Abstract
CD151 is a member of the tetraspanin family, which is involved in diverse cellular processes, including proliferation, motility, and invasion. However, the role of CD151 in breast cancer especially luminal and basal-like subtype breast cancer remains obscure. Here, we report the role of CD151 in the biological behaviors of luminal and basal-like subtype cell lines and the underlying molecular mechanism. A eukaryotic expression vector expressing both CD151 shRNA and GFP was transfected into MCF-7 and MDA-MB-468 cells. The CD151 gene-silencing effect is authenticated by real-time PCR and Western blot. Our data show that the capacity for proliferation, migration, and invasion of two kinds of cells is diminished after Knockdown of CD151 via lentivirus-mediated CD151 specific shRNA. Tumor cells are arrested in G0/G1 phase. Apoptosis is increased. Moreover, we also demonstrate that the expressions of mmp26 and CD147 are inhibited by knockdown of CD151. But the inhibition depends on the cell type. We can conclude that silencing gene CD151 inhibits expression of properties that are associated with the malignant phenotype of MCF-7 and MDA-MB-468 cells. It may become a potential target in breast cancer therapy especially for luminal and basal subtypes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer remains the most common cancer in women and threatens women’s health. Because of the high heterogeneity between and within tumors, the experts reached a consensus on breast cancerous molecular subtype in the 12th St Gallen International Breast Cancer conference [1] Luminal A (ER+/PR+, HER-2-, KI-67 < 14 %), Luminal B (ER+/PR+, HER-2-, KI-67 ≥ 14 %; ER+/PR+, HER-2+), HER−2 overexpression (ER−, HER-2+), Basal-like (ER−, PR−, HER-2,CK5/6 and (or) EGFR+) were included in the new molecular subtype. The correct characterization for different molecular subtypes is the foundation of the personalized treatment. Basal-like breast cancers (BLBC) are associated with the worst prognostic and clinical profile especially if not detected at an early stage. The name by its origin is derived from basal epithelial cells of breast tissue and thus typified by high expression of basal epithelial cytokeratins (CK5/6, CK14, and EGFR) [2]. A higher frequency of basal cancers was also demonstrated in Chinese women [3, 4]. Development of novel biotherapeutics target for this clinically intractable type of tumor is very important.
Tetraspanins are trans-membrane proteins involved in a variety of biological processes including fertilization, infectious processes, and tumor progression [5]. Tetraspanin proteins perform several functions depending on family member or interacting partners such as signaling molecules, integrins, and immunoglobulin superfamily members through forming complexes between them [6]. CD151 is one of 33 members of tetraspanins in humans. CD151 contributes to integrin-dependent cell adhesion and motility by directly interacting with laminin-binding integrins (a3b1, a6b1, a6b4, and a7b1) [7, 8]. Upregulation and overexpression of CD151 are found in many tumor types, for example, non-small cell lung, pancreatic, colon cancers, hepatocellular, oesophageal, and endometrial cancers. CD151 overexpression was associated with poor prognosis [9–14]. In addition, there have been several evidences supporting the contribution of CD151 in tumor progression [15, 16]. To date, there have been no reported studies that address CD151 expression in the breast tumors among Chinese patients. In this study, first we used antibodies against the CD151 protein to detect the subtypes of invasive breast cancer and the normal tissue to ascertain the expression level of CD151. Then we looked for detection of the CD151 expression in the breast cancer cell lines. After detecting, we investigated the effects of CD151 silencing on the growth and invasion of breast cancer cells by constructing recombinant shRNA-expressing lentiviral vector targeting CD151 in vitro. To reveal the possible mechanism, we also showed the effects of CD151 shRNA on the expression of mmp26, CD147 in MCF-7, and MDA-MB-468 cells in vitro.
