Abstract
Over the course of the torpor-arousal cycle, hibernators must make behavioral, physiological, and molecular rearrangements in order to keep a very low metabolic rate and retain organ viability. 13-lined ground squirrels (Ictidomys tridecemlineatus) remain immobile during hibernation, and although the mechanisms of skeletal muscle survival are largely unknown, studies have shown minimal muscle loss in hibernating organisms. Additionally, the ground squirrel heart undergoes cold-stress, reversible cardiac hypertrophy, and ischemia–reperfusion without experiencing fatal impairment. This study examines the role of the Janus kinase–signal transducer and activator of transcription (JAK-STAT) signaling pathway in the regulation of cell stress in cardiac and skeletal muscles, comparing euthermic and hibernating ground squirrels. Immunoblots showed a fivefold decrease in JAK3 expression during torpor in skeletal muscle, along with increases in STAT3 and 5 phosphorylation and suppressors of cytokine signaling-1 (SOCS1) protein levels. Immunoblots also showed coordinated increases in STAT1, 3 and 5 phosphorylation and STAT1 inhibitor protein expression in cardiac muscle during torpor. PCR analysis revealed that the activation of these pro-survival signaling cascades did not result in coordinate changes in downstream genes such as anti-apoptotic B-cell lymphoma-2 (Bcl-2) family gene expression. Overall, these results indicate activation of the JAK-STAT pathway in both cardiac and skeletal muscles, suggesting a response to cellular stress during hibernation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
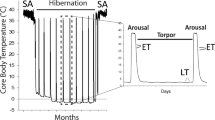
In order to survive cold environmental temperatures and/or severely depleted resources, many small mammals reduce their metabolic expenditures, hibernating for up to 9 months per year [1]. Mammalian hibernation consists of many deep torpor bouts (up to 1–3 weeks during midwinter) interrupted by short arousal periods (6–24 h, depending on the species) [1]. During these torpor bouts, the body temperature (T b) of ground squirrels decreases to 0–5 °C, heart beat decreases from 200–300 to 5–10 beats/min, metabolic rate decreases to 1–5 % of the normal resting rate at 37 °C, breathing rate decreases from over 40 breaths/min to less than 1 breath/min, and cerebral blood flow decreases by 90 % compared with euthermic levels [1–3]. By entering hibernation, ground squirrels save approximately 88 % of the energy they would need to maintain a body temperature of 37 °C during the winter months [1]. During the late summer months, ground squirrels prepare for hibernation by entering a phase of hyperphagia, with weight gains of up to 50 %. As a result, fatty acids mobilized from white adipose tissue becomes the main metabolic fuel source during hibernation [4]. The model organism of the current study, the 13-lined ground squirrel (Ictidomys tridecemlineatus), lives in the central grasslands of North America and endures harsh winter conditions by hibernating underground.
Of particular interest in the present research are the specific adaptations exhibited by cardiac and skeletal muscle that ensure that organ integrity can be maintained during hibernation. Heart rate and cardiac output decrease to 3–5 and ~2 % of the euthermic level, respectively [3]. However, the force of cardiac contraction must dramatically increase in order to maintain some organ perfusion at a very low T b, when blood is very cold and viscous [3]. Studies have shown that the force of cardiac contraction can increase two to fivefold in Richardson’s ground squirrels when body temperatures decrease from 38 to 5 °C [3]. Similar to cardiomyocytes, skeletal muscle also exhibits important metabolic and molecular changes that are essential to ensure muscle health. These changes include a preference for fatty acid oxidation, a decrease in the activity of glycolytic enzymes, enhanced stress tolerance (e.g., antioxidant defenses), and muscle-specific sarcomeric and structural changes [3, 5, 6]. As a result of many months spent in inactive torpor, skeletal muscle is also at risk for disuse atrophy. This is of particular concern given the need for well-developed capacity for shivering thermogenesis that is an important contributor to rewarming the body during arousal and to ensure motility upon emergence from hibernation in the spring. Consequently, hibernators preserve muscle mass throughout long periods of muscle disuse and starvation, unlike non-hibernators, who would suffer from relatively more muscle atrophy and degeneration. Due to the importance of maintaining cardiac and skeletal muscle health, we are interested in the mechanisms that promote cell survival during hibernation, with a particular focus on transcription factor regulation.
To achieve metabolic rate depression while maintaining homeostasis during hibernation, various changes must occur at the molecular level, including changes in gene expression, post-translational modifications, and alterations in enzyme kinetics [7]. We have identified various transcription factors which are essential to this process by regulating key genes that are essential for hibernation survival [8–10]. One such transcription factor family is the signal transducers and activators of transcription (STAT) protein family, which includes seven members: STAT1, STAT2, STAT3, STAT4, STAT5a, STAT5b, and STAT6 [11]. STATs 2, 4, and 6 are involved in cell-mediated immunity and interferon (IFN)-gamma signaling, whereas STATs 1, 3, and 5 are involved in the cell cycle, apoptosis, IFN-gamma signaling, and embryogenesis, among other processes [12]. Due to the importance of the latter processes in the viability of cardiac and skeletal muscle during torpor, STAT1, STAT3, and STAT5 will be the focus of this study.
Although STAT proteins are structurally similar, they each have distinct functions [13]. STAT1 is commonly implicated in pro-apoptotic functions due to its ability to up-regulate pro-apoptotic genes like caspase-1, FAS, p21, and p53, among others [13]. In contrast, STAT3 and STAT5 are involved in pro-survival gene expression. Furthermore, in skeletal muscle, STAT3 has been associated with myocyte proliferation in vivo and in vitro [14], while STAT5 phosphorylation has been shown to protect starving skeletal muscle [15]. In cardiac muscle, STAT3 and STAT5 are known to function as cardioprotective agents in the heart during ischemia/reperfusion by limiting cell death and necrosis, and by promoting hypertrophy and cardiac angiogenesis [11, 13, 16, 17]. Additionally, STAT3 has been implicated in cytoprotective and survival signaling pathways in damaged heart tissue due to oxidative stress [18].
The upregulation of certain genes/proteins during hibernation is important because hibernation is the time of energy conservation. Therefore, the use of metabolic fuel for the transcription/translation of particular proteins likely has importance during torpor [7]. Also, given the important role of STAT proteins in cellular survival, we hypothesized that transcriptional changes occurring due to the regulation of STAT proteins could contribute to ground squirrel survival during hibernation. In the present study, we evaluate the phosphorylation level of pro-survival STATs and their respective upstream controls including Janus kinases (JAKs), protein inhibitors of activated STATs (PIAS), and suppressors of cytokine signaling (SOCS) proteins during hibernation. Finally, we measured anti-apoptotic myeloid cell leukemia 1 (Mcl-1), B-cell lymphoma extra-large (Bcl-xL), and B-cell lymphoma 2 (Bcl-2) mRNA during hibernation, since these genes/proteins help maintain the integrity of cardiac and skeletal muscle tissue, two essential hibernator organs. In cardiac muscle, the phosphorylation of each STAT increased and there was an increase in the protein level of STAT1 inhibitors. In skeletal muscle, there were increases in the relative phosphorylation of pro-survival STATs and changes in SOCS1 protein levels which could modulate STAT gene expression. Bcl-2 family gene expression remained constant or decreased in both cardiac and skeletal muscle of hibernating ground squirrels. These results suggest that different pro-survival pathways are stimulated in skeletal and cardiac muscle to best protect these essential organs, although the direct downstream transcriptional consequences require further studies.
