Abstract
Estrogen is synthesized from cholesterol and high cholesterol levels are suggested to be associated with increased risk of estrogen receptor(ER)-positive breast cancer. The cholesterol metabolite 27-hydroxycholesterol (27-OHC) was recently identified as a selective estrogen receptor modulator (SERM) and may therefore impact breast cancer progression. However, the mechanisms by which 27-OHC may contribute to breast cancer are not all known. We determined the extent to which 27-OHC regulates cell proliferation in MCF7 ER-positive breast cancer cell line involving the tumor suppressor protein p53. We found that treatment of MCF7 cells with 27-OHC resulted reduced p53 transcriptional activity. Conversely, treatment of the ER-negative MDA-MB 231 cells with 27-OHC induced no significant change in p53 activity. Exposure of MCF7 cells to 27-OHC was also associated with increased protein levels of the E3 ubiquitin protein ligase MDM2 and decreased levels of p53. Moreover, 27-OHC also enhanced physical interaction between p53 and MDM2. Furthermore, 27-OHC-induced proliferation was attenuated using either the p53 activator Tenovin-1 or the MDM2 inhibitor Nutlin-3 and Mdm2 siRNA. Taken together, our results indicate that 27-OHC may contribute to ER-positive breast cancer progression by disrupting constitutive p53 signaling in an MDM2-dependent manner.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is the most common cancer amongst American women, with one in every eight women developing this disease [1]. The causes of breast cancer are multi-factorial, including environmental agents and genetic susceptibilities likely playing a role in the pathogenesis of this disease. Estrogen Receptor (ER) alpha and its agonists, estradiol (and estradiol-like compounds), play a significant role in the progression of ER-positive breast cancer forms. Several therapeutic strategies have been developed against ER, however, a significant number of ER-positive breast carcinoma patients experience drug resistance [2–4].
27-hydroxycholesterol (27-OHC) has been characterized as an endogenous ligand for ER [5–7]. 27-OHC is an oxysterol formed from cholesterol through the enzyme CYP27A1. This oxysterol is the most abundant cholesterol metabolite in plasma, and also accumulates in macrophages. In vitro, 27-OHC elicits a signaling response via ER at concentrations as low as 0.1 µM [6]. In the plasma of healthy human subjects, 27-OHC is found at concentrations of 0.2–0.6 µM, and these concentrations can increase dramatically under conditions such as hypercholesterolemia [8]. Thus, fluctuations in 27-OHC may modulate ER and potentially contribute to the pathogenesis of ER+ breast cancers. While it has been established that 27-OHC is an endogenous selective ER modulator (SERM) and that it exacerbates breast cancer pathophysiology [5, 9, 10], its role in molecular events following ER activation in the context of breast cancer pathogenesis is not fully understood.
The tumor suppressor protein, p53 plays an important role in apoptosis, cell cycle and senescence. Under normal conditions, wild type p53 is in “stand by” mode. Under genotoxic stress, p53 is activated to prevent anomalous cell proliferation and neoplastic development. Hence, p53 has been extensively studied as an anticancer target and as a cancer prognostic tool to diagnose and treat several types of cancers [11–17]. Levels of p53 are regulated by the E3 ubiquitin ligase, Mouse Double Minute 2 protein (MDM2). MDM2 tags p53 to undergo ubiquitination and subsequently proteasomal degradation [18–20]. In this report, we determined the effects of 27 OHC on cell proliferation in the context of p53 and MDM2 regulation. We found that 27-OHC via ER inhibits p53 transcriptional activity in an MDM2-dependent manner, resulting in cell proliferation.
Methods
Reagents
27-OHC was purchased from Santa Cruz Biotechnologies (Santa Cruz, CA), the p53 activator, tenovin-1 from Tocris Bioscience (Ellisville, MO), Nutlin-3 and Fulvestrant from Cayman Chemicals (Ann Arbor, MI), the reporter constructs encoding p53 response elements conjugated to the firefly luciferase gene from SA Biosciences (Frederick, MD), and β-estradiol from Sigma-Aldrich (St. Louis, MO). All cell culture reagents, with the exception of fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA) were from Invitrogen (Carlsbad, CA). Human MCF7 and MDA-MB 231 cell lines were purchased from ATCC (Manassas, VA).
Cell culture
The human ER-positive breast cancer cells MCF7 and ER-negative breast cancer cells MDA-MB-231 were grown in phenol red-free DMEM/F12 medium containing 10 % charcoal dextran stripped fetal bovine serum (FBS) and 1 % antibiotic/antimycotic mix. Cells were maintained at 37 °C in a saturated humidity atmosphere containing 95 % air and 5 % CO2.
Cell proliferation assays
Proliferation assays were conducted on black 96 well plates using CyQUANT Direct Cell Proliferation Assay purchased from Invitrogen (Carlsbad, CA), which quantifies cell number using DNA content and membrane integrity. Cells were processed for proliferation as per manufacturer’s protocol and read using Spectra MAX GEMINI EM (Molecular Devices).
