Abstract
Ovarian cancer is one of the greatest causes of cancer death in women. The association of TMEM49 and ovarian cancer is poorly defined. Here, we reported that TMEM49 was significantly increased in ovarian tumor tissues compared to ovarian normal tissues. Furthermore, down-regulation of TMEM49 through RNA interference inhibited cell proliferation and arrested G1/S transition in two ovarian cancer cell lines, OVCAR3 and A2780. More importantly, TMEM49 silencing induced cell apoptosis. Additionally, down-regulation of TMEM49 in ovarian cancer notably repressed cell invasion and adhesion. Further gene set enrichment analysis suggested that apoptosis and metastasis up related signal pathways were associated with the TMEM49 expression. Western blot revealed that the expression of Caspase3, Bad, and Bax were increased, while expression of MMP2, KLF10, and CXCL12 were reduced by TMEM49 knockdown. Since expression of TMEM49 seems to be associated with the apoptosis and metastasis up signaling pathways of ovarian cancer, and suppression of its expression can inhibit cancer cell growth and metastasis, TMEM49 may be a potential therapeutic target in human ovarian cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ovarian cancer has the third incidence and the highest mortality among the female genital malignant tumors [1]. Because the ovarian cancer clinical symptom is not obvious and it is lack of reliable early detection methods, about 70 % of the patients has been diagnosed with advanced tumor and widespread metastases, and the overall 5-year survival rate is around 40 % [2]. The current standard treatment for newly diagnosed ovarian cancer is a combination of optimal cytoreductive surgery and platinum-based chemotherapy. In recent years, the development of radical surgery and chemotherapy regimens has greatly improved the treatment of ovarian cancer. However, the advances in suitable treatment for ovarian cancer have been limited because the mechanisms underlying the development of this disease are far from fully understood. Molecular genetic analysis of ovarian cancer has identified the genetic alterations of several genes, such as c-erb-B2, c-myc, and p53 [3]. Genomic rearrangements studies revealed that additional genes involved in ovarian tumor progression correlate to clinical parameters [4]. Therefore, elucidating the molecular mechanism for the ovarian cancer development is essential to develop more rational and effective therapies.
TMEM49, also known as VMP1 (Vacuole membrane protein 1), is a 406amino-acid multi-pass membrane protein that localizes to the endoplasmic reticulum-Golgi intermediate compartment membrane. Autophagy is a degradation process of cytoplasmic cellular constituents. VMP1 had been reported to mediate autophagy in human pancreatic cancer to promote cell death [5]. VMP1 has been reported as an integral autophagosomal membrane protein and a new interactor of the autophagy-specific PtdIns3K complex in mammalian cells [6]. Over-expression of TMEM49, a stress-induced protein, results in the formation of intracellular vacuoles followed by cell death, suggesting that TMEM49 plays an important role in the maintenance of cellular integrity [7]. TMEM9 was identified to be involved in cancer-relevant processes, and has been shown to inhibit proliferation and pulmonary metastasis of hepatocellular carcinoma [8]. Additionally, TMEM49 may be involved in the early stages of acute pancreatitis [9]. Another finding suggested that TMEM49 plays a role in the promotion of apoptosis in pancreatic cell lines [10]. TMEM49 is expressed at high levels in kidney cancer, down-regulation of TMEMP49 by siRNA significantly increases the invasiveness of kidney cells, and TMEM49 expression is also decreased in the invasive breast cancer cells [11]. However, evidence for the function of TMEM49 in human malignancies is still limited.
Little is known about the expression pattern and biological function of TMEM49 in ovarian cancer. We carried out the present study to determine the over-expression of TMEM49 in ovarian tissues as well as cell lines. Then, we investigated the role of TMEM49 in ovarian cells by knocking down its expression with siRNA. Collectively, our studies provide the biological functions of TMEM49 in ovarian cancer and it may be an effective therapeutic target for this disease.