Methods
Animals
Thirteen-lined ground squirrels (Ictidomys tridecemlineatus) weighing approximately 150–300 g, were wild-captured by a United States Department of Agriculture-licensed trapper (TLS Research, Bloomingdale, IL) and transported to the Animal Hibernation Facility, National Institute of Neurological Disorders and Stroke (NIH, Bethesda, MD). All hibernation experiments were conducted by the laboratory of Dr. J. M. Hallenback. Guidelines set by the NINDS animal care and use committee (ACUC) concerning animal housing and experimental procedures were followed. Each thirteen-lined ground squirrel was fitted with a sensor chip (IPTT-300; Bio Medic Data Systems) injected subcutaneously while anesthetized with 5 % isoflurane and was housed individually in a shoebox cage at 21 °C. Animals were fed a standard rodent diet and water ad libitum until they gained sufficient lipid stores to enter hibernation. Animals were transferred to an environmental chamber at ~5 °C in constant darkness to enable a natural transition into torpor. Body temperature (T b), time, and respiration rate were monitored to determine sampling points. Hallmarks of hibernation include a body temperature that declines to slightly above ambient temperature and a respiratory rate that changes from continuous to intermittent breathing such that breaths are taken only every 30 s to 6 min [3, 19]. All thirteen-lined ground squirrels had been through a series of torpor-arousal bouts prior to sampling. Animals were sampled as in McMullen and Hallenbeck [20]. Tissue samples were shipped to Carleton University on dry ice. The tissues were stored at −80 °C until use. EC designates euthermic, cold room; these euthermic animals had a stable T b (~37 °C) in the 5 °C cold room, were able to enter torpor but had not done so in the past 72 h. LT designates late torpor; animals that were continuously in deep torpor for at least 5 days with T b values of 5–8 °C.
Protein extraction for Western blot analysis
All total protein samples from ground squirrel skeletal muscle and heart tissues were prepared as follows: tissues from 3 to 4 different animals from both sampling points were separately weighed and ground up using a mortar and pestle under liquid nitrogen. The sample was homogenized 1:2 w/v using a Polytron PT10 in ice-cold homogenization buffer (20 mM HEPES, 200 mM NaCl, 0.1 mM EDTA, 10 mM NaF, 1 mM Na3VO4, and 10 mM β-glycerophosphate, pH 7–8.), along with a few crystals of phenylmethanesulfonyl fluoride and 1 µL of Sigma protease inhibitor. The samples were stored on ice up until they were centrifuged at 4 °C at 10,000×g for 15 min. Immediately after centrifugation, the supernatant was removed and stored on ice. The BioRad assay method was used to determine the protein concentration and sample concentrations were standardized to 4 µg/mL. Samples used for Western blots were combined with 2X sodium dodecyl sulfate (SDS) buffer (100 mM Tris base, 4 % w/v lauryl sulfate (SDS), 20 % v/v glycerol, 0.2 % w/v bromophenol blue, and 10 % v/v 2-mercaptoethanol) to a final concentration of 2 µg/µL, boiled for 5–10 min, vortexed, and stored at −40 °C until use.
Protein extraction for Luminex® assay
Frozen cardiac and skeletal muscle tissue samples from EC and LT ground squirrels were used to prepare protein extracts according to the manufacturer’s directions and as previously described [33]. Briefly, 50 mg of frozen tissue from 4 different animals was weighed for each time point and homogenized 1:5 w/v with a glass homogenizer in ice-cold lysis buffer (1 mM Na3VO4, 10 mM NaF, 10 mM β-glycerophosphate, and 10 µL/mL Sigma protease inhibitor). The homogenized samples were incubated on ice for 30 min with periodic vortexing. Then, samples were centrifuged at 10,000 rpm for 20 min at 4 °C. The BioRad protein reagent was used to determine the concentration of soluble protein found in the supernatant. Assay Buffer 2 (EMD Millipore; Cat#43-041) was used to dilute skeletal muscle samples 0.04 μg/μL and cardiac muscle samples to 1 μg/μL. Positive and negative controls provided by the manufacturer were used in each assay, and were prepared according to the manufacturer’s instructions. Unstimulated Jurkat cell lysate (Cat#47-206) was used as a negative control, whereas Jurkat cell lysate: Anisomycin (Cat#47-207) was used as a positive control. The appropriate amount of assay buffer was added to the reconstituted lysates, as described in the manufacturer’s instructions.
Western blotting
Equal amounts of protein were loaded into each well of SDS-PAGE gels (STAT: 10 µg, 8–10 % gel; PIAS: 10–20 µg, 8 % gels; JAK: 20–30 µg, 8 % gels; SOCS: 20 µg, 15 % SDS-PAGE gels, and 15 % Tris-Tricine gels). The SDS-PAGE resolving gels were 8–15 % acrylamide, and used as a Tris–glycine buffer with a pH of 8.8. The 5 % stacking gels used a Tris–glycine buffer with a pH of 6.8. The SDS-PAGE gels were run for 45–75 min in a Tris–glycine running buffer, at 180 V, using the BioRad Mini Protean III system. The separated proteins were transferred onto a 0.45 micron polyvinylidene fluoride (PVDF) paper, at 160 mA for 60–150 min in a Tris–glycine transfer buffer. The resulting PVDF membranes were washed for 15 min in TBST (20 mM Tris base, pH 7.6, 140 mM NaCl, 0.05 % v/v Tween-20). In order to reduce unspecific binding, a blocking buffer containing either 2.5 % w/v milk, 1 mg/mL high molecular weight polyvinyl alcohol (70–100 kDa PVA), or low molecular weight PVA (30–70 kDa), were made with TBST. The membranes were probed with primary antibody (1:1000 v/v with TBST) on a rocker in a 4 °C fridge overnight, or on a rocker at room temperature for 4 h. The phospho-STAT1,3,5, SOCS1-3, and PIAS1,3,4 primary antibodies used in this study were purchased from Cell Signaling (p-STAT1 Y701 cat no. 7649, p-STAT3 Y727 cat. no. 9134, p-STAT3 Y705 cat. no. 9145, p-STAT5 Y694 cat. no. 4322, SOCS1 cat. no. 3950, SOCS2 cat. no. 2779, SOCS3 cat. no. 2932, PIAS1 cat. no. 3550, PIAS3 cat. no. 4164, PIAS4 cat. no. 4392) and the p-JAK antibodies were purchased from Santa Cruz Biotechnology (p-JAK1 Y1022/1023 cat. no. sc-16773-R, p-JAK2 Y1007/1008 cat. no. sc-16566-R, p-JAK3 Y980 cat. no. sc-16567-R). After the removal of primary antibodies, the membranes were washed in TBST for 15–20 min and were exposed to an HRP-linked anti-rabbit goat IgG secondary antibody (diluted with TBST to 1:8000–1:4000 v/v) for 40–60 min (with the exception of PIAS4 blots, which were exposed to an HRP-linked anti-rabbit sheep IgG secondary antibody). The membranes were washed for at least 30 min in TBST and then exposed using Luminol and hydrogen peroxide in the chemiluminescence visualizing system (Syngene). The total amount of protein in each lane was assessed using Coomassie Blue staining (0.25 % w/v Coomassie brilliant blue, 7.5 % v/v acetic acid, 50 % methanol) of the PVDF membranes.
