Abstract
Bone cells respond to various mechanical stimuli including fluid shear stress (FSS) in vitro. Induction of cyclooxygenase-2 (COX-2) is thought to be important for the anabolic effects of mechanical loading. Recently, extracellular-signal-regulated kinase 5 (ERK5) has been found to be involved in multiple cellular processes. However, the relationship between ERK5 and the induction of COX-2 is still unknown. Here, we investigated the potential involvement of ERK5 in the response of pre-osteoblastic MC3T3-E1 cells upon FSS. MC3T3-E1 cells were subjected to 12 dyn/cm2 FSS. Then, we established a ERK5 small interfering RNA (siRNA) transfected cell line using the MC3T3-E1 cells. After the successful transfection confirmed by real-time reverse transcription-polymerase chain reaction and Western blotting, the expression of COX-2, cAMP response element-binding protein (CREB), and nuclear factor kappa B cells (NF-κB) were assayed for downstream effectors of activated ERK5 under FSS by Western blotting. Our results showed that FSS could stimulate COX-2 activity, and induce the phosphorylation of ERK5, CREB, and NF-κB. When the MC3T3-E1 cells were transfected using siRNA before exposure to FSS, COX-2 activity was suppressed, and the phosphorylation of CREB and NF-κB was significantly downregulated. In summary, we demonstrated that ERK5 pathway is essential in the induction of COX-2 gene.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The skeleton is continuously remodeled throughout the lifetime of an individual, and mechanical loading of the skeleton is crucial for maintaining bone mass and strength [1]. Reduced mechanical loading by long-term bed rest or microgravity conditions in space has been shown to cause significant bone loss and mineral changes, whereas increased loading promotes bone formation [2]. As a major factor for affecting bone cell metabolism [3], fluid shear stress (FSS) activates various signal transduction pathways and initiates an anabolic response in osteoblasts, leading to increased cell proliferation and differentiation [4].
The mitogen-activated protein kinase (MAPK) family is one of the cellular signal pathways responding to distinct extracellular stimuli such as growth factors, oxidative stresses, and mechanical stimulation [5, 6]. The classical mammalian MAPK pathways consist of extracellular signal-regulated kinase (ERK), c-Jun Nterminal kinase (JNK), and p38 pathways. These members of the MAPK family have been implicated in the regulation of osteoblastic cell proliferation and differentiation [7, 8]. ERK5 is a new member of the MAPK family that is activated by the upstream MEK5 kinase in response to different growth factors and cellular stressors [9], and is associated with a diverse range of cellular processes including cellular proliferation [10], migration [11], and differentiation [12].
Prostaglandin E2 (PGE2) plays an important role in bone formation [13] and are known to be involved in bone’s adaptive response to FSS [14]. The limiting enzyme in the conversion of membrane-released arachidonic acid to PGE2 is cyclooxygenase 2 (COX-2) [15]. COX-2 is the major isozyme responsible for production of PGE2 response to mechanical loading in bone [16]. Studies [17, 18] have shown that COX-2 are rapidly released in response to mechanical loading, and a selective inhibitor to COX-2 can block mechanically induced bone formation in vivo [16]. In vitro, a number of signaling pathways have been reported to be involved in the induction of COX-2 by FSS in osteoblastic cells, such as ERK1/2 signalling pathway [6]. Further, some transcription factors including cAMP regulatory element-binding protein (CREB) [19] and nuclear factor kappa B (NF-κB) [20] also play an important role in the COX-2 upregulation.
Our previous work [21] has showed that FSS significantly induced ERK5 phosphorylation through the reorganization of the cytoskeleton and phosphorylated ERK5 will translocated to the nucleus to regulate gene expression. However, whether ERK5 was involved in effects of mechanical loading on COX-2 gene expression was unknown. In this study, we characterized FSS-induced COX-2 expression in MC3T3-E1 osteoblastic cells and investigated the role of ERK5 signaling in induction of COX-2 under FSS. Furthermore, we also explored whether CREB and NF-κB, as the promoter of COX-2, are mediated by ERK5 signalling pathway.
Materials and methods
Materials and antibodies
Cell culture reagents, culture dishes, and flasks were purchased from Life Technologies (Carlsbad, CA, USA) and Sigma (St. Louis, MO, USA). Rabbit anti-phospho-ERK5 (Thr218/Tyr 220) (1: 1000), rabbit anti-pCREB (Ser133) (1: 800), rabbit anti-p-NF-κB-p65 (S536) (1: 750), and rabbit anti-ERK5 (1: 800) were purchased from Cell Signaling Technology (Beverly, MA, USA). Rabbit anti-COX-2 antibody (1: 1000) was purchased from Abcam (Cambridge, UK). Rabbit anti-β-actin (1: 800) monoclonal antibody was purchased from Sigma. All secondary antibodies were purchased from Invitrogen (Paisley, UK). Other chemicals of the highest purity were obtained from Sigma and Millipore (Benford, MA, USA).