Materials and methods
Patients and tissue collection
We retrospectively analyzed 614 cases of primary breast cancer treated at clinical pathological diagnostic center of Qiqihar medical University from January 2008 till January 2011. 478 cases of invasive breast cancer were chosen. Approval from institutional research and ethical review committee was taken antecedently to conduct the study. Patients after provisional diagnosis of breast cancer underwent trucut biopsies, modified radical mastectomy, and breast conservative surgeries. None of the patients had been treated with any preoperative chemotherapy or radiotherapy or hormones. Histologic type of tumors was determined by WHO classification of breast tumors and graded by Modified Bloom-Richardson grading system. All of the sections used were 4 μm paraffin sections stained with hematoxylin and eosin. Immunohistochemical testing for ER, PR, Her2neu, Ki67, EGFR, and CK5/6 was applied on all cases. A total of 98 out of 478 cases were negative with ER and PR and Her2neu. According to the currently accepted BLBC (Basal-like breast cancer) immune phenotype diagnosis standard (ER/PR, Her2 negative, CK5/6 and (or) EGFR positive), 65 cases of BLBC were screened. As control, 100 cases of tissues samples 5 cm from the tumor margin were used.
Immunohistochemistry assay
Immunohistochemistry (IHC) was performed on 4 μm paraffin sections as described previously using a peroxidase ABC elite kit and DAB peroxidase substrate kit in accordance with the manufacturer’s recommendations. The antibodies and dilutions used for IHC were monoclonal mouse anti-human CD151 antibody at 1:100 (Santa Cruz, USA). The monoclonal mouse anti-ER, PR, Her2neu, Ki67, EGFR, and CK5/6 antibodies were ready-to-use antibodies of Fuzhou Maixin Biotech.
CD151 immunoreactivity was assessed as follows: (1) negative = no staining or faint membrane and cytoplasm positivity in <10 % of tumor cells; (2) positive = weak-to-strong complete membrane and cytoplasmic staining observed in >10 % of tumor. Ki-67 immunoreactivity was recorded as continuous variables, based on the proportion of positive tumor cells (0–100 %). Levels of Ki-67 were quantified as high Ki-67 (immunostaining ≥ 30 %), low (immunostaining < 14 %), and intermediate (between 14 to 30 %) approach adopted by St Gallen International Expert Consensus. Two pathologists independently scored the immunohistochemical staining and were blinded with respect to the results of the markers and the outcome data.
Lentivirus construction and target screening for RNAi
The human breast cancer cell lines MCF-7 and MDA-MB-468, which were purchased from the American Type Culture Collection (ATCC), were cultured according to the ATCC protocols. MCF7 cells were used to represent luminal (receptor positive) samples, and MDA-MB-468cell line was used to represent basal-like samples. Four different sequences (namely 3462, 3463, 3464, and 3465) of CD151 gene were designed as the target for RNA interference (RNAi) (Genepharma, Shanghai, China). The sequence of No. 3462 was 5′-GCTGGAGATCATCGCTGGTAT-3′. The sequence of No. 3463 was 5′- GCCCTCAAGAGTGACTACATC-3′. The sequence of No. 3464 was 5′- GAGACCATGCCTCCAACATCT-3′. The sequence of No.3465 was 5′- GCATCACCAAGTTGGAGACCT-3′. As a control, the scrambled sequence (negative shRNA) was 5′-TTCTCCGAACGTGTCACGT-3′. Then the amplified shRNA expression cassettes were subcloned into a lentiviral vector backbone (pHIV7-GFPPL) between the EcoRI and BamHI restriction enzyme sites. The orientation of the insert was confirmed by sequencing analysis. A lentivirus was produced by cotransduction of the shRNA expression pShuttle vector pGLV3 with packaging vectors pLV/helper-SL3, pLV/helper-SL4, and pLV/helper-SL5 by liperfectin 2000-mediated into the 293FTcells. Then lentiviruses were used to transfect MCF-7 and MDA-MB-468. MCF-7 and MDA-MB-468 cells were collected for the following polymerase chain reaction (PCR) and Western blot experiment at 72 and 96 h after transduction, respectively. Nonlentivirus and lentivirus containing mock shRNA transduction served as controls.