Total RNA extraction
Briefly, approximately 50 mg of frozen cardiac or skeletal muscle tissue was weighed for each biological replicate and then homogenized in 1 mL Trizol (Invitrogen) using a Polytron PT1200 homogenizer. Next, 200 µL of chloroform was added and mixed before samples were centrifuged at 10,000×g for 15 min at 4 °C. The upper aqueous phase containing the RNA fraction was transferred and precipitated by the addition of 750 µL of 2-propanol. Each sample was centrifuged at 12,000×g for 15 min at 4 °C, washed with 1 mL of 70 % ethanol, and then centrifuged again for 12,000×g for 5 min. The supernatants were removed and the RNA pellets were allowed to air dry for 15–30 min at room temperature. The pellets were then resuspended in 30 µL RNase-free water. RNA purity was assessed by measuring the absorbance at 260 and 280 nm. RNA integrity was assessed by visualizing 18S and 26S ribosomal bands on a 1 % agarose gel with SybrGreen staining.
cDNA synthesis and PCR amplifications
Firstly, 3 µg RNA from each sample was incubated with 1 µL of Oligo-dT (200 ng/µL) and placed in a thermocycler at 65 °C for 5 min. Next, the samples were chilled on ice for 5 min. Reverse transcription was performed with the following reagents: 4 µL of 5X first-strand buffer (Invitrogen), 2 µL of 0.1 M DTT (Invitrogen), 1 µL of 10 mM dNTPs (BioShop), and 1 µL of MMLV Reverse transcriptase (Invitrogen). Each sample was spun down before incubating at 42 °C for 45 min. Ten-fold serial dilutions of the cDNA samples were prepared for relative quantification of select mRNAs and reference genes. PCR reagents were prepared and qPCR was performed as previously described [21] using a BioRad MyIQ2 Detection System (BioRad, Hercules, CA, USA). The primers used for PCR, synthesized by Integrated DNA Technologies (Coralville, Iowa, USA), were designed using the 13-lined ground squirrel genome from the ensemble database (http://www.ensembl.org) and using Primer BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). The primer sequences for each gene are listed in Table 1.
All PCR runs underwent melt-curve analysis and dilution curve testing to identify which amplified more than one PCR product or amplified non-mRNA products, as indicated by more than one melt curve or by no change in C t values between serial dilutions, respectively. Raw C t values obtained from each PCR run were converted to a linear form using 2-C t calculations and were normalized against the reference gene. The chosen reference gene was considered acceptable by determining that its expression did not change between control and torpor conditions, as previously described [22]. For this experiment, actin γ2 was used as the reference gene in skeletal muscle and tubulin β1 was used in cardiac muscle. Standardized values for each experimental condition were expressed as mean ± SEM, where n = 3–4 biological replicates. Differences between control and hibernation conditions were considered statistically significant when the Student’s t test yielded a result of p < 0.05.
Luminex® assay
A Luminex multiplex panel was used to measure cleaved PARP and active Caspase-3 levels in heart and muscle samples (EMD Millipore; Human Late Apoptosis Magnetic Bead Kit, Cat#48-670MAG). Targets included cleaved PARP and active Caspase-3. The protocol for multiplex analysis was performed as instructed by the manufacturer. Briefly, the premixed antibody capture beads that were supplied with the kit (EMD Millipore; Cat#42-670MAG) were sonicated, vortexed, and diluted as required. After incubation with assay buffer, sample or control lysates were individually combined with the premixed beads, followed by a 2-h incubation on a shaker at 4 °C in the dark. The microplate was placed on a Handheld Magnetic Separator Block (Cat#40-285) for 60 s and the wells were emptied of the lysates. Assay Buffer was used to wash the wells twice. The magnetic separation block was attached and the assay buffer was decanted. The Milliplex Map Detection Antibody (EMD Millipore, Cat#44-670KMG) was vortexed, diluted, and added to each well as described by the manufacturer. Then, the microplate was sealed, covered, and shaken on a rocker for 1 h at room temperature. The wells were decanted, washed, and incubated with appropriately prepared Streptavidin–Phycoerythrin (EMD Millipore; Cat#45-001H) for 15 min at room temperature on a plate shaker protected from light. Milliplex Map Amplification Buffer (EMD Millipore; Cat#43-024A) was then added to each well and rocked for 15 min at room temperature in the dark. The buffer was then decanted, the beads were resuspended in Assay Buffer, and the microplate rocked at room temperature for 5 min before taking measurements. Median fluorescence intensity (MFI) was collected using a Luminex® 200 instrument and xPonent software (Luminex® Corporation). As with PCR and Western blot data, standardized values for each experimental condition were expressed as mean ± SEM, where n = 3–4 biological replicates. Differences between control and hibernation conditions were considered statistically significant when the Student’s t test yielded a result of p < 0.05.
Results
The response of p-STAT proteins to deep torpor in cardiac and skeletal muscle
The relative expression of phosphorylated STAT1, STAT3, and STAT5 transcription factors associated with cell proliferation, differentiation, and apoptosis were measured in cardiac and skeletal muscle by immunoblotting comparing euthermic control squirrels (EC) with animals in deep torpor (LT) [23]. STAT transcriptional activity is regulated by post-translational modifications such as protein phosphorylation at several distinct sites. Transcriptional activity is enhanced when STAT1 is phosphorylated at Y701, STAT3 at Y705/S727, and STAT5 at Y694; therefore, antibodies that recognize these phosphorylation sites were used to monitor changes in the activity state. As compared with controls, relative protein levels of p-STAT1 Y701 did not change in skeletal muscle tissue during torpor (Fig. 1). Conversely, cardiac muscle showed significant changes in p-STAT1 (Y701) levels during deep torpor (increased by 2.0-fold), as compared to controls (Fig. 2). As compared with EC controls, levels of the p-STAT3 (Y705) transcription factor did not change in cardiac and skeletal muscle tissues during deep torpor. In contrast, the relative level of p-STAT3 (S727) phosphorylation in both cardiac and skeletal muscle tissues increased significantly in deep torpor (by 1.4-fold and by 1.9-fold, respectively). Also, the relative amount of p-STAT5 (Y694) increased in both cardiac and skeletal muscle tissues during LT as compared to EC (values were 2.0- and 2.1-fold higher in LT for cardiac and skeletal muscle, respectively).