Dual luciferase assays
Dual Luciferase Reporter Assay System (Promega; Madison,WI) was used to determine the effect of 27-OHC on p53 activity. MCF7 and MDA-MB-231 cells were incubated for 18 h with transfection ready p53 response element conjugated with firefly luciferase construct and constitutively expressing Renilla Luciferase construct (SA biosciences; Valencia,CA) using Lipofectamine 2000 (Invitrogen; Carlsbad, CA) as per the manufacturer’s recommendations. Firefly luciferase readings were normalized against constitutive Renilla luciferase readings.
Western blot analysis
Treated MCF7 cells were washed with PBS, trypsinized and centrifuged at 5000 g. The pellets were washed with PBS and homogenized in M-PER tissue protein extraction reagent (Thermo Scientific; Waltham, MA) supplemented with protease and phosphatase inhibitors. Denatured proteins (5 µg) were separated in 10 or 12.5 % SDS-PAGE gels, transferred to a PVDF membrane (Millipore; Billerica, MA) and incubated with antibodies to p53 (1:1000, Thermo Scientific; Waltham, MA), or MDM2 (1:1000, Santa Cruz; Dallas, TX). β-actin was used as a gel loading control for the whole cell homogenates. The blots were developed with enhanced chemiluminescence (ECL Clarity kit, Bio-Rad; Hercules, CA). Bands were visualized on a PVDF membrane and analyzed by LabWorks 4.5 software on a UVP Bioimaging System (Upland, CA).Quantification of results was performed by densitometry and the results analyzed as total integrated densitometric values (arbitrary units).
Co-immunoprecipitation
Co-immunoprecipitation (Co-IP) in cell homogenates was performed for p53 and MDM2 using “Catch and Release” immunoprecipitation kit (Millipore; Billerica, MA) according to the manufacturer’s protocol. Briefly, 3 × 106 MCF7 cells were homogenized in Mammalian Protein Extraction Reagent (MPER) supplemented with protease and phosphatase inhibitors (Thermo Scientific; Waltham, MA). The homogenates containing the equivalent to 500 μg of total protein content were incubated with 2 μg of p53 mouse antibody (1:1000; Thermo Scientific; Waltham, MA) or 2 μg of MDM2 mouse antibody (1:1000; Santa Cruz; Dallas, TX) overnight in the spin columns followed by elution. 5 μL of the eluate from p53 antibody precipitated protein-antibody complex was resolved on a SDS-PAGE gel followed by transfer onto a polyvinylidene difluoride (PVDF) membrane (Bio-Rad; Hercules, CA) and incubated with MDM2 antibody followed by development with enhanced chemiluminescence ECL clarity (Bio-Rad; Hercules, CA). Analogously, 5 μL of the eluate from anti-MDM2 antibody precipitated protein-antibody complex was resolved on a SDS-PAGE gel followed by transfer onto a PVDF membrane and incubated with p53 antibody followed by development with enhanced chemiluminescence ECL clarity. Bands were visualized on a PVDF membrane and analyzed by LabWorks 4.5 software on a UVP Bioimaging System.
Double immunofluorescence staining
Coverslip seeded cells were rinsed with PBS and fixed in cold acetone, blocked with 10 % normal goat serum and incubated overnight at 4 °C with p53 antibody (anti-rabbit) and MDM2 (anti-mouse). p53 was conjugated to Texas Red and MDM2 to Alexa Fluor 488. All coverslips were washed and mounted with Vectashield containing DAPI and visualized with a Zeiss LSM 510 META confocal system coupled to a Zeiss Axiophot 200 inverted epifluorescence microscope (Carl Zeiss Microscopy; Dublin, CA). Quantification of percent overlap was determined using MetaMorph Microscopy Automation and Image Analysis Software (Molecular Devices).
Small interfering RNA
The cells were transfected with MDM2 siRNA using Lipofectamine 2000 (Invitrogen; Carlsbad, CA) and incubated for 48 h, followed by their respective treatments. Non-silencing control (scrambled) siRNA was obtained from Santa Cruz Biotechnologies (sc-37007; Dallas, TX). The siRNA to MDM2 sense and antisense strands were GCUUCGGAACAAGAGACCC and GGGUCUCUUGUUCCGAAGC (Santa Cruz; Dallas, TX).
Statistical analysis
All the assays were carried out in triplicates. The significance of differences was assessed by unpaired t test and one-way analysis of variance (One-Way ANOVA) followed by Tukey’s post hoc test. Statistical analysis was performed with GraphPad Prism software 4.01. Quantitative data for all experimental analyses are presented as mean values ± SEM with unit value assigned to control and the magnitude of differences among the samples being expressed relative to the unit value of control.