Real-time quantitative RT-PCR (qRT-PCR)
Total RNA extraction was performed using Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instruction. The purity and integrity of the RNA were assessed. Then qRT-PCR was performed using Primer Assay and SYBR Green RTPCR Kit (Takara, JAPAN) on MX3000P Instrument (Stratagene, US). The detection and quantification involved the following steps: reverse transcription at 42 °C for 30 min, initial activation at 85 °C for 3 min, followed by 40 cycles of denaturation at 95 °C for 3 min, annealing at 62 °C for 40 s, and extension at 72 °C for 30 s. All assays were performed in triplicates. The forward and reverse primers were as follows: CD151, 5′-GACCATGCCTCCAACATCTACAAG-3′, 5′-GGTGCTCCTGGATGAAGGTCTC-3′, all of which will give a 73-bp band; β-actin, 5′- CCCTGGCACCCAGCAC-3′, 5′-GCCGATCCACACGGAGTAC-3′, all of which will give a 70-bp band. All values obtained were normalized to β-actin and relative expression analysis involved the 2−ΔΔCT method.
Western blotting analysis
Whole-cell lysate (20–40 μg) was resolved by SDS-PAGE and then transferred onto PVDF membranes. PVDF membranes were washed briefly in Tris-buffered saline and 0.1 % Tween-20 (TBST) and blocked in a solution of TBST containing 5 % nonfat dry milk for 15 min with constant agitation. After blocking, the PVDF membrane was incubated with the following primary antibodies overnight at 4 °C: mouse monoclonal CD151 (1:200 dilutions in TBST) antibody, β-actin (1:500 dilutions in TBST) antibody, GAPDH(1:1000 dilutions in TBST) antibody, rabbit polyclonal MMP26(1:200 dilutions in TBST) antibody, and CD147(1:100 dilutions in TBST) antibody. Membranes were washed in TBST (3 times for 15 min) and were incubated for 1 h with horseradish peroxidase-conjugated secondary antibodies at a 1:2000 dilution at room temperature with constant agitation before enhanced chemiluminescence using a Pierce ECL Western blotting substrate and exposure to film. The housekeeping gene β-actin was quantified as an internal control.
Cell proliferation assays
The Cell Counting Kit-8 (CCK-8; Dojindo, Rockville, USA) was used to assess cell proliferation according to the manufacturer’s protocol. Tumor cells were seeded in 96-well culture plates, and treated with 10 % FBS and incubated at 37 °C. The optical density at 450 nm was measured at 24, 48, 72, 96, and 120 h after virus transfection. The data shown are representative of 3 independent experiments and are presented as the mean ± S.D.
Cell cycle analysis
Ninety-six hour after transfection, cell cycle data were obtained by analyzing of PI-stained cells using fluorescence-activated cell sorting (FACS) with a FACS Calibur flow cytometer. For each sample, at least 3 × 106 cells were counted, and the data were analyzed with BD CellQuest software. The data shown are representative of 3 independent experiments and are presented as the mean ± S.D.
Apoptosis analysis
Tumor cells (approximately 5 × 106) were stained with 500 μl of Annexin V-PE and 7AAD (KeyGen, Nanjing, China) at room temperature and then analyzed by flow cytometry (Becton–Dickinson) within 1 h. The Annexin V (+)/7AAD (–) cells were regarded as apoptotic cells.
Wound healing assay and cell invasion assays
Cells (5 × 105) were seeded on a six-well plate and cultured for 24 h. A scratch was made on the cell monolayer with a 20-μL pipette tip and monitored with a microscope every 12 h. The migration was determined by the rate of cells filling the scratched area. The normalized wound area was calculated by the software TScratch. The Transwell chamber and polycarbonate membrane precoated with matrigel (BD Biosciences) was used to evaluate invasion ability of the cancer cell. 500 μL of cell suspension (3 × 105 cells/mL) per experiment was seeded into the upper well of the chamber containing serum-free media. The lower chamber had been filled with DMEM with 10 % BSA. After 48 h of incubation, cells that had invaded the membrane were fixed with 70 % ethanol and stained with 0.1 % crystal violet and sealed on slides. Subsequently the cells were counted under the microscope. Statistical significance was calculated from at least three independent experiments.
Statistical analysis
Statistical treatment was performed using SPSS 17.0 software. One-way analysis of variance (ANOVA) and the χ2 test were used to analyze the significance between groups. Multiple comparisons between the experimental group and control groups were made using Tukey’s HSD when the probability for ANOVA was statistically significant. All data represent mean ± SD. The statistical significance was set at P < 0.05.