Materials and methods
Tissue samples and cell lines
All investigation described in this study were done after informed consent was obtained and in accordance with the guidelines of the ethics committee of Fujian University of Traditional Chinese Medicine. Fresh frozen primary ovarian tumor tissue samples and adjacent ovarian tissues were collected from the Affiliated People’s Hospital of Fujian University of Traditional Chinese Medicine.
The human ovarian cell lines, CAOV3, SKOV3, HO-8910, A2780, 3AO, and OVCAR3 were purchased from cell bank of Shanghai biology Institute. All cells were cultured in DMEM medium with 10 % fetal bovine serum and 1 % penicillin/streptomycin at 37 °C in 5 % CO2 atmosphere.
Immunohistochemistry
Paraffin tissue sections (5 μm) of ovarian tumor and ovarian normal specimens were used for TMEM49 immunohistochemical studies. Slides were deparaffinized, and endogenous peroxidase activity was blocked by incubation with 1 % H2O2. After antigen retrieval, anti-TMEM49 (Abcam) was then applied overnight at 4 °C. After rinsing with PBS, the biotinylated secondary IgG antibody was applied for 30 min at room temperature. Immunoperoxidase staining was performed using the Vectastatin Elite ABC kit (Vector Laboratories), and sections were counterstained with hematoxylin.
Silencing of TMEM49 by small interfering RNA
Three siRNAs targeting position TMEM49-siRNA-1 559-581 (5′-GAAGGAGTGCATCAACAGTAT-3′), TMEM49-siRNA-2 925-947 (5′-GAACCAGATGATGAAGAGTAT-3′), and TMEM49-siRNA-3 1212-1234 (5′-CAAGCACATAGTGGAGCAAAT-3′) of human TMEM49 mRNA were synthesized. A non-specific scramble siRNA sequence was used as negative control (NC). The siRNAs were transfected into OVCAR3 and A2780 cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instruction. Assays were performed after transfection 48 h.
Real-time PCR
Total RNA was extracted from cultured cells or tissue samples using TRIzol Reagent (Invitrogen) according to the manufacturer’s instructions. Real-time quantitative PCR was performed using a standard SYBR Green PCR kit on an ABI 7300 Thermocycler Real-time machine. GADPH was served as an internal control. All data represent the average of three replicates. The primers for TMEM49 were 5′-GAGACAGCCGCTCATTAC-3′ (forward), 5′-CCAAGCCCAACAGAAGAC-3′ (reverse), and GADPH were 5′-CACCCACTCCTCCACCTTTG-3′ (forward), 5′-CCACCACCCTGTTGCTGTAG-3′ (reverse).
Western blot
Treated and untreated OVCAR3 and A2780 cells were harvest. Proteins from cell samples were separated by SDS-PAGE, followed by electron transfer to a nitrocellulose membrane by means of a transfer cell. The proteins were then transferred to nitrocellulose membrane. The blots were blocked with 5 % skim milk, followed by incubation with antibodies against TMEM49 (Abcam, ab69224), Caspase3 (Abcam, ab32351), Bad (CST, #9292), Bax (Santa, sc-493), MMP2 (Abcam, ab110186), KLF10 (Santa, sc-23158), CXCL12 (Santa, sc-28876), and GADPH (CST, #5174). Blots were then incubated with goat anti-mouse or anti-rabbit secondary antibody and visualized using enhanced chemiluminescence (ECL, Millpore, USA).
Cell proliferation assay
Cell proliferation was measured by using Cell Counting Kit-8 (CCK-8) (Donjindo, Japan) according to the manufacturer’s protocol. Briefly, treated and untreated cells were seeded into 96-well plates at a density of 5 × 103 cells in 200 μl medium per well. At indicated time point 0, 24, 48, and 72 h culture cells, CCK8 solution was added to each well and incubated for 1 h. The absorbance rate at 450 nm was read in microplate reader. All experiments were performed in triplicates and repeated at least three times.