Results
27-OHC increases proliferation in ER+ breast cancer cells
27-OHC has been reported to be a novel SERM and an agent that can promote ER+ breast cancer growth [5, 6, 9, 10]. As cell proliferation is considered a hallmark of tumor growth and cancer progression [21], we measured proliferation with and without 27-OHC treatment in ER-positive MCF7 and ER-negative MDA-MB 231 cell lines. Treatment of MCF7 with 0.1 or 1 µM 27-OHC increased cell proliferation by about 80 % compared to treatment with vehicle (Fig. 1a). Treatment with estradiol of MCF7 cells also increased proliferation in a magnitude comparable to that of 27-OHC. On the other hand, the ER-negative cells MDA-MB-231 did not exhibit significant proliferation when treated with 27-OHC (Fig. 1b). These result are in accordance with the recent discovery that 27-OHC binds to ER and exacerbates ER-positive breast cancers [6, 7].
27-OHC induces proliferation in breast cancer cell lines. a Proliferation assay in the MCF7 cells shows an increase in proliferation 48 h after treatment with 0.1 or 1 μM of 27-OHC and 2 nM Estradiol (E2) compared to treatment with vehicle. b Conversely to MCF7 cells, the MDA-MB 231 cells exhibit no change in proliferation in the presence of 27-OHC either at 0.1 or 1 μM concentration for 48 h. Data are expressed as Mean ± SEM. ***p < 0.001 versus vehicle
27-OHC reduces transcriptional activity of p53
In breast cancers, p53 is often mutated implicating a disruption and deficiency in its activity, contributing to breast cancer progression. Moreover, p53 is a promising target in breast cancers [16, 22]. MCF7 is one of the few breast cancer cells which has wild type p53, most ER+ breast cancer cell lines have mutated p53 [23, 24]. We used MCF7 cell line which is also very commonly used and has shown promise in breast cancer therapeutic studies as a reference cell model [25, 26]. Published work suggests that 27-OHC stimulates cell proliferation in breast cancer cells but not in normal breast epithelial cells [5, 6, 9]. Since MCF7 expresses wild type p53 it is vital to examine the impact of 27 OHC on p53 activity [23, 24, 27, 28]. Subsequently, we transfected cells with a luciferase reporter linked to a p53 receptor element and treated with 27-OHC. Treatment of MCF7 with 0.1 or 1 µM 27-OHC significantly decreased p53-driven transcription by ~25 % compared to incubation with vehicle (Fig. 2a). Interestingly, treatment with estradiol did not induce significant changes in p53 activity compared to vehicle (Fig. 2a). In the ER-negative MDA-MB 231 cells, 27-OHC, either at 0.1 or 1 µM, exerted no significant effect on p53 activity (Fig. 2b). To corroborate if the action by 27-OHC which reduced p53-mediated transcription was via ER, we used fulvestrant, an ER inhibitor. We found that when 27-OHC was concomitantly treated with fulvestrant, the 27-OHC-induced p53 inactivation was attenuated (Fig. 2c). This result suggests that inhibitory actions of 27-OHC on p53 are ER mediated.
27-OHC reduces p53 activation. a Luciferase reporter assay in MCF7 cells demonstrates a decrease in p53 activity in the presence of 0.1 μM 27-OHC, 1 μM 27-OHC, or 2 nM Estradiol (E2) for 24 h. b No change in p53 activity was detected with the luciferase reporter assay in the ER-negative MDA-MB 231 cells treated with 0.1 μM or 1 μM 27-OHC for 24 h. c Luciferase reporter assay in MCF7 cells demonstrates that fulvestrant, an ER inhibitor, attenuates 27-OHC-induced p53 inactivity when treated with 1 µM 27-OHC and/or 5 µM of fulvestrant. Data are expressed as Mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 versus vehicle. ## p < 0.01 versus 27-OHC only. RLU Relative luciferase units
27-OHC regulates p53 and MDM2 expression
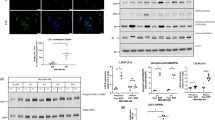
Regulation of p53 degradation is important to maintain its activity. MDM2 plays a critical role in regulating p53 levels by enhancing p53 inactivity and degradation. MDM2 catalyzes p53 degradation by flagging it for destruction. In contrast, during DNA damage MDM2 undergoes self-ubiquitination and downregulates its protein expression, which leads to the upregulation of the DNA damage response through p53 [18, 19]. MDM2 is overexpressed in human cancers where it causes the disruption of p53 signaling and potentially other oncogenic pathways [18, 19]. We determined the effect of 27-OHC on p53 and MDM2 expression levels using Western blot analyses. We found that treatment of cells with 27-OHC decreased p53 expression levels by ~50 % and increased MDM2 expression levels by ~28 % compared to vehicle (Fig. 3a–c). This result suggests that 27-OHC downregulates p53 expression and upregulates MDM2 expression.