Relative levels of phosphorylated STATs in 13-lined ground squirrel muscle tissue. Sampling points are euthermic controls (EC) and late torpor (LT). Histograms in (a) show mean relative expression levels of p-STAT (Y701), p-STAT3 (Y705), p-STAT3 (S727), and p-STAT5 (Y694) (±SEM, n = 4 independent protein isolations from different animals, except for p-STAT3 (S727) and p-STAT5 (Y694) where n = 7 and n = 3 were used, respectively). Representative Western blots are shown in (b). Data were analyzed using a Student’s t test. Asterisks above the LT bar indicate statistical significance, as compared with the EC time point (p < 0.05)
Relative levels of phosphorylated STATs in 13-lined ground squirrel heart tissue. Histograms in (a) show mean relative expression levels of p-STAT (Y701), p-STAT3 (Y705), p-STAT3 (S727), and p-STAT5 (Y694) (±SEM, n = 4 independent protein isolations from different animals). Representative Western blots are shown in (b). Data were analyzed using a Student’s t test. Asterisks above the LT bar indicate statistical significance with respect to the EC time point (p < 0.05)
The response of p-JAKs to deep torpor in skeletal and cardiac muscle
Immunoblotting was used to determine the relative levels of phosphorylation of p-JAKs1-3, comparing EC and LT in skeletal and cardiac muscles. In cardiac muscle, the relative levels of p-JAK1 (Y1022/1023), p-JAK2 (Y1007/1008), and p-JAK3 (Y908) were unchanged during LT as compared to EC (Fig. 3). The relative phosphorylation levels of p-JAK1 and p-JAK2 in skeletal muscle were also unchanged; however, p-JAK3 phosphorylation levels decreased fivefold during torpor as compared to controls (Fig. 4).
Relative levels of phosphorylated JAKs in 13-lined ground squirrel heart tissue. Histograms in (a) show mean relative expression levels of p-JAK1 (Y1022/1023), p-JAK2 (Y1007/Y1008), and p-JAK3 (Y908) (±SEM, n = 4 independent protein isolations from different animals). Representative Western blots are shown in (b). Data were analyzed using a Student’s t test. Asterisks above the LT bar indicate statistical significance with respect to the EC time point (p < 0.05)
Relative levels of phosphorylated JAKs in 13-lined ground squirrel muscle tissue. Histograms in (a) show mean relative expression levels of p-JAK1 (Y1022/1023), p-JAK2 (Y1007/Y1008), and p-JAK3 (Y908) (±SEM, n = 4 independent protein isolations from different animals). Representative Western blots are shown in (b). Data were analyzed using a Student’s t test. Asterisks above the LT bar indicate statistical significance with respect to the EC time point (p < 0.05)
The response of SOCS and PIAS proteins to deep torpor in skeletal and cardiac muscle
Relative levels of SOCS1-3 and PIAS1, 3 and 4 proteins were analyzed in skeletal and cardiac muscle comparing EC and LT. The relative protein levels of PIAS1, PIAS3, and PIAS4 were unchanged in skeletal muscle but increased by 3.6- and 1.9-fold for PIAS1 and PIAS4, respectively, in cardiac muscle of hibernating animals (Fig. 5). As compared with controls, relative protein levels of SOCS1, 2, and 3 did not change in cardiac muscle tissue during torpor. The relative protein levels of SOCS2 and SOCS3 did not change in skeletal muscle tissue, but there was a significant increase in the amount of SOCS1 protein in skeletal muscle tissue during LT (by 1.5-fold) (Fig. 6).
Relative levels of PIAS and SOCS proteins in 13-lined ground squirrel heart tissue. Histograms in (a) show mean relative expression levels of PIAS1, PIAS3, PIAS4, SOCS1, SOCS2, and SOCS3 (±SEM, n = 4 independent protein isolations from different animals). Representative Western blots are shown in (b). Data were analyzed using a Student’s t test. Asterisks above the LT bar indicate statistical significance with respect to the EC time point (p < 0.05)
Relative levels of PIAS and SOCS proteins in 13-lined ground squirrel muscle tissue. Histograms in (a) show mean relative expression levels of PIAS1, PIAS3, PIAS4, SOCS1, SOCS2, and SOCS3 (±SEM, n = 4 independent protein isolations from different animals except for PIAS3 where n = 3 was used for the LT condition only). Representative Western blots are shown in (b). Data were analyzed using a Student’s t test. Asterisks above the LT bar indicate statistical significance with respect to the EC time point (p < 0.05)
Changes in expression of STAT-mediated genes during deep torpor in skeletal and cardiac muscle
To determine whether the changes in STAT expression and phosphorylation pattern have an influence on the expression of STAT downstream genes, we assessed mRNA expression levels of Mcl-1, Bcl-2, and Bcl-xL in both euthermic and torpid cardiac and skeletal muscle tissues of the 13-lined ground squirrel. Mcl-1, Bcl-2, and Bcl-xL transcript levels did not change during LT with respect to EC in skeletal muscle (Fig. 7). Similarly, Mcl-1 and Bcl-2 transcript levels did not change between EC and LT time points in cardiac muscle. In contrast, Bcl-xL transcript levels in cardiac muscle decreased during LT to less than 65 % of the value at EC (p < 0.05) (Fig. 8).
Expression of mRNAs in skeletal muscle tissue samples from the euthermic control (black bar) and torpid ground squirrels (gray bar) was evaluated by RT-PCR. Relative expression of indicated mRNAs was normalized to the expression of actin gamma 2 mRNA from the same sample. The relative mRNA expression in the torpid animals was further normalized to that in the euthermic control, which was arbitrarily set as 1.0. Data are mean ± SEM (n = 3 independent trials from different EC animals and n = 4 independent trials from different LT animals). Significant difference in mRNA expression in torpid compared to that of the euthermic control according to Student’s t test was indicated with asterisks (p < 0.05)
Expression of mRNAs in cardiac muscle tissue samples from the euthermic control (black bar) and torpid ground squirrels (gray bar) was evaluated by RT-PCR. Relative expression of indicated mRNAs was normalized to the expression of tubulin beta mRNA from the same sample. The relative mRNA expression in the torpid animals was further normalized to that in the euthermic control, which was arbitrarily set as 1.0. Data are mean ± SEM (n = 4 independent trials from different animals). Significant difference in mRNA expression in torpid compared to that of the euthermic control according to Student’s t test was indicated with asterisks (p < 0.05)
The response of activated pro-apoptotic proteins to deep torpor in skeletal and cardiac muscle
A Luminex® assay was used to determine the relative levels of cleaved poly-(ADP-Ribose) polymerase (PARP) and active caspase-3, comparing EC and LT in skeletal and cardiac muscle. In skeletal muscle, cleaved PARP levels decreased to 60 % of euthermic levels during LT but caspase-3 levels remained constant (Fig. 9). There were no changes in the levels of cleaved PARP or active caspase-3 during LT in heart (Fig. 10).