Cell cycle distribution analysis
Propidium iodide (PI) staining was used to analyze DNA content. Treated and untreated cells were harvested and labeled with PI. Briefly, cells were re-suspended in PBS, incubated with PI at room temperature in the dark for 30 min. DNA content was then analyzed using flow cytometry (BD Biosciences). The percentage of cells in the G0/G1, S, and G2/M phases was determined.
Cell apoptosis assay
Cells were harvested for 48 h with or without transfection and treated with annexin V-fluorescein isothiocyanate and apoptosis rates were analyzed using flow cytometry (FACSCalibur, BD Biosciences).
Invasion and adhesion assay
For invasion assay, the upper well of the Boyden Chamber was coated with 10 mg/ml Matrigel. The Matrigel was allowed to harden at 37 °C in a 5 % CO2 incubator for 1 h.
For cell adhesion assay, treated and untreated cells were serum starved for 24 h, 5 × 104 cells were plated into the upper well of the chambers. After 24 h of incubation, cells on the top of the membrane were removed. Cell number was counted in 10 fields for each condition. The experiments were performed in triplicate.
Statistical analysis
All experiments were performed in triplicate, and the results were expressed as the mean ± SD. Statistical significance was determined using the GraphPad Prism 5.0 (GraphPad, La Jolla, CA). The two-tailed Student’s t test was used to evaluate statistical differences between two groups. ** indicates P < 0.05.
Results
Over-expression of TMEM49 in ovarian cancer cells
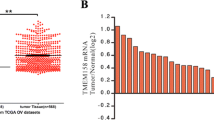
We evaluated TMEM49 mRNA level in ovarian tumor tissues (n = 30) by real-time PCR. Our data suggested that TMEM49 expression was significantly elevated in ovarian tumor tissues compared with that in ovarian normal tissues (Fig. 1a). Further, we analyzed TMEM49 expression in ovarian tumor tissues using high-throughput RNA-sequencing data of the ovarian cancer cohort of The Cancer Genome Atlas (TCGA) and also found a significant increase of TMEM49 expression in ovarian tumor tissues compared with ovarian normal tissues (Fig. 1b, **P < 0.01). The over-expression of TMEM49 was further confirmed by immunohistochemical staining (Fig. 1c).
Over-expression of TMEM49 in ovarian cancer. a The mRNA level of TMEM49 in ovarian and normal tissues. b TMEM49 expression was significantly increased in ovarian tumor tissues when compared with normal tissues of patients from TCGA dataset. c TMEM49 expression was evaluated by immunohistochemistry in ovarian and adjacent tissues
Knockdown of TMEM49 suppressed the proliferation of ovarian cancer cells
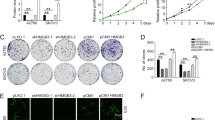
We then measured the protein and mRNA expression levels of TMEM49 in 6 ovarian tumor cell lines, CAOV3, SKOV3, HO-8910, A2780, 3AO, and OVCAR3 cells, by Western blot and real-time PCR. Protein and mRNA levels of TMEM49 were higher in A2780 and OVCAR3 cells than in the other 4 cell lines (Fig. 2a).
To investigate the functions of TMEM49 over-expression on ovarian cancer, we knockdown its expression in ovarian tumor cells by RNAi. A2780 and OVCAR3 cells were chosen for RNAi experiment because of their higher expression level of TMEM49. Three siRNAs targeting human TMEM49 (TMEM49-siRNA) and negative control were synthesized and used to transfect A2780 and OVCAR3 cells. All the siRNAs were able to efficiently suppress TMEM49 expression in ovarian tumor cells. TMEM49-siRNA1 was the most effective one and used for the following assays (Fig. 2b, c).