Results
CD151 is upregulated in human breast cancer tissues and cell lines
First we compared the expressions of CD151 in breast cancer tissues and the corresponding normal tissues. Immunohistochemical examination revealed that CD151 was located in the cytoplasm and membrane of breast cancer cells and was expressed significantly more strongly in breast tumors compared to paracancerous tissue (Table 1). In normal breast tissue, CD151 positioned at the basal-myoepithelial cell layer surrounding both ducts and lobular alveolae. Some cases showed CD151 faint expression in the alveolar and ductal epithelial cell (Fig. 1a). 163 cases (34.1 %) were identified as CD151-positive expression, and 315 cases were classified as CD151-negative expression. Then we detected the expressions of CD151 in breast cancer cell lines and normal breast cell line by Western blot analysis. As shown in Fig. 1b, higher expression of CD151 was detected in MCF-7 and MDA-MB-468 cells compared with normal cell HBL-100, therefore those two cell lines were selected in the following experiments.
CD151 expression is associated with clinicopathological features
The correlations between CD151 expression in breast cancer and the 6 clinicopathological characteristics of patients (age, tumor size, lymph node metastasis, pathological type, breast cancer molecular subtype, and Ki-67 labeling index) are shown in Table 2. When compared among the breast cancer subtypes (Luminal A, Luminal B, HER2, and BLBC), CD151 overexpression varied significantly according to the breast cancer subtype (P = 0.009). The Luminal A subtype had a lower incidence in tumors with CD151 expression. CD151 overexpression was most frequent in the BLBC subtype (52.31 %). Furthermore, CD151 overexpression was significantly associated with tumor size (P < 0.001), lymph node involvement (P < 0.001), and Ki-67 labeling index (P < 0.001). There were no significant differences in histological type (P = 0.145).
Lentivirus-mediated shRNA inhibited CD151 mRNA and protein expression in breast cancer cell lines
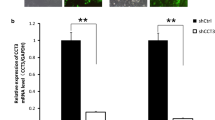
The MCF-7 and MDA-MB-468 cell lines were transfected with different lentivirus expressing CD151 shRNAs, and gene-silencing analysis showed that No.3463 lentivirus was the most effective vector in blocking CD151 expression. As shown in Fig. 2, CD151 knockdown clones 3463 exhibited 85 and 93 % reduction in mRNA expression in MDA-MB-468, and 72 and 85 % in MCF-7 after transfected 72 and 96 h, respectively, 38 and 86 % reduction in protein expression in MDA-MB-468 and 21 and 84 % in MCF-7, respectively. Then clone 3463 lentivirus and mock lentivirus were selected and produced at a viral titer of 1 × 109 TU/mL (Genepharma, Shanghai, China) for further studies.
Knockdown of CD151 by lentivirus-mediated shRNA. a The expression levels of CD151 mRNA were measured by qRT-PCR. There was a dramatic decrease of CD151 mRNA in the 3463-shRNA group. b The expression levels of CD151 protein were measured by Western blot. The protein level in the 3463-shRNA group decreased significantly compared to the other groups. The β-actin gene and protein were used as the internal controls for qRT-PCR and Western blot analysis, respectively
CD151 silencing suppressed luminal and basal-like breast cancer cells proliferation and accelerated apoptosis
Cell proliferation was determined using cell Counting Kit-8 assay once daily for 5 days. Shown in Fig. 3a, CD151 silencing inhibited MCF-7 cell proliferation in a time-dependent manner. Compared with the blank group and scrambled shRNA group, A value of the cells in CD151-shRNA group was significantly reduced on day 5 [blank group: (2.72 ± 0.05), scrambled shRNA: (2.64 ± 0.05), and CD151-shRNA: (1.63 ± 0.02), F = 615.167, P<0.001]. Similar results were found in MDA-MB-468 cells, as shown in Fig. 3a [blank group: (2.68 ± 0.04), scrambled shRNA: (2.65 ± 0.06), CD151-shRNA: (1.60 ± 0.05), F = 442.169, P < 0.001].
Cell apoptosis was detected by Annexin V-PE Apoptosis Kit, followed by flow cytometry. This is shown in Fig. 3b. CD151 Silencing accelerated both MCF-7 cells and MDA-MB-468 cells apoptosis (in MCF-7 cells, P < 0.001; in MDA-MB-468 cells, P < 0.001).