Relative levels of cleaved PARP and active caspase-3 proteins in 13-lined ground squirrel muscle tissue. Histogram shows mean relative expression levels of cleaved PARP and active caspase-3 (±SEM, n = 4 independent protein isolations from different animals except for active caspase-3 where n = 3 was used for the EC condition only). Data were analyzed using a Student’s t test. Asterisks above the LT bar indicate statistical significance with respect to the EC time point (p < 0.05)
Relative levels of cleaved PARP and active caspase-3 proteins in 13-lined ground squirrel heart tissue. Histogram shows mean relative expression levels of cleaved PARP and active caspase-3 (±SEM, n = 4 independent protein isolations from different animals except for cleaved PARP where n = 3 was used for the EC condition and except for active caspase-3 where n = 3 was used for the LT condition). Data were analyzed using a Student’s t test. Asterisks above the LT bar indicate statistical significance with respect to the EC time point (p < 0.05)
Discussion
During hibernation, ground squirrel skeletal and cardiac muscles must make substantial molecular rearrangements, while keeping a very low metabolic rate, in order to avoid cell death and retain organ viability. Muscle atrophy is the reorganization and loss of muscle fiber that can result from muscle disuse, a common occurrence during extended bedrest, spaceflight, and cessation of exercise [24]. Hibernating 13-lined ground squirrels remain immobile during hibernation without loss of muscle mass, and this is of particular importance given the role of skeletal muscle in shivering thermogenesis during arousal and the importance of muscle fitness upon emergence from hibernation [5, 25]. Hibernating ground squirrel hearts must respond to cold winter conditions by increasing the force of contraction to pump viscous blood throughout the body [3, 7]. Additionally, the ground squirrel serves as an excellent model organism for cardiomyocyte viability during reversible cardiac hypertrophy, ischemia–reperfusion, and cold-stress [6, 7, 26]. Since regulatory mechanisms acting on signal transduction pathways, transcription factors, and gene expression have been associated with cell survival in hibernating mammals [27–29], the current study aimed to further our understanding of the regulation of the JAK-STAT pathway in the 13-lined ground squirrel striated muscle during hibernation.
Signal transducers and activators of transcription (STATs) are important transcription factors that control the expression of genes involved in cell cycle progression, apoptosis, T-cell development, IFN-gamma signaling, embryogenesis, and mammary gland development [30, 31]. In particular, STATs 2, 4, and 6 control T-cell development and IFN-gamma signaling, while STATs 1, 3, and 5 have roles in cell survival, growth, and development [31]. The survival of cardiac and skeletal muscle cells is crucial in order for hibernating 13-lined ground squirrels to endure harsh winter conditions, so STATs 1, 3, and 5 were decidedly the focus of our study. Using Western blotting, it was found that the levels of phosphorylated forms of STAT1 (Y701), STAT3 (Y727), and STAT5 (Y694) increased during LT in ground squirrel cardiac muscle (Fig. 2). In contrast, p-STAT3 (Y705) levels did not change during LT with respect to EC. These results are consistent with the findings of other research groups, who have monitored STAT expression and activity in vitro and in vivo during various forms of cardiac stress, including ischemia–reperfusion stress, hypoxia, hypertrophy, tissue remodeling, myocardial infarction, and oxidative stress [13]. For example, STAT3 was determined to be cardioprotective by various research groups because its phosphorylation leads to the expression of cardioprotective genes including Bcl-xL, Bcl-2, manganese-dependent superoxide dismutase (MnSOD), cyclooxygenase-2 (Cox-2), and nitric oxide synthase (iNos) [11, 13, 18], its absence leads to increased myocardial infarct size and cardiac cell apoptosis in mice hearts subjected to ischemia–reperfusion stress [13], and its upregulation in rat hearts preconditioned to ischemia–reperfusion stress is associated with smaller infarct sizes, superior ventricle function post-ischemia, and fewer cardiac cell deaths [18]. Thus, an increase in the level of phosphorylated forms of STAT3 in the cardiac muscle of hibernating ground squirrels could suggest an elevated need for cardioprotective gene expression.
STAT3 has two major phosphorylation sites—Y705 and S727. Phosphorylation at Y705 is required for STAT dimerization, nuclear translocation, and DNA binding [32]. Serine phosphorylation has been found to be equally important in DNA binding [33–35]. S727A mutants were found to be approximately 50 % as effective as their wild-type counterpart at initiating transcription, likely because this phosphorylation site aides in the recruitment of cofactors required for STAT activation of transcription [33, 35]. Therefore, an increase in p-STAT3 (S727) in both skeletal and cardiac muscle during hibernation could be important to maintain adequate DNA-binding affinity [1]. Phosphorylation at S727 is also required for mitochondrial STAT3 to augment electron transport chain (ETC) activity in order to maintain cellular homeostasis [36, 37]. Since hibernating ground squirrels dramatically decrease their metabolism in order to conserve energy, STAT3 activity at the mitochondria could support some level of ATP supply for vital metabolic processes during hibernation or may be essential during arousal when metabolic activity increases dramatically over a short period of time. Alternatively, p-STAT3 (Y705) levels did not change between euthermic and deep torpor conditions in cardiac or skeletal muscle, but maintaining basal Y705 phosphorylation levels could be important to keep the transcription factor in the nucleus during torpor (Figs. 1, 2).
In contrast to STAT3, STAT1 is known to transcribe pro-apoptotic genes, such as BAK1 and BAX [38]. Mice suffer from larger myocardial infarct sizes when STAT1 is constitutively expressed in cardiac tissue [13]. STAT1 expression is also associated with more cardiomyocyte death following ischemia–reperfusion injury [11]. The increase in the phosphorylated form of STAT1 in cardiac tissue is consistent with the results of other research groups, who have found increases in STAT1 following conditions such as ischemia–reperfusion and hypoxia, analogous stressors that the 13-lined ground squirrel endure during hibernation [11, 13]. STAT1 and STAT3 are upregulated together following various cardiac stressors, including virally induced cardiac damage and ischemia–reperfusion injury [11, 13, 39]. In each study, it was concluded that the pro-apoptotic effects of STAT1 were diminished by the anti-apoptotic effects of STAT3 [11, 13]. Specifically, STAT3 expression was shown to inhibit STAT1-mediated cardiomyocyte cell death [11]. This study shows a concurrent upregulation of both p-STAT1 (Y701) and p-STAT3 (S727) in cardiac muscle during torpor, suggesting that pro-survival signaling cascades may compete with cell death signals during deep torpor. Additionally, STAT activity is regulated by JAK phosphorylation and inhibitor expression (Fig. 11). Following docking to a receptor, dimerized and phosphorylated JAK kinases present SH2 docking sites for STATs. Activated JAKs phosphorylate STATs at tyrosine residues, which allows them to dimerize and translocate to the nucleus [40]. Suppressors of cytokine signaling (SOCS) and protein inhibitors of activated STAT (PIAS) proteins can block JAK-STAT signaling at the JAK and STAT levels, respectively. In cardiac muscle, there were no changes in phosphorylated JAK or total SOCS protein levels between euthermic and torpid ground squirrels, but PIAS1 and PIAS4 expressions increased (Figs. 3, 5). PIAS1 and PIAS3 proteins inhibit STAT1 and STAT3, respectively, by blocking their DNA-binding ability, while PIAS4 functions as a recruiter of co-repressor molecules that inhibit the transcriptional activity of STAT1 [57]. Therefore, PIAS1 and PIAS4 may be upregulated to block unfavorable effects of STAT1 upregulation during torpor.