27-OHC reduces p53 and increases MDM2 levels. a Representative Western blot and densitometric analysis of MCF7 cells showing a substantial decrease in p53 levels (b) and a significant increase in MDM2 levels (c) following treatment with 1 µM of 27-OHC for 24 h. Data are expressed as Mean ± SEM. *p < 0.05 versus vehicle
27-OHC enhances p53 and MDM2 dimerization
The relationship between p53 and MDM2 has been widely studied. MDM2 is known to bind to p53 and shuttle it to the cytoplasm for degradation [18, 29]. To determine whether 27-OHC enhances this interaction between MDM2 and p53, we first performed a co-immunoprecipitation assay. We found that treatment with 27-OHC increased MDM2 and p53 binding. When MDM2 was blotted against p53 immuno-precipitated lysate or when p53 was blotted against MDM2 immuno-precipitated lysate, an increase in binding was observed (Fig. 4c). We also used double immunofluorescence staining to observe p53-MDM2 co-localization. Subsequently, we found that upon 27-OHC treatment, a significant increase in overlap of labeled p53 and MDM2 at the nuclear envelope was observed (Fig. 4a, b). This result indicates 27-OHC enhances physical interaction between p53 and MDM2.
27-OHC promotes p53-MDM2 interaction in MCF7 cells. a Representative confocal microscopy photomicrographs showing increased intensity of MDM2 staining (green) and reduced intensity of p53 staining (red) following treatment of MCF7 cells with 1 µM 27-OHC. Arrows indicate co-localization of p53 and MDM2 in MCF7 cells treated with 1 µM 27-OHC for 24 h. b Representative graph showing increased percent of overlap between MDM2 and p53 in the confocal images. c Representative non-denatured Western blots for p53 IP blotted against MDM2 and MDM2 IP blotted against p53 demonstrating an increase in p53-MDM2 binding in of MCF7 cells treated for 24 h as follows: lane 1 Rabbit Serum (negative control); lane 2 lysate only; lane 3, 4 vehicle only; lane 5,6 1 µM 27-OHC. Data are expressed as Mean ± SEM. *p < 0.05 versus vehicle; Bar 10 µm. (Color figure online)
27-OHC increased proliferation via MDM2 mediated p53 inactivity
To determine whether 27-OHC promotes cell proliferation via p53 inactivation; we treated cells with the p53 activator, Tenovin-1 [30]. We found that Tenovin-1 significantly increased p53 transcriptional activity (Fig. 5a) and inhibited 27-OHC-induced proliferation (Fig. 5b). This result suggests that p53 inactivation is necessary for 27-OHC-induced cell proliferation. Given that upon stimulating cell proliferation with 27-OHC, p53 levels decreased, MDM2 levels increased and p53-MDM2 binding increased (Figs. 3, 4). We determined if 27-OHC-induced proliferation was due to MDM2-dependent p53 degradation. We treated cells with the MDM2-p53 interaction inhibitor, Nutlin-3 [31]. We found that Nutlin-3 markedly blocked 27-OHC-induced cell proliferation comparable to that of basal levels (Fig. 5c). This result suggests that MDM2-p53 binding is required for 27-OHC-induced cell proliferation. To investigate if specifically MDM2 expression is necessary for 27-OHC-induced cell proliferation, we knocked down MDM2 using siRNA. We determined that upon knocking down MDM2, 27-OHC-induced cell proliferation reduced to basal levels of proliferation (Fig. 5d, e). This data strongly suggests that 27-OHC requires MDM2-mediated p53 degradation to induce cell proliferation.
27-OHC induces proliferation via MDM2-mediated p53 inactivation. a Luciferase reporter assay demonstrates an increase in p53 activity in MCF7 in the presence of 10 μM Tenovin-1 for 48 h. b Cell proliferation assay demonstrates that 27-OHC-induced increase in proliferation is attenuated by Tenovin-1 in MCF7 cells. Cells were treated vehicle, 1 μM 27-OHC, 10 μM Tenovin-1, or 1 μM 27-OHC + 10 μM Tenovin-1 for 48 h. c In MCF7, 1 μM 27-OHC-induced increase in proliferation is also attenuated by the MDM2-p53 interaction inhibitor Nutlin-3. Cells were treated with vehicle, 1 μM 27-OHC, 5 μM Nutlin-3, or 1 μM 27-OHC + 5 μM Nutlin-3 for 48 h. d Representative blot demonstrates MDM2 knock down efficiency in MCF7. e 1 μM 27-OHC-induced increase in proliferation is also attenuated by MDM2 siRNA. Cells were incubated with respective treatments for 48 h. Data are expressed as Mean ± SEM versus vehicle. Data are expressed as Mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 versus vehicle, and ### p < 0.001 versus 27-OHC only. RLU relative luciferase units
Discussion
The goal of this study was to elucidate cellular mechanisms involved in 27-OHC-induced proliferation in breast cancer cells. Our results demonstrate that 27-OHC decrease p53 activity and protein levels, effects that are at the origin of the observed increase in cell proliferation. Our study also shows that 27-OHC increases MDM2 levels and enhances the interaction between p53 and MDM2. We further show that 27-OHC-induced proliferation is dependent on MDM2-mediated p53 degradation. Our results are the first showing that 27-OHC through the ER, exacerbates breast cancer cell proliferation via the p53-MDM2 axis.