To explore the effects of TMEM49 silencing on the proliferation of ovarian cells, we examined cell proliferation by using CCK-8 assay. TMEM49-siRNA notably impaired cell proliferation of A2780 (Fig. 3a) and OVCAR3 (Fig. 3b) cells compared to the vector cells. These results indicated that TMEM49 promoted the proliferation in ovarian cells.
Knockdown of TMEM49 arrested cell cycle and induced apoptosis of ovarian cancer cells
The possible effect of TMEM49 knockdown on cell cycle progression was then assessed by PI staining and flow cytometry analysis. Knockdown of TMEM49 in A2780 (Fig. 4a) and OVCAR3 (Fig. 4b) resulted in an increase of cells in the G1 phase and a corresponding decrease of cells in the S phase, which suggested that silencing of TMEM49 prevented ovarian cells from entering the S phase. And, we evaluated the apoptotic function of TMEM49 in A2780 and OVCAR3 cells by Annexin V-FITC/PI staining assay. The results showed that knockdown of TMEM49 notably induced cell apoptosis in ovarian cancer cells compared with the vector (Fig. 5).
Knockdown of TMEM49 inhibited invasion and adhesion of ovarian cancer cells
To investigate the involvement of TMEM49 in cell invasion, transwell assays were carried out to determine the effect of TMEM49 knockdown on cell invasion. The invasion cells were viewed and counted at ×200 magnification. As shown in Fig. 6a, b, invasion cells with TMEM49 knockdown were decreased compared to control cells. We also explored whether TMEM49 affected the adhesion ability of ovarian cells. By in vitro adhesion assay, we found that adhesion cells with TMEM49 knockdown were decreased compared to control cells (Fig. 6c, d).
Effects of TMEM49 siRNA on invasion and adhesion of ovarian cancer cells. a The invasive ability of OVCAR3 cells was identified after siRNA-TMEM49 transfection. b The invasive ability of A2780 cells was identified. c The adhesion ability of OVCAR3 cell was identified. d The adhesion ability of A2780 cells was identified. **P < 0.01 compared with the mock cells
Identified signaling pathways related to TMEM49 expression in ovarian cancer cells
To probe the TMEM49 associated pathways on an unbiased basis, we performed GSEA based on KEGG database. Among all the “KEGG pathways” gene sets, the apoptosis and metastasis up pathways were identified with significant association with TMEM49 expression in the TCGA ovarian cancer dataset (Fig. 7a). The effects of TMEM49 siRNA on the protein expression of Caspase3, Bad, Bax, MMP2, KLF10, and CXCL12 were investigated. As a result, TMEM49 siRNA treatment significantly increased the expression of Caspase3, Bad and Bax, and decreased MMP2, KLF10, and CXCL12 protein expression compared with the control group (Fig. 7b).
Western blot results. a Gene set enrichment analysis (GSEA) identified the apoptosis and metastasis up signaling pathways significant association with TMEM49 expression. b The protein expression of caspase3, Bad, Bax, MMP2, KLF10, and CXCL12 in OVCAR3 cells was analyzed by Western blot. c The protein expression of caspase3, Bad, Bax, MMP2, KLF10, and CXCL12 in A2780 cells was analyzed. **P < 0.01 compared with the mock cells
Discussion
Cancer is a major public health problem in the world [12]. Ovarian cancer is one of the greatest causes of cancer death in women. TMEM49 expression triggers autophagy and precedes apoptotic cell death in pancreas beta cells [13]. TMEM49 induction by gemcitabine promotes the autophagy pathway and apoptotic cell death in pancreatic cancer cells [5]. To date, several reports have demonstrated anti-tumor functions of TMEM49 [14]. In this study, we focused on investigating the function of TMEM49 on ovarian by knockdown of its expression in two ovarian cell lines, A2780 and OVCAR3. The TCGA dataset showed that TMEM49 was highly expressed in ovarian cancer patients. Experiments in vitro showed that knockdown of TMEM49 in ovarian cells inhibited cell growth and metastasis, and induced cell apoptosis. The apoptosis and metastasis up pathways were also activated in TMEM49 knockdown condition. Thus, TMEM49 may serve as a potential target for treatment of ovarian cancer.