Cell culture
Cells from the MC3T3-E1 mouse osteoblastic cell line were obtained from the Chinese Academy of Medical Sciences. The cells were grown on Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F-12) containing 10 % fetal bovine serum (FBS), 100 U/ml penicillin G and 100 U/ml streptomycin, and were maintained in a 5 % CO2 humidified environment at 37 °C. Before being subjected to FSS, cells were seeded onto type I collagen-coated (200 μg/ml) glass cover slips (20 × 50 mm) at a density of 4000 cells/cm2. Cell medium was changed every 3 days. The cells were serum-starved for 24 h in DMEM/F-12 without FBS before FSS experiment.
Transfection with small interfering RNA (siRNA)
MC3T3-E1 cells were transfected with siRNA against ERK5 or CREB and non-silencing siRNA (Santa Cruz Biotechnology, CA, United States) constructs using the commercial transfection reagent (Santa Cruz Biotechnology, CA, United States) according to the manufacturer’s instructions. Following transfection, cells were incubated at 37 °C in a CO2 incubator for 48 h before being harvested for the assays described below. To assess the specific effect of ERK5 siRNA on silencing ERK5 expression, the protein levels of ERK5 and ERK1/2 were detected by Western blotting analysis 48 h post-transfection.
FSS experiment
FSS experiment was conducted as described previously [21, 22]. Cells were subjected to 12 dynes/cm2 FSS for 45 min in the presence or absence of siRNA ERK5. The flow system was maintained at 37 °C, and was filled with 25 ml of serum free medium that was aerated with 5 % CO2/95 % air.
Real-time polymerase chain reaction (RT-PCR)
Total RNA was extracted from the culture of MC3T3 cells using the RNAiso plus reagent (TaKaRa Biotechnology Co, Dalian, China) and then reverse transcribed into complimentary DNA (cDNA) by the Primescript™ reverse transcription Master Mix (TaKaRa Biotechnology Co, Dalian, China) according to the manufacturer’s instructions. Reverse transcription reaction was performed at 37°C for 15 min followed by 85 °C for 5 s. The primers used in the PCR reactions are provided in Table 1. Quantitative RT-PCR was performed by the Applied Biosystems 7500/7500 Fast Real-Time PCR Software (Applied Biosystems, CA, Unite States) using the SYBR® Premix Ex Taq™II(TaKaRa Biotechnology Co., Dalian, China). The qRT-PCR was conducted at 95 °C for 30 s, then by 40 cycles of 95 °C for 5 s and 60 °C for 34 s. Relative quantity of mRNA level was performed by using comparative C T method (△△ C T) with β-actin as internal reference.
Western blot analysis
After each treatment, the cells were washed with cold PBS twice and lysed on ice with RIPA buffer (Beyotime Biotechnology, Haimen, China) including proteinase and phosphatase inhibitors supplemented with 1 mmol/l PMSF. The lysates were centrifuged at 12,000 rpm for 10 min at 4 °C, and the supernatants were collected. The protein concentration was measured by BCA protein assay system (Beyotime Biotechnology, Haimen, China). The cell lysate and sample buffer were mixed and then boiled for 5 min. Equal amounts of protein (30–40 ug) were separated by 8–12 % SDS/PAGE and transferred onto PVDF membranes (Millipore). The membranes were blocked in TBST (Tris-buffered saline with 0.1 % Tween-20) and 5 % non-fat dried milk for 2 h at room temperature and incubated with appropriate primary antibodies for 2–4 h at 37 °C. After being washed with TBST for three times, the membrane was incubated with horseradish peroxidase (HRP)-conjugated secondary antibody and HRP-conjugated antibiotin antibody for 1.5 h at a room temperature. The protein bands were detected using the Super Signal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific Inc., Rockford, IL, United States) and imaged using a VersaDoc Imaging System (Bio-Rad Laboratories Co., Ltd. Hercules, CA, United States). Densitometric analysis was performed using Quantity One Software v4.62 (BioRad Laboratories Co., Ltd. Hercules, CA, United States) and the results were presented as the mean of three independent experiments.
Statistical analysis
All data were presented as the mean ± SD of each group. Statistical analyses were performed using a one-way analysis of variance followed by the Bonferroni correction. P < 0.05 was considered to be significant.