P53 is a highly regulated protein in the cell and has a short half-life [32]. Loss of p53 function and/or perturbations in its signaling pathways via mutations plays an important role in several cancers [33–35]. p53 plays an important role in “guarding the genome” from genotoxic stress and regulates apoptosis, cell cycle, senescence and metabolism. Lack of functioning p53 leads to abnormalities in cell cycle and apoptosis which may lead to cancer progression. Approximately, 23 % of breast cancers exhibit a p53 mutation and loss of p53 function via mutation is still one of the main molecular characteristics of breast carcinomas, while other more spontaneous cancers such as ovarian, intestinal, and lung cancers have higher incidences of p53 mutations [16, 36]. In this report, we show that the cholesterol oxidation metabolite 27-OHC dysregulates p53 expression and function.
We report that 27-OHC, a SERM, inhibited wild type p53 activity, while interestingly, estradiol, a standard and endogenous ligand for ER had no effect on p53 activity. This demonstrates that 27-OHC may contribute to ER-positive breast cancer progression via different mechanisms compared to known estrogens. To verify that 27-OHC exerted it effects via ER, we treated cells with fulvestrant, an ER inhibitor, concomitantly with 27-OHC and found that it blocked the p53 transcriptional inactivation effects of 27-OHC when treated alone. This demonstrates that while 27-OHC and estradiol activate the same receptor (ER), the downstream events appear to be distinct. Estrogen level regulation in humans has been extensively studied and are primarily targeted for hormone therapy using interventions such as aromatase inhibitors and ovarian ablation [37]. Although hormone therapy is relatively effective, such therapy is challenged by endocrine resistance and recurrence of breast cancers. Hormone therapy is designed specifically to reduce estrogen levels only [38, 39]. The recent discovery that 27-OHC also activates ER and promotes ER-positive breast cancer progression, may explain why hormone therapy may not be as effective. Furthermore, a recent study by Nelson and colleagues demonstrated that the gene expression profile in MCF-7 breast cancer cells grown in the presence of 27-OHC was remarkably different relative to estradiol-treated MCF-7 cells, with 788 unique genes in the 27-OHC profile that are different from the estradiol profile of 8141 genes [5]. In the above mentioned study, 27-OHC and estradiol regulated 1511 shared genes [5]. Understanding the role of 27-OHC in ER-positive breast cancer may reveal novel molecular mechanisms that play a role in breast cancer progression, resistance, and recurrence of breast cancers.
There is a positive correlation between cholesterol and 27-OHC levels in humans. Patients with hypercholesterolemia have higher levels of 27-OHC and thus may be at a greater risk for developing ER-positive breast cancer [5]. 27-OHC is synthesized from cholesterol through hydroxylation by a cytochrome P-450 enzyme, sterol 27 hydroxylase (CYP27A1), localized in the inner mitochondrial membrane of the liver. 27-OHC is a substrate for bile acid synthesis, and when bile acid levels are adequate, excess levels of 27-OHC are catabolized by CYP7B1 [40, 41]. In support of hypercholesterolemia as a risk factor for developing breast cancers, the use of cholesterol lowering agents, such as HMG-CoA reductase inhibitors (statins), has been associated with better prognosis and breast cancer survival rates [42]. Furthermore, Cruz et al. showed that in ER-positive mammary tumor cells, simvastatin blocked 27-OHC-induced cell proliferation [9].
Our results add further insights into the potential cellular mechanisms by which 27-OHC influences cancer cell progression. We show that 27-OHC increases levels of the E3 ubiquitin ligase MDM2 and its interaction with p53. MDM2 is known to regulate p53 activity by flagging it for ubiquitination and proteasomal degradation [29]. Since DNA damage is a potent activator of p53, prior to DNA damage MDM2 binds to both p53 and ribosomal protein RPL26 resulting in ubiquitination and proteasomal degradation. Upon DNA damage, p53 and MDM2 undergo posttranslational modifications, inhibiting their interaction and subsequently activating p53 activity [12, 18]. In the presence of 27-OHC, MDM2, and p53 interaction is enhanced, resulting in p53 inactivation and degradation. It is suggested that MDM2 by itself may be an oncogene [43, 44]. MDM2 is overexpressed in various cancers including sarcoma, leukemia, breast cancers, melanoma, and glioblastoma [43, 45].
We also demonstrate that 27-OHC-induced cell proliferation in the ER-positive breast cancer MCF7 cells is dependent on the inactivation of p53. Treatment with Tenovin-1, a potent p53 activator, attenuated the 27-OHC-induced cell proliferation. This supports the notion that 27-OHC-induced p53 inactivation promotes breast cancer cell proliferation. Since p53 can undergo inactivation and degradation through multiple mechanisms [18, 46, 47], it was vital to test the role of MDM2 specifically in 27-OHC-induced p53 inactivation that led to cell proliferation. To verify whether p53 inactivation is MDM2-dependent, we treated cells with Nutlin-3, an MDM2-p53 interaction inhibitor [48, 49]. We found that Nutlin-3 prevents MDM2 mediated and 27-OHC-induced degradation of p53. We also found that upon knocking down MDM2, 27-OHC-induced cell proliferation was attenuated, suggesting that inactivation of p53 is MDM2-dependent. Such data may indicate that 27-OHC exacerbates ER-positive breast cancer progression by inactivating and degrading p53 via MDM2.