Among several ovarian cell lines, TMEM49 was found expressing notably in OVCAR3 and A2780 cells. Therefore, OVCAR3 and A2780 cell lines were determined for further investigation. Then, we examined the effect of siRNA-TMEM49 on proliferation, cell cycle, and apoptosis in OVCAR3 and A2780 ovarian cells. We found that knockdown of TMEM49 effectively inhibited proliferation and induced cell apoptosis. In addition, cell cycle analysis showed that G1/S cell cycle transition was promoted by TMEM49 knockdown.
As so far, the pathology of ovarian cancer is more complex, and the mechanism also remains to be elucidated. Herein, we represented that the reduction of the TMEM49 expression in ovarian cells could decrease their invasion and adhesion capability. GSEA is a method of analyzing and interpreting microarray and such data using biological knowledge [15]. Data of TMEM49 in KEGG pathway showed that TMEM49 was associated with apoptosis and metastasis up pathways. Caspases are crucial mediators of programed apoptosis. Among them, Caspase3 is a frequently activated death protease and indispensable for chromatin condensation and DNA fragmentation [16]. The activity of Bad, a pro-apoptotic Bcl-2 family member, is regulated by phosphorylation in integrating pathways of apoptosis [17]. Bax is a pro-apoptotic protein [18]. MMP2 is a key protein involved in cancer metastasis [19]. KLF10, a Kruppel-like transcriptional factor induced by TGF-β, BMP-2, and EGF, has been implicated in cell differentiation, as a target gene for a variety of signaling pathways [20]. CXCL12 promotes invasion of cells through extracellular matrix [21]. Western blot revealed that Caspase3, Bad, and Bax protein expression were up-regulated, and MMP2, KLF10, and CXCL12 expression were decreased when compared to the control group. These results strongly supported our notion that knockdown TMEM49 may affect apoptosis and metastasis up signalings in ovarian cancer.
In summary, our study provides for the first time that TMEM49 played an important role in the proliferation, apoptosis, and metastasis of ovarian cells, and TMEM49 might regulate biological progress through regulating apoptosis and metastasis up related signal pathways. Inhibition of TMEM49 expression in tumor tissues might provide a therapeutic approach in ovarian cancer.
References
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F (2015) Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136(5):E359–E386
Banerjee S, Kaye SB (2013) New strategies in the treatment of ovarian cancer: current clinical perspectives and future potential. Clin Cancer Res 19(5):961–968
Aunoble B, Sanches R, Didier E, Bignon Y (2000) Major oncogenes and tumor suppressor genes involved in epithelial ovarian cancer (review). Int J Oncol 16(3):567–643
Suzuki S, Moore DH, Ginzinger DG, Godfrey TE, Barclay J, Powell B, Pinkel D, Zaloudek C, Lu K, Mills G (2000) An approach to analysis of large-scale correlations between genome changes and clinical endpoints in ovarian cancer. Cancer Res 60(19):5382–5385
Pardo R, Lo Ré A, Archange C, Ropolo A, Papademetrio DL, Gonzalez CD, Alvarez EM, Iovanna JL, Vaccaro MI (2010) Gemcitabine induces the VMP1-mediated autophagy pathway to promote apoptotic death in human pancreatic cancer cells. Pancreatology 10(1):19–26
Molejon MI, Ropolo A, Vaccaro MI (2013) VMP1 is a new player in the regulation of the autophagy-specific phosphatidylinositol 3-kinase complex activation. Autophagy 9(6):933–935