Results
FSS induces phosphorylation of ERK5 and expression of COX-2 gene in MC3T3-E1 osteoblastic cells
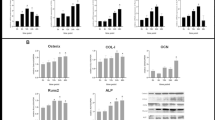
We sought to determine whether ERK5 and COX-2 are activated by FSS in our experimental system. After MC3TC-EI cells were subjected to 12 dyn/cm2 FSS for 0–60 min, the phosphorylation of ERK5 and the expression of COX-2 were significantly elicited. We found that ERK5 phosphorylation was markedly (P < 0.05) increased at 15 min and reached a peak at 45 min (Fig. 1) while the change of ERK5 was not detected (Fig. 1). Similar to ERK5 phosphorylation, the expression of COX-2 was also markedly upregulated under FSS and reached peak within 45 min (Fig. 1). Besides, we also investigated the expression of COX-2 mRNA when MC3TC-EI cells were subjected to 12 dyn/cm2 FSS for 0–60 min; the results were similar with findings by Western blotting.
Effects of ERK5 siRNA on ERK5 gene
ERK5 siRNA was successfully transfected into MC3TC-EI cells (Fig. 2A). Compared with a control siRNA or transfection agent alone, transfection of ERK5 siRNA at 48 h significantly reduced the expression of ERK5 gene in mRNA level (to 25.81 ± 8.90 %, P < 0.05, Fig. 2B) and protein level (to 20.01 ± 4.85 %, P < 0.05, Fig. 2C, D), while ERK5 siRNA had no effect on ERK1/2 protein level (Fig. 2C, D).
Verification of successful transfection and knockdown of ERK5 (A). Cells were transfected with 60 nM ERK5-specific siRNA, 60 nM non-silencing siRNA or left untransfected. Cells were lysed 48 h after transfection, RT-PCR and Western blot illustrates the effect of ERK5 siRNA on the expression of ERK5 (B, C, D) and ERK1/2 (C, D) in osteoblastic cells. *P < 0.05 versus control group and non-silence siRNA group
ERK5 is essential in FSS-induced COX-2 expression
To investigate the role of ERK5 in the induction of COX-2 by FSS, we examined the expression of COX-2 in each group under FSS for 45 min. The results revealed that the expression of COX-2 gene (1.8 fold, P < 0.05, Fig. 3A, B) was significantly increased upon FSS for 45 min. When osteoblasts were incubated with ERK5 siRNA before exposure to FSS, the induction of COX-2 (to 34.69 + 7.89 %, P < 0.05, Fig. 3A, B) was all significantly decreased, compared with FSS group.
Effects of ERK5 siRNA on FSS induction of COX-2 mRNA and protein level in MC3T3-E1 cells. Western blot analysis was performed for COX-2, p-ERK5 and β-actin (A). Cells were subjected to FSS (12 dynes/cm2) with and without 10 nM ERK5 siRNA for 45 min. B for quantification of COX-2 and p-ERK5, versus β-actin, data were presented as mean ± SD from at least three independent experiments. *P < 0.05 versus control, *P < 0.05 versus control group, # P < 0.05 versus control group, **P < 0.05 versus FSS group
Transcription factors CREB and NF-κB in the role of ERK5-COX-2 pathway under FSS
Previous studies [19, 23] have confirmed that the COX-2 promoter included CREB and NF-κB, which are important for the induction of COX-2 under FSS in osteoblast. To evaluate the involvement of the ERK5 signaling pathway in the FSS-induced CREB and NF-κB, we explored the expression of p-CREB and p-NF-κB after each treatment. As shown in Fig. 4, compared with control group, FSS could significantly upregulated the phosphorylation of CREB (11.6 fold, P < 0.05) and NF-κB (2.34 fold, P < 0.05). Compared with FSS group, knockdown of ERK5 by siRNA in osteoblasts before exposure to FSS completely attenuated the phosphorylation of CREB (to 31.56 + 9.23 %, P < 0.05) and NF-κB (28.64 + 8.92 %, P < 0.05) (Fig. 4A, B).
ERK5 regulates p-CREB and p-NF-κB expression under FSS. After various treatments, cells were lysed and measured by Western blot (A, C). B and D for quantification of p-CREB and p-NF-κB, versus β-actin; data were presented as mean ± SD from at least three independent experiments. *P < 0.05 versus control, # P < 0.05 versus FSS group, **P < 0.05 versus FSS + CREB siRNA group, ## P < 0.05 versus FSS + PDTC group
Then, we investigated the role of CREB and NF-κB in the induction of COX-2 under FSS. As shown in Fig. 4, compared with FSS group, knockdown of CREB by siRNA or inhibit the NF-κB phosphorylation by PDTC in osteoblasts before exposure to FSS completely attenuated the expression of COX-2 gene (Fig. 4C, D).