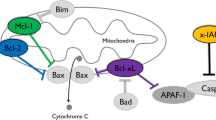
Schematic of JAK-STAT signaling cascade. Receptor tyrosine kinases (RTKs) dimerize and activate Janus kinases (JAKs) upon cytokine/growth factor/hormone docking. Phosphorylated JAKs become docking sites for signal transducers and activators of transcription (STATs), which are themselves phosphorylated. STAT transcription factors dimerize and translocate into the nucleus upon phosphorylation. STAT dimers bind DNA to initiate transcription of important genes such as Bcl-2, myc, and p21. Suppressors of cytokine signaling (SOCS) proteins inhibit JAKs by limiting their activity, by competing with STATs for JAK binding sites, and by targeting JAKs for proteasomal degradation. Protein inhibitors of activated STATs (PIAS) proteins inhibit STATs by blocking STAT DNA-binding ability or by recruiting co-repressor molecules
STAT5 also appears to be cardioprotective because STAT5a is upregulated following ischemia–reperfusion preconditioning and is associated with smaller myocardial infarct size and cardiomyocyte cell death [41], is activated by ischemic injury and its upregulation has been associated with angiotensinogen gene expression, followed by smaller myocardial infarct sizes and fewer cardiomyocyte deaths [17], and STAT5a knockout mice were unable to precondition their hearts following short ischemia–reperfusion cycles, but STAT6 knockout mice were unable to [11, 41]. In the current study, levels of p-STAT5 (Y694) doubled during deep torpor relative to euthermic controls in cardiac muscle (Fig. 2). This suggests that the signaling cascade induced by any of the various stressors that affect the hibernator heart (hypoxia, ischemia/reperfusion, hypertrophy, etc.) could activate STAT5 in order to transcribe selective genes involved in the protection from these stressors. One mechanism of STAT3- and STAT5-mediated pro-survival effects in cells is through binding to the promoters of anti-apoptotic genes (Mcl-1, Bcl-xL, Bcl-2, and x-IAP), which prevents apoptosis by inhibiting mitochondrial outer membrane permeabilization (MOMP) and subsequent caspase activation [42, 43]. PCR analyses were performed using primer sets specific for Mcl-1, Bcl-2, and Bcl-xL mRNA. All mRNA levels remained constant during LT with respect to EC in both tissues, except Bcl-xL in cardiac muscle, which decreased during LT to just over 60 % of the EC value (Figs. 7, 8). Interestingly, similar trends in relative Mcl-1, Bcl-2, and Bcl-xL protein levels were found in I. tridecemlineatus cardiac and skeletal muscle comparing euthermic conditions and late torpor [27]. To determine if unchanging levels of Bcl-2 family members had an impact on hibernator heart and skeletal muscle cell viability, a Luminex assay was performed to measure the levels of active caspase-3 and cleaved PARP. Active caspase-3 functions as a protease and the N-terminal domain of cleaved PARP blocks the DNA repair activity of uncleaved PARP in order to facilitate apoptosis [44]. The levels of these markers for apoptosis did not change in heart during LT, suggesting that apoptosis is not prevalent during torpor (Fig. 10). These results are consistent with the results of Rouble et al. who found unchanging levels of p-p53 (S46), a phosphorylated protein present when the cell is programed for apoptosis, in the heart and brain of hibernating 13-lined ground squirrels [28]. Together, these results suggest that increases in the phosphorylated forms of STAT do not increase the expression of the Bcl-2 family of anti-apoptotic proteins during torpor, and must protect skeletal and cardiac muscle by activating other cellular pathways. For example, the study by Rouble et al. found significant increases of caspase inhibitors c-IAP1/2 and x-IAP in 13-lined ground squirrel heart and muscle, respectively [28]. STAT-controlled upregulation of hypoxia-induced genes for iNOS and COX-2 could also protect the heart from ischemia–reperfusion injury prompted by increased breathing rate and rapid influx of reactive oxygen species upon rewarming [13]. These data suggest that STAT1, 3 and 5 are activated in 13-lined ground squirrel and may have pro-survival effects, but they are not involved in the expression of the anti-apoptotic Bcl-2 family genes during torpor.
Similar to the changes in phosphorylation levels found in cardiac muscle tissue, p-STAT3 (S727) and p-STAT5 (Y694) significantly increased while p-STAT3 (Y705) remained the same between control and torpor, in skeletal muscle tissue (Fig. 1). We anticipated increases in the levels of phosphorylated forms of STAT3 and STAT5 to lead to protective effects in hibernating ground squirrel muscle, which is at risk of apoptosis as a result of muscle disuse, muscle atrophy, as well the conditions that affect all tissues during hibernation including nutrient deprivation, hypoxia, DNA damage, ion imbalances, etc. [42]. Active caspase-3 levels did not change during LT compared to EC, but cleaved PARP levels decreased to 60 % of EC during LT (Fig. 9). This suggests that anti-apoptotic pathways are activated in skeletal muscle to ensure cell viability throughout torpor. Although there were no changes in the mRNA levels of anti-apoptotic genes in skeletal muscle between euthermic and torpid ground squirrels, STAT3 and 5 could be involved in upregulating other protective molecular pathways. STAT3 signaling cascades also control embryonic development, muscle generation, and muscle regrowth following injury [45]. Depending on the cofactors with which it is associated, STAT3 enables myoblast differentiation or myogenic proliferation during muscle development [45–47]. Additionally, STAT5 is one of the most important transcription factors in skeletal muscle tissue and mediates the expression of genes like IGF-1, pyruvate dehydrogenase 4 (PDK4), and fatty acid synthase in order to maintain proper skeletal muscle function [48, 49]. Therefore, increases in the levels of phosphorylated STAT3 and STAT5 could prevent muscle loss (from muscle disuse and starvation) in hibernating 13-lined ground squirrels by transcribing genes involved in caspase inhibition, energy metabolism, and/or cellular growth.
In skeletal muscle, a decrease in p-JAK3 (Y908) was observed during LT, with respect to EC (Fig. 4). Interestingly, p-JAK1 and p-JAK3 heterodimerization can result in the phosphorylation of STAT3 and STAT5 [50]. Several studies have described the various outcomes of JAK1-3 activation in skeletal muscle. For example, JAK3 inhibition promotes myogenic differentiation via STAT1 and STAT3 activity [46]. Therefore, the increase in p-STAT3 (S727) phosphorylation could indeed be the result of the decrease in p-JAK3 (Y908) phosphorylation in skeletal muscle. In contrast, JAK1-3 phosphorylation levels did not change during torpor with respect to euthermic controls in cardiac muscle, which suggests alternative mechanisms of p-STAT1 (Y701), p-STAT3 (S727), and p-STAT5 (Y694) phosphorylation in heart tissue. For example, STATs can be phosphorylated by non-receptor tyrosine kinases such as Src kinases, Abelson (Abl) kinases, and by transmembrane G-protein-coupled receptors like serotonin 5-HT2A receptor and angiotensin II receptor [51–53]. JAK1 and Abl kinases are moderately to highly expressed in skeletal and cardiac muscle. Consequently, a larger role for the more highly expressed kinases (JAK1 and Abl) in the activation of STATs in these tissues could provide a possible explanation for STAT activation despite no changes in JAK2/3 phosphorylation in heart [11, 50]. The discord between STAT phosphorylation and JAK activation may also be a result of JAK phosphorylation during early torpor or rapid de-phosphorylation during LT, such that it is not detected at the exact sampling time of LT animals. Further analysis is required in order to understand STAT-mediated activation. STAT signaling in skeletal muscle was also found to be regulated by inhibitors. SOCS1 and SOCS3 can inhibit both JAK1 and JAK2 [39, 50, 54, 55]. Thus, the increase in SOCS1 correlates with unchanging JAK1 and JAK2 phosphorylation levels between control and torpor, and could regulate the JAK-STAT pathway to ensure that only certain effector genes are transcribed during hibernation, when ATP and nutrients are limited (Fig. 6).