After performing cell cycle analysis by flow cytometry (Fig. 4), we found that inhibition of CD151 increased significantly the number of cells in G0/G1 phase and decreased the number of cells in S phase in MCF-7 and MDA-MB-468 cells (in MCF-7 cells, F = 6.829, P = 0.028; in MDA-MB-468 cells, F = 6.094, P = 0.036). In comparison to blank group, there was no significant difference in scrambled shRNA group (in MCF-7 cells, P = 0.384; in MDA-MB-468 cells, P = 0.929).
CD151 silencing suppressed luminal and basal-like breast cancer cells motility and invasion
The cell motility of breast cancer cells was evaluated by wound healing assay. The migration was determined by the rate of cells filling the scratched area. The normalized wound area was calculated by the software TScratch. As shown in Fig. 5a, compared to cells transduced with blank and scrambled shRNA, the cells transduced with CD151-shRNA showed a wider wound area 48 h after wound generation. The normalized wound area was calculated by the software TScratch [17]. It indicated that knockdown of CD151 inhibits cell motility in breast cancer cells. We further investigated the cell invasiveness using matrigel-coated transwell assay in Fig. 5b. The number of invaded cells was quantified. Consistent with the findings in wound healing assay, cells transduced with CD151-shRNA showed a significant reduction in cell invasion ability when compared with blank and scrambled shRNA-treated cells. The number of invaded MCF-7cells transduced with blank, scrambled and CD151-shRNA were 103 ± 15, 94 ± 18 and 48 ± 17, respectively (F = 9.347, P = 0.014). Similar findings were observed in MDA-MB-468 cells (109 ± 12, 104 ± 8 and 55 ± 6, F = 32.840, P = 0.001). Taken together, these results indicated that silencing of CD151 decreased the invasive properties of breast cancer cells.
CD151 silencing suppressed cell motility and invasion in vitro using wound healing assay and transwell invasion assay. a Monolayers of cells were mechanically wounded with a pipette tip and monitored with a microscope every 12 h. The migration was determined by the rate of cells filling the scratched area. B: Cell invasion was determined by matrigel-coated transwell assay. Cells crossed the Matrigel-coated filter were fixed, stained, and counted. Cell invasion was significantly less pronounced in CD151shRNA group than in the blank control and NC groups. Data represent the means ± SD of three independent experiments (*P < 0.05)
MMP26 and Emmprin/CD147 expression after the inhibition of CD151
As shown in Fig. 6, the expression level of MMP26 decreased significantly in the KD group of luminal breast cancer cell line MCF-7 (F = 86.692, P < 0.001). But not in that of Basal-like breast cancer cell line MDA-MB-468 (F = 4.589, P = 0.062). The expression level of emmprin/CD147 was just the opposite (in MCF-7 cells, F = 4.138, P = 0.074; in MDA-MB-468 cells, F = 79.796, P < 0.001).
Expression Levels of MMP26 and CD147 after inhibition of CD151. a Western blot analysis was performed using MMP26 and CD147 specific antibodies in the blank control and NC group control, CD151shRNA infected MCF-7 cells, and MDA-MB-468 cells. Typical bands in the Western blot assay showed that the protein levels MMP26 and CD147 were regulated in a cell-type specific manner after the inhibition of CD151. b Quantitative analysis of MMP26 and CD147 expressions. The experiment was repeated three times, and the data were expressed as the means ± SD (*P < 0.05)
Discussion
As a potential therapy, RNA interference mediated gene silencing is already being tested in clinical trials for a number of diseases [17, 18]. Lentiviral vectors are considered the most efficient transduction tools of RNA interference to deliver a gene for basic research and gene therapy because of the ability to transduce non-dividing and dividing cells, stable transgene expression, and minimal cytotoxicity [19]. In the present study, we constructed recombinant shRNA-expressing lentiviral vector targeting CD151, and then evaluated the effects of CD151 silencing on the cellular growth and invasion in Luminal and Basal-Like Breast Cancer cell lines. CD151 gene knockdown via lentivirus-mediated specific shRNA resulted in a significant decrease in the growth and invasion of breast cancer cells. The results demonstrate that lentivirus-mediated RNAi might be an effective and convenient approach for the treatment of breast cancer.