Although the prognostic value of p53 in ER-positive breast cancers is controversial, we propose that in the presence of excess 27-OHC, p53 function is compromised via MDM2, causing an exacerbation of ER-positive breast cancer progression. Subsequently, activation of p53 has been considered to be an important therapeutic target in tumorigenesis and cancer progression. A number of small molecules have been developed to activate wild type p53 and reactivate mutant p53. Interestingly, both DNA based and dendritic cell delivered p53 vaccines have been developed to activate and upregulate p53 [15, 50]. In addition, gene therapy has also been used against breast carcinomas [51–54]. Recently, Rejeeth et al. used silica nanoparticle supplemented with transferrin to administer p53 to MCF-7 and showed that such a treatment reduced cell growth by 60.7 % [53].
In summary, we demonstrate that 27-OHC exacerbates ER-positive MCF7 cancer cell proliferation by disrupting wild type p53 activity. We propose that 27-OHC activates ER, an effect that leads to activation of MDM2 and subsequent inactivation of p53. Fulvestrant, which is an ER-specific antagonist, blocks 27-OHC activation of ER, resulting in disinhibition of Mdm2-mediated p53 transcriptional activity. It has been shown that ER activation enhances MDM2 expression and that fulvestrant attenuates MDM2 upregulation by inhibiting ER [20]. Since MDM2 is upregulated by ER activation and regulates p53 by inactivating it, we measured p53 activity and found that fulvestrant inhibited 27-OHC (through ER)-induced Mdm2-mediated p53 inactivation (Fig. 6). Our data indicates that 27-OHC through ER can disrupt p53 response. We suggest that individuals with high levels of 27-OHC may have an increased risk of developing ER-positive breast cancers via loss of constitutive p53 activity. We also demonstrate that 27-OHC enhances MDM2-p53 interaction resulting in loss of wild type p53 expression and activity. Such results strongly suggest MDM2 involvement in the 27-OHC-induced p53 inactivity and subsequent cell proliferation (Fig. 6). Our findings provide a novel cellular mechanism for 27-OHC that may contribute in the pathophysiology of ER-positive breast cancers. Understanding MDM2-p53 interplay in the presence of 27-OHC may reveal innovative therapeutic avenues that can challenge ER-positive breast cancer progression.
Proposed mechanism of action of 27-OHC in ER+ breast cancer. 27-OHC activates ER which upregulates MDM2 and mediates p53 inactivation and degradation. This results in an increase in cell proliferation, thus exacerbating ER+ breast cancer progression. Interactions depicted as activation are denoted by arrows; those depicted as inhibitions are indicated by a bar
References
How many women get breast cancer? http://www.cancer.org/cancer/breastcancer/overviewguide/breast-cancer-overview-key-statistics. Accessed 7 Jul 2014
Buzdar AU (2004) Phase III study of letrozole versus tamoxifen as first-line therapy of advanced breast cancer in postmenopausal women: analysis of survival and update of efficacy from the international letrozole breast cancer group. J Clin Oncol 22:3199–3200. doi:10.1200/JCO.2004.99.058
Baum M, Budzar AU, Cuzick J et al (2002) Anastrozole alone or in combination with tamoxifen versus tamoxifen alone for adjuvant treatment of postmenopausal women with early breast cancer: first results of the ATAC randomised trial. Lancet 359:2131–2139
Franco S, Perez A, Tan-Chiu E et al (2004) Response to fulvestrant in heavily pretreated postmenopausal women: a single-center experience. Breast Cancer Res Treat 88:103–108. doi:10.1007/s10549-004-0748-7
Nelson ER, Wardell SE, Jasper JS et al (2013) 27-Hydroxycholesterol links hypercholesterolemia and breast cancer pathophysiology. Science 342:1094–1098. doi:10.1126/science.1241908
Wu Q, Ishikawa T, Sirianni R et al (2013) 27-Hydroxycholesterol promotes cell-autonomous, ER-positive breast cancer growth. Cell Rep 5:637–645. doi:10.1016/j.celrep.2013.10.006
Umetani M, Shaul PW (2011) 27-Hydroxycholesterol: the first identified endogenous SERM. Trends Endocrinol Metab 22:130–135. doi:10.1016/j.tem.2011.01.003
Duane WC, Javitt NB (1999) 27-Hydroxycholesterol: production rates in normal human subjects. J Lipid Res 40:1194–1199