Conclusion
The current data suggest that the activation of select JAK-STAT pathway proteins in cardiac and skeletal muscle could help preserve the integrity of these organs throughout hibernation. Increases in pro-survival STATs and STAT1 inhibitors could limit the pro-apoptotic influence of STAT1. Increases in STAT3 and STAT5 phosphorylation could ensure cardiac and skeletal muscle viability by upregulating expression of caspase inhibitors c-IAP1/2 and x-IAP, upregulation hypoxia-induced genes, and genes involved in muscle proliferation. The change in the expression of certain activators of STATs (JAKs, Src kinases, Abl kinases, etc.) or inhibitors of STATs (PIAS and SOCS proteins) provide additional regulation of this complex pathway. These seemingly small metabolic changes may be paramount when low temperature, muscle disuse, hypoxia, ischemia–reperfusion, as well as the many other physiological challenges associated with torpor are present and are able to stimulate damaging pro-apoptotic pathways. The 13-lined ground squirrel serves as a great model of cellular stress and of the metabolic changes required to maintain organ viability throughout the winter season.
References
Storey KB, Storey JM (2004) Mammalian hibernation: biochemical adaptation and gene expression. In: Storey KB (ed) Functional metabolism: regulation and adaptation. Wiley, Hoboken, pp 443–472
Storey KB (2010) Out cold: biochemical regulation of mammalian hibernation—a mini-review. Gerontology 56:220–230. doi:10.1159/000228829
Wang LCH, Lee TF (1996) Torpor and hibernation in mammals: metabolic, physiological, and biochemical adaptations. In: Handbook of physiology: environmental physiology, pp 507–532
Sheriff MJ, Fridinger RW, Tøien Ø et al (2013) Metabolic rate and prehibernation fattening in free-living arctic ground squirrels. Physiol Biochem Zool 86:515–527. doi:10.1086/673092
Cotton CJ, Harlow HJ (2010) Avoidance of skeletal muscle atrophy in spontaneous and facultative hibernators. Physiol Biochem Zool 83:551–560. doi:10.1086/650471
Tessier SN, Storey KB (2012) Myocyte enhancer factor-2 and cardiac muscle gene expression during hibernation in thirteen-lined ground squirrels. Gene 501:8–16. doi:10.1016/j.gene.2012.04.004
Storey KB, Storey JM (2000) Gene expression and protein adaptations in mammalian hibernation. In: Heldmaier G, Klingenspor M (eds) Life in the cold. Springer, New York, pp 303–313
Luu BE, Tessier SN, Duford DL, Storey KB (2015) The regulation of troponins I, C and ANP by GATA4 and Nk2–5 in heart of hibernating thirteen-lined ground squirrels, Ictidomys tridecemlineatus. PLoS ONE 10:e0117747. doi:10.1371/journal.pone.0117747
Wu CW, Storey KB (2014) FoxO3a-mediated activation of stress responsive genes during early torpor in a mammalian hibernator. Mol Cell Biochem. doi:10.1007/s11010-014-1969-7
Allan ME, Storey KB (2012) Expression of NF-kB and downstream antioxidant genes in skeletal muscle of hibernating ground squirrels, Spermophilus tridecemlineatus. Cell Biochem Funct 30:166–174. doi:10.1002/cbf.1832
Boengler K, Hilfiker-Kleiner D, Drexler H et al (2008) The myocardial JAK/STAT pathway: from protection to failure. Pharmacol Ther 120:172–185. doi:10.1016/j.pharmthera.2008.08.002
Siveen KS, Sikka S, Surana R et al (2014) Targeting the STAT3 signaling pathway in cancer: role of synthetic and natural inhibitors. Biochim Biophys Acta 1845:136–154. doi:10.1016/j.bbcan.2013.12.005
Barry SP, Townsend PA, Latchman DS, Stephanou A (2007) Role of the JAK-STAT pathway in myocardial injury. Trends Mol Med 13:82–89. doi:10.1016/j.molmed.2006.12.002
Trenerry MK, Gatta ADP, Cameron-Smith D (2011) JAK/STAT signaling and human in vitro myogenesis. BMC Physiol 11:6. doi:10.1186/1472-6793-11-6
Vendelbo MH, Jørgensen JO, Pedersen SB et al (2010) Exercise and fasting activate growth hormone-dependent myocellular signal transducer and activator of transcription-5b phosphorylation and insulin-like growth factor-I messenger ribonucleic acid expression in humans. J Clin Endocrinol Metab 95:64–68. doi:10.1210/jc.2010-0689
Cambi GE, Lucchese G, Djeokeng MMH et al (2012) Impaired JAK2-induced activation of STAT3 in failing human myocytes. Mol BioSyst 8:2351. doi:10.1039/c2mb25120e
Mascareno E, El-Shafei M, Maulik N et al (2001) JAK/STAT signaling is associated with cardiac dysfunction during ischemia and reperfusion. Circulation 104:325–329. doi:10.1161/01.CIR.104.3.325
Hattori R, Maulik N, Otani H et al (2001) Role of STAT3 in ischemic preconditioning. J Mol Cell Cardiol 33:1929–1936. doi:10.1006/jmcc.2001.1456
Nelson CJ, Otis JP, Carey HV (2009) A role for nuclear receptors in mammalian hibernation. J Physiol 587:1863–1870. doi:10.1113/jphysiol.2008.167692
McMullen DC, Hallenbeck JM (2010) Regulation of Akt during torpor in the hibernating ground squirrel, Ictidomys tridecemlineatus. J Comp Physiol B Biochem Syst Environ Physiol 180:927–934. doi:10.1007/s00360-010-0468-8
Pellissier F, Glogowskib CM, Heinemannb SF et al (2006) Lab assembly of a low-cost, robust SYBR green buffer system for quantitative real-time polymerase chain reaction. Anal Biochem 350:310–312. doi:10.1016/j.ab.2005.12.002
Schmittgen TD, Zakrajsek BA (2000) Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods 46:69–81. doi:10.1016/S0165-022X(00)00129-9
Dorritie KA, Redner RL, Johnson DE (2014) STAT transcription factors in normal and cancer stem cells. Adv Biol Regul. doi:10.1016/j.jbior.2014.05.004
Wickler SJ, Hoyt DF, van Breukelen F (1991) Disuse atrophy in the hibernating golden-mantled ground squirrel, Spermophilus lateralis. Am J Physiol 261:R1214–R1217
Ivakine EA, Cohn RD (2014) Maintaining skeletal muscle mass: lessons learned from hibernation. Exp Physiol 99:632–637. doi:10.1113/expphysiol.2013.074344
Grabek KR, Karimpour-Fard A, Epperson LE et al (2011) Multistate proteomics analysis reveals novel strategies used by a hibernator to precondition the heart and conserve ATP for winter heterothermy. Physiol Genomics 43:1263–1275. doi:10.1152/physiolgenomics.00125.2011
Biggar KK, Wu C-W, Tessier SN et al (2015) Modulation of gene expression in key survival pathways during daily torpor in the gray mouse lemur, Microcebus murinus. Genomics Proteomics Bioinform 13:111–118. doi:10.1016/j.gpb.2015.03.001