The tetraspanin CD151 is an important member of the tetraspanin family. These proteins in tetraspanin family serve as scaffolds for multiprotein complexes (called TEMs or Tetraspanin-Enriched Microdomains) where they associate with molecules such as integrins, growth factor receptors, and matrix metalloproteases, modifying their functions in various cellular processes [20]. Previous studies have shown that many cancer cell lines and cancer tissues had high expressions of CD151, such as breast cancer, esophageal squamous cell carcinoma, endometrial cancer, and pancreatic ductal adenocarcinoma, et al. Increased expression of CD151 correlates with advanced tumor grades and poor prognosis. However, not all studies show the same results. In this study, we confirmed that expression of CD151 increased markedly in breast cancer tissues compared with that in normal breast tissues. This result is consistent with previous relevant reports [21]. There is no effective treatment for basal-like breast cancer (BLBC) because it lacks ER and PR and HER2 expression, thus precluding hormone treatment and trastuzumab treatment [22]. Effective molecular target of basal-like subtype breast cancer has been focused on. In our study, CD151 overexpression was found in 52.31(34/65) of the BLBC subtype. Our data indicated that the BLBC subtype had the highest CD151 expression, Luminal subtype, and HER2 subtype had low proportion of CD151 expression. These differences may be controversial with relevant report [23]. However, the clinicopathological characteristics of CD151 overexpression cases were similar in these cohorts.
CD151 plays a variety of roles in multiple stages of human cancer development and progression. Many studies have shown that knockdown/-out or mutation of CD151 decreases cell motility and/or decreases experimental metastasis. Our results of in vitro experiments revealed clearly the lower mobility and invasiveness which is consistent with reports. Whether CD151 affects the growth of cancer cell or not is controversial in some transplant tumor model. For example, Séverine Roselli used MMTV/PyMT model showed that CD151 deletion significantly impaired tumor occurrence and growth [16]. But Ben indicated that Knocking-out Cd151 had no significant effect on prostate cancer initiation. Furthermore, CD151 does not affect proliferation, apoptosis, or angiogenesis in the primary tumor. However, it did significantly reduce the incidence of metastases [24]. In our study growth inhibition in vitro, apoptotic abnormalities were observed after CD151 inhibition in the examined cell lines. The intrinsic inhibition of CD151 arrested tumor cells in the G0/G1 phase in vitro was also observed. These findings suggest that CD151 positively regulates apoptosis, the cell cycle and subsequently promotes tumor cell proliferation. So the results of our study clearly indicate that CD151 positively regulates cell proliferation, tumorigenesis, apoptosis, and cell invasiveness.
The regulatory mechanism of CD151 is complicated. Several studies have demonstrated that CD151 can form tight complexes with the laminin-binding integrins (α3β1, α6β1, and α6β4) to promote tumor cell adhesion, migration, and spreading and drug sensitivity. In tumor invasion, proteolytic activity is crucial for destruction and remodeling of surrounding stroma. MMPs are the key enzymes responsible for tumor invasion, controlled in part by integrin-mediated pathways [25]. A study using melanoma cells showed that transfected CD151 increases motility and MMP-9 expression by c-Jun binding to AP-1 [26]. A study using Human epidermal carcinoma cell line HSC5, established from the squamous cell carcinoma of the auricle skin indicated that the expression levels of MMP2, MMP7, and MMP9 were downregulated by CD151 knockdown, and CD151 was directly associated with MMP7 [27]. MMP-26, also known as endometase or matrilysin-2, was recently identified as the smallest member of MMP family [28].MMP-26 is able to process the lacent MMP-9, to generate its active form [29, 30]. Therefore, we investigated the changes in the levels of MMP-26 in the examined cell lines. However, our results demonstrated that the decreased protein level of MMP-26 after inhibition of CD151 was only presented in MCF-7cells. There was no difference in MDA-MB-468 cells. The results show that the expressions of mmp26 are perhaps regulated by CD151 in a cell-type specific manner. CD147 (also named EMMPRIN), a glycosylated cell surface trans-membrane protein of the immunoglobulin superfamily (IgSF) is capable of inducing the expression of several matrix metalloproteinases (MMPs), including MT1-MMP, MMP-1, MMP-2, and MMP-9 [31, 32]. In this study, we found CD147 expression significantly decreased in MDA-MB-468 cells after CD151 knockout, but there is no obvious change in MCF-7 cells. The result shows that the expressions of CD147 are also perhaps regulated by CD151 in a cell-type specific manner.
