
Liu C, Takahashi M, Li Y, Song S, Dillon TJ, Shinde U, Stork PJ (2008) Ras is required for the cyclic AMP-dependent activation of Rap1 via Epac2. Mol Cell Biol 28:7109–7125. doi:10.1128/MCB.01060-08
Sehrawat S, Cullere X, Patel S, Italiano J Jr, Mayadas TN (2008) Role of Epac1, an exchange factor for Rap GTPases, in endothelial microtubule dynamics and barrier function. Mol Biol Cell 19:1261–1270. doi:10.1091/mbc.E06-10-0972
Cheng X, Ji Z, Tsalkova T, Mei F (2008) Epac and PKA: a tale of two intracellular cAMP receptors. Acta Biochim Biophys Sin 40:651–662
Grandoch M, Rose A, ter Braak M, Jendrossek V, Rubben H, Fischer JW, Schmidt M, Weber AA (2009) Epac inhibits migration and proliferation of human prostate carcinoma cells. Br J Cancer 101:2038–2042. doi:10.1038/sj.bjc.6605439
Rangarajan S, Enserink JM, Kuiperij HB, de Rooij J, Price LS, Schwede F, Bos JL (2003) Cyclic AMP induces integrin-mediated cell adhesion through Epac and Rap1 upon stimulation of the beta 2-adrenergic receptor. J Cell Biol 160:487–493. doi:10.1083/jcb.200209105
Lorenz R, Aleksic T, Wagner M, Adler G, Weber CK (2008) The cAMP/Epac1/Rap1 pathway in pancreatic carcinoma. Pancreas 37:102–103. doi:10.1097/MPA.0b013e318160748f
Onodera Y, Nam JM, Bissell MJ (2014) Increased sugar uptake promotes oncogenesis via EPAC/RAP1 and O-GlcNAc pathways. J Clin Invest 124:367–384. doi:10.1172/JCI63146
Honore S, Pasquier E, Braguer D (2005) Understanding microtubule dynamics for improved cancer therapy. Cell Mol Life Sci 62:3039–3056. doi:10.1007/s00018-005-5330-x
Pasquier E, Kavallaris M (2008) Microtubules: a dynamic target in cancer therapy. IUBMB Life 60:165–170. doi:10.1002/iub.25
Desai A, Mitchison TJ (1997) Microtubule polymerization dynamics. Annu Rev Cell Dev Biol 13:83–117. doi:10.1146/annurev.cellbio.13.1.83
Komarova YA, Vorobjev IA, Borisy GG (2002) Life cycle of MTs: persistent growth in the cell interior, asymmetric transition frequencies and effects of the cell boundary. J Cell Sci 115:3527–3539
Witczak O, Skalhegg BS, Keryer G, Bornens M, Tasken K, Jahnsen T, Orstavik S (1999) Cloning and characterization of a cDNA encoding an A-kinase anchoring protein located in the centrosome, AKAP450. EMBO J 18:1858–1868. doi:10.1093/emboj/18.7.1858
Rempel N (2001) Centrosomes as scaffolds: the role of pericentrin and protein kinase A-anchoring proteins. Einstein Q J Biol Med 18:54–58
Sehrawat S, Ernandez T, Cullere X, Takahashi M, Ono Y, Komarova Y, Mayadas TN (2011) AKAP9 regulation of microtubule dynamics promotes Epac1-induced endothelial barrier properties. Blood 117:708–718. doi:10.1182/blood-2010-02-268870
Frank B, Rigas SH, Bermejo JL, Wiestler M, Wagner K, Hemminki K, Reed MW, Sutter C, Wappenschmidt B, Balasubramanian SP, Meindl A, Kiechle M, Bugert P, Schmutzler RK, Bartram CR, Justenhoven C, Ko YD, Bruning T, Brauch H, Hamann U, Pharoah PP, Dunning AM, Pooley KA, Easton DF, Cox A, Burwinkel B (2008) The CASP8-652 6 N del promoter polymorphism and breast cancer risk: a multicenter study. Breast Cancer Res Treat 111:139–144. doi:10.1007/s10549-007-9752-z
Milne RL, Lorenzo-Bermejo J, Burwinkel B, Malats N, Arias JI, Zamora MP, Benitez J, Humphreys MK, Garcia-Closas M, Chanock SJ, Lissowska J, Sherman ME, Mannermaa A, Kataja V, Kosma VM, Nevanlinna H, Heikkinen T, Aittomaki K, Blomqvist C, Anton-Culver H, Ziogas A, Devilee P, van Asperen CJ, Tollenaar RA, Seynaeve C, Hall P, Czene K, Liu J, Irwanto AK, Kang D, Yoo KY, Noh DY, Couch FJ, Olson JE, Wang X, Fredericksen Z, Nordestgaard BG, Bojesen SE, Flyger H, Margolin S, Lindblom A, Fasching PA, Schulz-Wendtland R, Ekici AB, Beckmann MW, Wang-Gohrke S, Shen CY, Yu JC, Hsu HM, Wu PE, Giles GG, Severi G, Baglietto L, English DR, Cox A, Brock I, Elliott G, Reed MW, Beesley J, Chen X, Investigators K, Fletcher O, Gibson L, dos Santos Silva I, Peto J, Frank B, Heil J, Meindl A, Chang-Claude J, Hein R, Vrieling A, Flesch-Janys D, Southey MC, Smith L, Apicella C, Hopper JL, Dunning AM, Pooley KA, Pharoah PD, Hamann U, Pesch B, Ko YD, Easton DF, Chenevix-Trench G (2011) 7q21-rs6964587 and breast cancer risk: an extended case-control study by the Breast Cancer Association Consortium. J Med Genet 48:698–702. doi:10.1136/jmedgenet-2011-100303
Tsalkova T, Mei FC, Li S, Chepurny OG, Leech CA, Liu T, Holz GG, Woods VL Jr, Cheng X (2012) Isoform-specific antagonists of exchange proteins directly activated by cAMP. Proc Natl Acad Sci U S A 109:18613–18618. doi:10.1073/pnas.1210209109