Rouble AN, Hefler J, Mamady H et al (2013) Anti-apoptotic signaling as a cytoprotective mechanism in mammalian hibernation. PeerJ 1:e29. doi:10.7717/peerj.29
Hindle AG, Grabek KR, Epperson LE et al (2014) Metabolic changes associated with the long winter fast dominate the liver proteome in 13-lined ground squirrels. Physiol Genomics 46:348–361. doi:10.1152/physiolgenomics.00190.2013
Stephanou A (2009) JAK-STAT pathway in disease. Landes Bioscience, Austin
Calò V, Migliavacca M, Bazan V et al (2003) STAT proteins: from normal control of cellular events to tumorigenesis. J Cell Physiol 197:157–168. doi:10.1002/jcp.10364
Wen Z, Darnell JE (1997) Mapping of Stat3 serine phosphorylation to a single residue (727) and evidence that serine phosphorylation has no influence on DNA binding of Stat1 and Stat3. Nucleic Acids Res 25:2062–2067. doi:10.1093/nar/25.11.2062
Shen Y, Schlessinger K, Zhu X et al (2004) Essential role of STAT3 in postnatal survival and growth revealed by mice lacking STAT3 serine 727 phosphorylation. Mol Cell Biol 24:407–419. doi:10.1128/MCB.24.1.407-419.2004
Gough DJ, Koetz L, Levy DE (2013) The MEK-ERK pathway is necessary for serine phosphorylation of mitochondrial STAT3 and ras-mediated transformation. PLoS ONE 8:1–9. doi:10.1371/journal.pone.0083395
Wen Z, Zhong Z, Darnell JE (1995) Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 82:241–250. doi:10.1016/0092-8674(95)90311-9
Redell MS, Ruiz MJ, Alonzo T, Tweardy DJ (2010) Abstract 1791: Stat3 signaling in acute myeloid leukemia: ligand-dependent and -independent activation, impact on prognosis, and induction of apoptosis by a novel Stat3 inhibitor. Cancer Res 70:1791. doi:10.1158/1538-7445.AM10-1791
Wegrzyn J, Potla R, Chwae Y et al (2009) Function of mitochondrial STAT3 in cellular respiration. Science (80-) 323:793–797. doi:10.1126/science.1164551
Satoh JI, Tabunoki H (2013) A comprehensive profile of ChIP-Seq-based STAT1 target genes suggests the complexity of STAT1-mediated gene regulatory mechanisms. Gene Regul Syst Bio 2013:41–56. doi:10.4137/GRSB.S11433
Tamiya T, Kashiwagi I, Takahashi R et al (2011) Suppressors of cytokine signaling (SOCS) proteins and JAK/STAT pathways: regulation of T-cell inflammation by SOCS1 and SOCS3. Arterioscler Thromb Vasc Biol 31:980–985. doi:10.1161/ATVBAHA.110.207464
Imada K, Leonard WJ (2000) The Jak-STAT pathway. Mol Immunol 37:1–11
Yamaura G, Turoczi T, Yamamoto F et al (2003) STAT signaling in ischemic heart: a role of STAT5A in ischemic preconditioning. Am J Physiol Heart Circ Physiol 285:H476–H482. doi:10.1152/ajpheart.00079.2003
Spierings D, McStay G, Saleh M et al (2005) Connected to death: the (unexpurgated) mitochondrial pathway of apoptosis. Science 310:66–67. doi:10.1126/science.1117105
Labi V, Erlacher M (2015) How cell death shapes cancer. Cell Death 6:e1675. doi:10.1038/cddis.2015.20
D’Amours D, Sallmann FR, Dixit VM, Poirier GG (2001) Gain-of-function of poly(ADP-ribose) polymerase-1 upon cleavage by apoptotic proteases: implications for apoptosis. J Cell Sci 114:3771–3778
Wang K, Wang C, Xiao F et al (2008) JAK2/STAT2/STAT3 are required for myogenic differentiation. J Biol Chem 283:34029–34036. doi:10.1074/jbc.M803012200
Jang Y-N, Baik EJ (2013) JAK-STAT pathway and myogenic differentiation. Jak-Stat 2:e23282. doi:10.4161/jkst.23282
Sun L, Ma K, Wang H et al (2007) JAK1-STAT1-STAT3, a key pathway promoting proliferation and preventing premature differentiation of myoblasts. J Cell Biol 179:129–138. doi:10.1083/jcb.200703184
Consitt LA, Wideman L, Hickey MS, Morrison RF (2008) Phosphorylation of the JAK2-STAT5 pathway in response to acute aerobic exercise. Med Sci Sports Exerc 40:1031–1038. doi:10.1249/MSS.0b013e3181690760
Klover P, Chen W, Zhu B-M, Hennighausen L (2009) Skeletal muscle growth and fiber composition in mice are regulated through the transcription factors STAT5a/b: linking growth hormone to the androgen receptor. FASEB J 23:3140–3148. doi:10.1096/fj.08-128215
Kiu H, Nicholson SE (2012) Biology and significance of the JAK/STAT signalling pathways. Growth Factors 30:88–106. doi:10.3109/08977194.2012.660936
Al Zaid Siddiquee K, Turkson J (2008) STAT3 as a target for inducing apoptosis in solid and hematological tumors. Cell Res 18:254–267. doi:10.1038/cr.2008.18
Subramaniam A, Shanmugam MK, Perumal E et al (2013) Potential role of signal transducer and activator of transcription (STAT)3 signaling pathway in inflammation, survival, proliferation and invasion of hepatocellular carcinoma. Biochim Biophys Acta: Rev Cancer 1835:46–60. doi:10.1016/j.bbcan.2012.10.002
Yu H, Jove R (2004) The STATs of cancer—new molecular targets come of age. Nat Rev Cancer 4:97–105. doi:10.1038/nrc1275
Valentino L, Pierre J (2006) JAK/STAT signal transduction: regulators and implication in hematological malignancies. Biochem Pharmacol 71:713–721. doi:10.1016/j.bcp.2005.12.017
Babon JJ, Kershaw NJ, Murphy JM et al (2013) Suppression of cyokine signalling by SOCS3: characterisation of the mode of inhibition and the basis of its specificity. Immunity 36:239–250. doi:10.1016/j.immuni.2011.12.015
Acknowledgments
We thank Dr. J. M. Hallenbeck at the NIH for providing ground squirrel tissues, Bryan E. Luu for advice and assistance with polymerase chain reaction and Luminex® assay preparation, and Jan Storey for editorial review of this manuscript. This work was supported by a Discovery Grant (#6793) from the Natural Sciences and Engineering Research Council of Canada and a grant from the Heart and Stroke Foundation of Canada (#G-14-0005874). KBS holds the Canada Research Chair in Molecular Physiology, SNT holds a NSERC Postdoctoral Fellowship, and SML held an NSERC Undergraduate Student Research Award.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Logan, S.M., Tessier, S.N., Tye, J. et al. Response of the JAK-STAT pathway to mammalian hibernation in 13-lined ground squirrel striated muscle. Mol Cell Biochem 414, 115–127 (2016). https://doi.org/10.1007/s11010-016-2665-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-016-2665-6