Cruz P, Torres C, Ramírez ME et al (2010) Proliferation of human mammary cancer cells exposed to 27-hydroxycholesterol. Exp Ther Med 1:531–536. doi:10.3892/etm_00000084
DuSell CD, Umetani M, Shaul PW et al (2008) 27-hydroxycholesterol is an endogenous selective estrogen receptor modulator. Mol Endocrinol 22:65–77. doi:10.1210/me.2007-0383
Miller LD, Smeds J, George J et al (2005) An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A 102:13550–13555. doi:10.1073/pnas.0506230102
Khoo KH, Hoe KK, Verma CS, Lane DP (2014) Drugging the p53 pathway: understanding the route to clinical efficacy. Nat Rev Drug Discov 13:217–236. doi:10.1038/nrd4236
Coates AS, Millar EK, O’Toole SA et al (2012) Prognostic interaction between expression of p53 and estrogen receptor in patients with node-negative breast cancer: results from IBCSG Trials VIII and IX. Breast Cancer Res 14:R143. doi:10.1186/bcr3348
Pharoah PD, Day NE, Caldas C (1999) Somatic mutations in the p53 gene and prognosis in breast cancer: a meta-analysis. Br J Cancer 80:1968–1973. doi:10.1038/sj.bjc.6690628
Van der Burg SH, de Cock K, Menon AG et al (2001) Long lasting p53-specific T cell memory responses in the absence of anti-p53 antibodies in patients with resected primary colorectal cancer. Eur J Immunol 31:146–155. doi:10.1002/1521-4141(200101)31:1<146:AID-IMMU146>3.0.CO;2-T
Walerych D, Napoli M, Collavin L, Del Sal G (2012) The rebel angel: mutant p53 as the driving oncogene in breast cancer. Carcinogenesis 33:2007–2017. doi:10.1093/carcin/bgs232
Ahn SH, Kim HJ, Han W et al (2013) Effect modification of hormonal therapy by p53 status in invasive breast cancer. J Breast Cancer 16:386–394. doi:10.4048/jbc.2013.16.4.386
Moll UM, Petrenko O (2003) The MDM2-p53 Interaction. Mol Cancer Res 1:1001–1008
Manfredi JJ (2010) The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev 24:1580–1589. doi:10.1101/gad.1941710
Dolfi SC, Jäger AV, Medina DJ et al (2014) Fulvestrant treatment alters MDM2 protein turnover and sensitivity of human breast carcinoma cells to chemotherapeutic drugs. Cancer Lett 350:52–60. doi:10.1016/j.canlet.2014.04.009
Gao C-F, Xie Q, Su Y-L et al (2005) Proliferation and invasion: plasticity in tumor cells. Proc Natl Acad Sci U S A 102:10528–10533. doi:10.1073/pnas.0504367102
Sørlie T, Perou CM, Tibshirani R et al (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 98:10869–10874. doi:10.1073/pnas.191367098
Wasielewski M, Elstrodt F, Klijn JGM et al (2006) Thirteen new p53 gene mutants identified among 41 human breast cancer cell lines. Breast Cancer Res Treat 99:97–101. doi:10.1007/s10549-006-9186-z
Okumura N, Saji S, Eguchi H et al (2002) Estradiol stabilizes p53 protein in breast cancer cell line, MCF-7. Jpn J Cancer Res 93:867–873. doi:10.1111/j.1349-7006.2002.tb01331.x
Lee AV, Oesterreich S, Davidson NE (2015) MCF-7 cells–changing the course of breast cancer research and care for 45 years. J Natl Cancer Inst 107:djv073. doi:10.1093/jnci/djv073
Comşa Ş, Cîmpean AM, Raica M (2015) The story of MCF-7 breast cancer cell line: 40 years of experience in research. Anticancer Res 35:3147–3154
Zheng L, Ren JQ, Li H et al (2004) Downregulation of wild-type p53 protein by HER-2/neu mediated PI3 K pathway activation in human breast cancer cells: its effect on cell proliferation and implication for therapy. Cell Res 14:497–506. doi:10.1038/sj.cr.7290253
Lim LY, Vidnovic N, Ellisen LW, Leong C-O (2009) Mutant p53 mediates survival of breast cancer cells. Br J Cancer 101:1606–1612. doi:10.1038/sj.bjc.6605335
Alarcon-Vargas D (2002) p53-Mdm2–the affair that never ends. Carcinogenesis 23:541–547. doi:10.1093/carcin/23.4.541
Lain S, Hollick JJ, Campbell J et al (2008) Discovery, in vivo activity, and mechanism of action of a small-molecule p53 activator. Cancer Cell 13:454–463. doi:10.1016/j.ccr.2008.03.004
Park EJ, Choi KS, Yoo YH, Kwon TK (2013) Nutlin-3, a small-molecule MDM2 inhibitor, sensitizes Caki cells to TRAIL-induced apoptosis through p53-mediated PUMA upregulation and ROS-mediated DR5 upregulation. Anticancer Drugs 24:260–269. doi:10.1097/CAD.0b013e32835c0311
Giaccia AJ, Kastan MB (1998) The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev 12:2973–2983. doi:10.1101/gad.12.19.2973
Sherr C (2004) Principles of tumor suppression. Cell 116:235–246. doi:10.1016/S0092-8674(03)01075-4
Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100:57–70. doi:10.1016/S0092-8674(00)81683-9
Sherr CJ, McCormick F (2002) The RB and p53 pathways in cancer. Cancer Cell 2:103–112. doi:10.1016/S1535-6108(02)00102-2
Yu X, Narayanan S, Vazquez A, Carpizo DR (2014) Small molecule compounds targeting the p53 pathway: are we finally making progress? Apoptosis. doi:10.1007/s10495-014-0990-3
Gonzalez-Angulo AM, Morales-Vasquez F, Hortobagyi GN (2007) Overview of resistance to systemic therapy in patients with breast cancer. Adv Exp Med Biol 608:1–22
Jankowitz RC, Davidson NE (2013) Adjuvant endocrine therapy for breast cancer: how long is long enough? Oncology (Williston Park) 27(1210–6):1224
Hasson SP, Rubinek T, Ryvo L, Wolf I (2013) Endocrine resistance in breast cancer: focus on the phosphatidylinositol 3-Kinase/Akt/Mammalian target of rapamycin signaling pathway. Breast Care (Basel) 8:248–255. doi:10.1159/000354757
Fu X, Menke JG, Chen Y et al (2001) 27-Hydroxycholesterol is an endogenous ligand for Liver X Receptor in cholesterol-loaded Cells. J Biol Chem 276:38378–38387
Ma D, Liu W, Wang Y (2014) ApoA-I or ABCA1 expression suppresses fatty acid synthesis by reducing 27-hydroxycholesterol levels. Biochimie. doi:10.1016/j.biochi.2014.04.010
Ahern TP, Pedersen L, Tarp M et al (2011) Statin prescriptions and breast cancer recurrence risk: a Danish nationwide prospective cohort study. J Natl Cancer Inst 103:1461–1468. doi:10.1093/jnci/djr291
Momand J (1998) The MDM2 gene amplification database. Nucleic Acids Res 26:3453–3459. doi:10.1093/nar/26.15.3453
Wang H, Zeng X, Oliver P et al (1999) MDM2 oncogene as a target for cancer therapy: an antisense approach. Int J Oncol 15:653–660
Jones SN, Hancock AR, Vogel H et al (1998) Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc Natl Acad Sci 95:15608–15612. doi:10.1073/pnas.95.26.15608
Tsvetkov P, Reuven N, Shaul Y (2010) Ubiquitin-independent p53 proteasomal degradation. Cell Death Differ 17:103–108. doi:10.1038/cdd.2009.67
Kruse J-P, Gu W (2009) Modes of p53 regulation. Cell 137:609–622. doi:10.1016/j.cell.2009.04.050
Miyachi M, Kakazu N, Yagyu S et al (2009) Restoration of p53 pathway by nutlin-3 induces cell cycle arrest and apoptosis in human rhabdomyosarcoma cells. Clin Cancer Res 15:4077–4084. doi:10.1158/1078-0432.CCR-08-2955
Poyurovsky MV, Katz C, Laptenko O et al (2010) The C terminus of p53 binds the N-terminal domain of MDM2. Nat Struct Mol Biol 17:982–989. doi:10.1038/nsmb.1872
Speetjens FM, Kuppen PJK, Welters MJP et al (2009) Induction of p53-specific immunity by a p53 synthetic long peptide vaccine in patients treated for metastatic colorectal cancer. Clin Cancer Res 15:1086–1095. doi:10.1158/1078-0432.CCR-08-2227
Sasaki R, Shirakawa T, Zhang ZJ et al (2001) Additional gene therapy with Ad5CMV-p53 enhanced the efficacy of radiotherapy in human prostate cancer cells. Int J Radiat Oncol Biol Phys 51:1336–1345
Song SU, Boyce FM (2001) Combination treatment for osteosarcoma with baculoviral vector mediated gene therapy (p53) and chemotherapy (adriamycin). Exp Mol Med 33:46–53. doi:10.1038/emm.2001.9
Rejeeth C, Kannan S (2014) p53 gene therapy of human breast carcinoma: using a transferrin-modified silica nanoparticles. Breast Cancer. doi:10.1007/s12282-014-0537-z
Obermiller PS, Tait DL, Holt JT (2000) Gene therapy for carcinoma of the breast: therapeutic genetic correction strategies. Breast Cancer Res 2:28–31. doi:10.1186/bcr26
Acknowledgments
We thank Dr. Bry on Grove and Sarah Abrahamson for their technical assistance with the confocal microscopy.
Disclaimer
The contents do not represent the views of the Department of Veterans Affairs or the United States Government.
Funding
University of North Dakota School of Medicine seed grant to OG.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors do not have any conflicts of interests to report.
Rights and permissions
About this article
Cite this article
Raza, S., Ohm, J.E., Dhasarathy, A. et al. The cholesterol metabolite 27-hydroxycholesterol regulates p53 activity and increases cell proliferation via MDM2 in breast cancer cells. Mol Cell Biochem 410, 187–195 (2015). https://doi.org/10.1007/s11010-015-2551-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-015-2551-7