Zhu Y, Chen H, Boulton S, Mei F, Ye N, Melacini G, Zhou J, Cheng X (2015) Biochemical and pharmacological characterizations of ESI-09 based EPAC inhibitors: defining the ESI-09 “therapeutic window”. Sci Rep 5:9344. doi:10.1038/srep09344
Almahariq M, Tsalkova T, Mei FC, Chen H, Zhou J, Sastry SK, Schwede F, Cheng X (2013) A novel EPAC-specific inhibitor suppresses pancreatic cancer cell migration and invasion. Mol Pharmacol 83:122–128. doi:10.1124/mol.112.080689
Song BW, Chang W, Hong BK, Kim IK, Cha MJ, Lim S, Choi EJ, Ham O, Lee SY, Lee CY, Park JH, Choi E, Song H, Jang Y, Hwang KC (2013) Protein kinase C activation stimulates mesenchymal stem cell adhesion through activation of focal adhesion kinase. Cell Transplant 22:797–809. doi:10.3727/096368912X656126
Fadok VA, Bratton DL, Guthrie L, Henson PM (2001) Differential effects of apoptotic versus lysed cells on macrophage production of cytokines: role of proteases. J Immunol 166:6847–6854
Koopman G, Reutelingsperger CP, Kuijten GA, Keehnen RM, Pals ST, van Oers MH (1994) Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood 84:1415–1420
Cullere X, Shaw SK, Andersson L, Hirahashi J, Luscinskas FW, Mayadas TN (2005) Regulation of vascular endothelial barrier function by Epac, a cAMP-activated exchange factor for Rap GTPase. Blood 105:1950–1955. doi:10.1182/blood-2004-05-1987
Fukuhara S, Sakurai A, Sano H, Yamagishi A, Somekawa S, Takakura N, Saito Y, Kangawa K, Mochizuki N (2005) Cyclic AMP potentiates vascular endothelial cadherin-mediated cell-cell contact to enhance endothelial barrier function through an Epac-Rap1 signaling pathway. Mol Cell Biol 25:136–146. doi:10.1128/MCB.25.1.136-146.2005
Funaki C, Hodges RR, Dartt DA (2010) Identification of the Raf-1 signaling pathway used by cAMP to inhibit p42/p44 MAPK in rat lacrimal gland acini: role in potentiation of protein secretion. Invest Ophthalmol Vis Sci 51:6321–6328. doi:10.1167/iovs.10-5690
Wertheimer E, Krapf D, de la Vega-Beltran JL, Sanchez-Cardenas C, Navarrete F, Haddad D, Escoffier J, Salicioni AM, Levin LR, Buck J, Mager J, Darszon A, Visconti PE (2013) Compartmentalization of distinct cAMP signaling pathways in mammalian sperm. J Biol Chem 288:35307–35320. doi:10.1074/jbc.M113.489476
Singhmar P, Huo X, Eijkelkamp N, Berciano SR, Baameur F, Mei FC, Zhu Y, Cheng X, Hawke D, Mayor F Jr, Murga C, Heijnen CJ, Kavelaars A (2016) Critical role for Epac1 in inflammatory pain controlled by GRK2-mediated phosphorylation of Epac1. Proc Natl Acad Sci USA 113:3036–3041. doi:10.1073/pnas.1516036113
Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW (2006) A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10:515–527. doi:10.1016/j.ccr.2006.10.008
Quail DF, Joyce JA (2013) Microenvironmental regulation of tumor progression and metastasis. Nat Med 19:1423–1437. doi:10.1038/nm.3394
Almahariq M, Mei FC, Wang H, Cao AT, Yao S, Soong L, Sun J, Cong Y, Chen J, Cheng X (2015) Exchange protein directly activated by cAMP modulates regulatory T-cell-mediated immunosuppression. Biochem J 465:295–303. doi:10.1042/BJ20140952
Acknowledgements
Seema Sehrawat is the recipient of BioCARe Award from Department of Biotechnology, Govt. of India and acknowledges the funding support. Shailja Singh acknowledges DBT PILOT project grant on cancer. Shiv Nadar Foundation is acknowledged for providing the Ph.D. fellowship to Mr. Naveen Kumar. Dr. Sonal Gupta is the recipient of University Grant Commission Postdoctoral fellowship For Women. We would like to thank Dr. Sunita Setlur, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA, USA and Dr. Chandan Kumar-Sinha, Michigan Center for Translational Pathology, University of Michigan, and Ann Arbor, MI, USA for their advice on TCGA analysis. The TCGA analysis was performed using the breast cancer data available for the cbioportal. The results presented in this study are partly based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/. We also sincerely thank the financial support received from LRE JNU and ICMR CAR 2016 -17.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Naveen Kumar and Sonal Gupta—equal first author.
Rights and permissions
About this article
Cite this article
Kumar, N., Gupta, S., Dabral, S. et al. Role of exchange protein directly activated by cAMP (EPAC1) in breast cancer cell migration and apoptosis. Mol Cell Biochem 430, 115–125 (2017). https://doi.org/10.1007/s11010-017-2959-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-017-2959-3