Dusetti NJ, Jiang Y, Vaccaro MI, Tomasini R, Samir AA, Calvo EL, Ropolo A, Fiedler F, Mallo GV, Dagorn J-C (2002) Cloning and expression of the rat vacuole membrane protein 1 (VMP1), a new gene activated in pancreas with acute pancreatitis, which promotes vacuole formation. Biochem Biophys Res Commun 290(2):641–649
Guo L, Yang LY, Fan C, Chen GD, Wu F (2012) Novel roles of Vmp1: inhibition metastasis and proliferation of hepatocellular carcinoma. Cancer Sci 103(12):2110–2119
Grasso D, Ropolo A, Lo Re A, Boggio V, Molejon MI, Iovanna JL, Gonzalez CD, Urrutia R, Vaccaro MI (2011) Zymophagy, a novel selective autophagy pathway mediated by VMP1-USP9x-p62, prevents pancreatic cell death. J Biol Chem 286(10):8308–8324. doi:10.1074/jbc.M110.197301
Ropolo A, Grasso D, Pardo R, Sacchetti ML, Archange C, Re AL, Seux M, Nowak J, Gonzalez CD, Iovanna JL (2007) The pancreatitis-induced vacuole membrane protein 1 triggers autophagy in mammalian cells. J Biol Chem 282(51):37124–37133
Sauermann M, Sahin Ö, Sültmann H, Hahne F, Blaszkiewicz S, Majety M, Zatloukal K, Füzesi L, Poustka A, Wiemann S (2008) Reduced expression of vacuole membrane protein 1 affects the invasion capacity of tumor cells. Oncogene 27(9):1320–1326
Siegel R, Ma J, Zou Z, Jemal A (2014) Cancer statistics, 2014. CA 64(1):9–29
Grasso D, Sacchetti ML, Bruno L, Lo Re A, Iovanna JL, Gonzalez CD, Vaccaro MI (2009) Autophagy and VMP1 expression are early cellular events in experimental diabetes. Pancreatology 9(1–2):81–88. doi:10.1159/000178878
Qian Q, Zhou H, Chen Y, Shen C, He S, Zhao H, Wang L, Wan D, Gu W (2014) VMP1 related autophagy and apoptosis in colorectal cancer cells: vMP1 regulates cell death. Biochem Biophys Res Commun 443(3):1041–1047
Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP (2007) GSEA-P: a desktop application for gene set enrichment analysis. Bioinformatics 23(23):3251–3253
Porter AG, Jänicke RU (1999) Emerging roles of caspase-3 in apoptosis. Cell Death Differ 6(2):99–104
Danial NN, Gramm CF, Scorrano L, Zhang C-Y, Krauss S, Ranger AM, Datta SR, Greenberg ME, Licklider LJ, Lowell BB (2003) BAD and glucokinase reside in a mitochondrial complex that integrates glycolysis and apoptosis. Nature 424(6951):952–956
Oltval ZN, Milliman CL, Korsmeyer SJ (1993) Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programed cell death. Cell 74(4):609–619
Gialeli C, Theocharis AD, Karamanos NK (2011) Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J 278(1):16–27
Subramaniam M, Hawse JR, Rajamannan NM, Ingle JN, Spelsberg TC (2010) Functional role of KLF10 in multiple disease processes. Biofactors 36(1):8–18
Scotton CJ, Wilson JL, Scott K, Stamp G, Wilbanks GD, Fricker S, Bridger G, Balkwill FR (2002) Multiple actions of the chemokine CXCL12 on epithelial tumor cells in human ovarian cancer. Cancer Res 62(20):5930–5938
Acknowledgments
Grant support was provided by Natural Science Foundation of Fujian Province, China (Grant No. 2013J01324).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial interests.
Additional information
Lili Zheng and Lingling Chen have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Zheng, L., Chen, L., Zhang, X. et al. TMEM49-related apoptosis and metastasis in ovarian cancer and regulated cell death. Mol Cell Biochem 416, 1–9 (2016). https://doi.org/10.1007/s11010-016-2684-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-016-2684-3