In summary, this study demonstrated that CD151 plays a crucial role in the development of breast cancer. CD151 overexpression in BC especially in BLBC and is associated with specific clinicopathological and immunohistochemical characteristics. Moreover, the inhibition of CD151 activity reduced cell survival and proliferation, induced apoptosis, and suppressed tumor invasiveness. CD151 can serve as a powerful, independent negative prognostic marker. Here for the first time, we report that CD147 and mmp26 might be regulated by CD151 in a cell-type specific manner. Further research of intrinsic mechanism responsible for these effects is necessary.
References
Goldhirsch A, Wood EP, Coates AS et al (2011) Strategies for subtypes dealing with the diversity of breast cancer: highlights of the St Gallen International Expert Consensus on the primary therapy of early breast cancer 2011. Ann Oncol 22:1736–1747. doi:10.1093/annonc/mdr304
The Cancer Genome Atlas Network (2012) Molecular portraits of human breast tumours. Nature 490:61–70. doi:10.1038/nature11412
Liu Y, Jiang Qiu-Y, Xin T et al (2012) Clinical significance of basal-like breast cancer in Chinese women in Heilongjiang Province. Asian Pacific J Cancer Prevention 13:2735–2738
Xue C, Wang X, Peng R et al (2012) Distribution, clinicopathologic features and survival of breast cancer subtypes in Southern China. Cancer Sci 103:1679–1687. doi:10.1111/j.1349-7006
Funakoshi T, Tachibana I, Hoshida Y et al (2003) Expression of tetraspanins in human lung cancer cells: frequent down regulation of CD9 and its contribution to cell motility in small cell lung cancer. Oncogene 22:674–687. doi:10.1038/sj.onc.1206106
Hemler ME (2008) Targeting of tetraspanin proteins-potential benefits and strategies. Nat Rev Drug Discov 7:747–758. doi:10.1038/nrd2659
Sincock PM, Fitter S, Parton RG et al (1999) PETA-3/CD151, a member of the transmembrane 4 superfamily, is localized to the plasma membrane and endocytic system of endothelial cells, associates with multiple integrins and modulates cell function. J Cell Sci 112:833–844
Zhang XA, Kazarov AR, Yang X et al (2002) Function of the tetraspanin CD151-alpha6beta1 integrin complexduring cellular morphogenesis. Mol Biol Cell 13:1–11
Tokuhara T, Hasegawa H, Hattori N et al (2001) Clinical significance of CD151 gene expression in non-small cell lung cancer. Clin Cancer Res 7:4109–4114
Hui Zhu G, Huang C, Jun-Qiu Zh et al (2011) Expression and prognostic significance of CD151, c-Met, and integrin alpha3/alpha6 in pancreatic ductal adenocarcinoma. Dig Dis Sci 56:1090–1098. doi:10.1007/s10620-010-1416-x
Hashida H, Takabayashi A, Tokuhara T et al (2003) Clinical significance of transmembrane 4 superfamily in colon cancer. Br J Cancer 89:158–167
Ke AW, Shi GM, Zhou J et al (2009) Role of overexpression of CD151 and/or c-Met in predicting prognosis of hepatocellular carcinoma. Hepatology 49:491–503. doi:10.1002/hep.22639
Shigemasa S, Tatsuya M, Naritaka T et al (2011) Prognostic significance of CD151 expression in esophageal squamous cell carcinoma with aggressive cell proliferation and invasiveness. Ann Surg Oncol 18:888–893. doi:10.1245/s10434-010-1387-3
Voss MA, Gordon N, Maloney S et al (2011) Tetraspanin CD151 is a novel prognostic marker in poor outcome endometrial cancer. Br J Cancer 104:1611–1618. doi:10.1038/bjc.2011
Baldwin LA, Hoff JT et al (2014) CD151-α3β1 integrin complexes suppress ovarian tumor growth by repressing slug-mediated EMT and canonical Wnt signaling. Oncotarget 5: 12203–12217
Mosig RA, Lin L, Senturk E et al (2012) Application of RNA-Seq transcriptome analysis: CD151 is an invasion/migration target in all stages of epithelial ovarian cancer. J Ovarian Res 5:1–9. doi:10.1186/1757-2215-5-4
Petrocca F, Lieberman J (2011) Promise and chal-lenge of RNA interference-based therapy for cancer. J Clin Oncol 29:747–754. doi:10.1200/JCO.2009.27.6287
Li Z, Tian T, Xiaopeng H et al (2014) Targeting Six1 by lentivirus-mediated RNA interference inhibits colorectal cancer cell growth and invasion. Int J Clin Exp Pathol 7:631–639
Manjunath N, Wu H, Subramanya S, Shankar P (2009) Lentiviral delivery of short hairpin RNAs. Adv Drug Deliv Rev 61:732–745. doi:10.1016/j.addr
Roselli S, Kahl RGS, Copeland BT et al (2014) Deletion of Cd151 reduces mammary tumorigenesis in the MMTV/PyMT mouse model. BMC Cancer 14:509–519. doi:10.1186/1471-2407-14-509
Yang XH, Richardson AL, Torres-Arzayus MI et al (2008) CD151 accelerates breast cancer by regulating A6 integrin function, signaling, and molecular organization. Cancer Res 68:3204–3212. doi:10.1158/0008-5472
Hashmi AA, Edhi MM, Naqvi H et al (2014) Clinicopathologic features of triple negative breast cancers: an experience from Pakistan. Diagn Pathol 9:43–52. doi:10.1186/1746-1596-9-43
Kwon MJ, Park S, Choi JY et al (2012) Clinical significance of CD151 overexpression in subtypes of invasive breast cancer. Br J Cancer 106:923–930. doi:10.1038/bjc
Copeland BT, Bowman MJ, Ashman LK (2013) Genetic ablation of the tetraspanin CD151 reduces spontaneous metastatic spread of prostate cancer in the TRAMP model. Mol Cancer Res 11:95–105. doi:10.1158/1541-7786
Gialeli C, Theocharis AD, Karamanos NK (2011) Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J 278:16–27. doi:10.1111/j.1742-4658.2010.07919.x
Hong IK, Jin YJ, Byun HJ et al (2006) Homophilic interactions of tetraspanin CD151 upregulate motility and matrix metalloproteinase-9 expression of human melanoma cells through adhesion-dependent c-Jun activation signaling pathways. J Biol Chem 281:24279–24292
Hasegawa M, Furuya M, Kasuya Y et al (2007) CD151 dynamics in carcinoma–stroma interaction: integrin expression, adhesion strength and proteolytic activity. Lab Invest 87:882–892
Park HI, Ni J, Gerkema FE, Liu D, Belozerov VE, Sang QX (2000) Identification and characterization of human endometase (Matrix metalloproteinase-26) from endometrial tumor. J Biol Chem 275:20540–20544
Qiu W, Bai SX, Zhao M et al (2005) Spatio-temporal expression of matrix metalloproteinase-26 in human placental trophoblasts and fetal red cells during normal placentation. Biol Reprod 72:954–959
Uria JA, Lopez-Otin C (2000) Matrilysin-2, a new matrix metalloproteinase expressed in human tumors and showing the minimal domain organization required for secretion, latency, and activity. Cancer Res 60:4745–4751
Xu T, Zhou M, Peng L et al (2014) Upregulation of CD147 promotes cell invasion, epithelial-to-mesenchymal transition and activates MAPK/ERK signaling pathway in colorectal cancer. Int J Clin Exp Pathol 7:7432–7441
Iacono KT, Brown AL, Greene MI, Saouaf SJ (2007) CD147 immunoglobulin superfamily receptor function and role in pathology. Exp Mol Pathol 83:283–295
Acknowledgments
This work was supported by the Major State Basic Research Development Program of China (973 Program) (81172499), the project of Heilongjiang Education Department(11511453,12521626), The Science and Technology Development Fund of Jilin Province (20076023), and the platform foundation from Jilin University (450060445657).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, T., wang, S., Wang, L. et al. Targeting CD151 by lentivirus-mediated RNA interference inhibits luminal and basal-like breast cancer cell growth and invasion. Mol Cell Biochem 407, 111–121 (2015). https://doi.org/10.1007/s11010-015-2459-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-015-2459-2