Discussion
Although regulation of the COX-2 gene has been extensively investigated in osteoblast under FSS, little is known about whether ERK5, a novel MAPK family is involved in the induction of COX-2. Here, we discovered that in MC3T3-E1 osteoblastic cells, ERK5 is activated under FSS and ERK5 siRNA mediated the expression of COX-2 gene under FSS. Furthermore, we found for the first time that the transcription factors CREB and NF-κB, all known to participate in the regulation of COX-2 gene, are regulated by ERK5.
ERK5 is activated by its only upstream kinase, MAPK kinase (MEK) 5, which substrates only ERK5 [24]. MEK5/ERK5 signalling pathway has been associated with a diverse range of cellular processes including cellular proliferation, migration, survival, and angiogenesis. Deletion of ERK5 or MEK5 genes in mice showed that the ERK5 is essential for normal cardiovascular development [25], and MEK5/ERK5 pathway has also been implicated in neuronal survival [26] and differentiation [27]. Furthermore, ERK5 appears to mediate the actions of oncogenes in some cancers, including breast cancer [28] and osteosarcoma [29]. Kim et al. [29] reported that a ERK5 was distinctly overexpressed in osteosarcoma cell and the expression of ERK5 regulates the invasion of osteosarcoma cells by inducing MMP-9 expression. Nevertheless, the physiological functions of ERK5 in the osteoblast have been poorly studied.
In this study, in subjecting MC3T3-E1 cells to 12 dyn/cm2 FSS, we observed that the ERK5 phosphorylation was up-regulated after 15 min and levels peaked at 45 min, which was consistent with our previous work [21, 30]. Similar to other studies [19, 20, 31], the induction of COX-2 protein by FSS was increased at 15 min and reach peak at 45 min. Previous studies showed that COX-2 induction are necessary for mechanically induced bone formation [15, 16], and some signalling pathways play an important role in this process. For instance, Wadhwa et al. [6] demonstrated that FSS-induced upregulation of COX-2 was largely mediated by ERK1/2 signaling pathway. Similar to ERK1/2, ERK5 also has a Thr–Glu–Tyr (TEY) sequence in its dual phosphorylation site [9]. It seems reasonable to suggest that ERK5 may also regulate the expression of COX-2 gene in osteoblast.
To investigate the role of ERK5 in FSS-induction of COX-2, we firstly knocked down ERK5 gene with siRNA. We observed that the expression of EKR5 was markedly decreased by transfection of ERK5 siRNA (60 nM) at 48 h, furthermore, the expression of ERK1/2 was not affected in this process. Then, we examined the role of ERK5 on FSS-induced COX-2 expression and down-regulation of COX-2 was observed. These data are the first report of a causal relationship between ERK5 activation and COX-2 expression induced by FSS.
What are the downstream targets that mediate ERK5 regulation of COX-2 in osteoblast? It was reported that as the promoter of COX-2, transcription factors including CREB [19] and NF-κB [20] played crucial roles in the FSS-induced COX-2 expression in osteoblasts. In osteoblastic MC3T3-E1 cells, Ogasawara et al. [19] investigated transcriptional regulatory elements responsible for the shear stress-induced COX-2 expression, and found that mutation of sites of CREB decreased in the FSS-induced COX-2 expression. Chen et al. [20] examined the role of NF-κB on COX-2 upregulation in osteoblasts in response to FSS and revealed that NF-κB translocation to the nucleus is essential for the FSS-induced increase in COX-2 expression. However, whether these transcription factors were mediated by ERK5 in osteoblast were still unknown. Our data showed that CREB and NF-κB are all activated when cells were subjecting to FSS for 45 min. Furthermore, inhibition of ERK5 by siRNA before exposure to FSS reduced the phosphorylation of CREB and NF-κB, suggesting that these transcription factors may be the possible downstreams of ERK5 in osteoblast. Besides, we confirmed that transcription factors CREB and NF-κB mediated the COX-2 expression under FSS, which was consistent with previous studies [19, 20].
Conclusions
In summary, we propose that ERK5 is activated under FSS in osteoblast and that it stimulates expression of COX-2. Besides, we also first reported that as the promoter of COX-2, transcription factors CREB and NF-κB are also activated by ERK5, suggesting that CREB and NF-κB are the possible downstream kinases of ERK5 in osteoblast.
References
Raisz LG (1999) Physiology and pathophysiology of bone remodeling. Clin Chem 45:1353–1358
Ozcivici E, Luu YK, Adler B, Qin YX, Rubin J, Judex S, Rubin CT (2010) Mechanical signals as anabolic agents in bone. Nat Rev Rheumatol 6:50–59
Basso N, Heersche JN (2002) Characteristics of in vitro osteoblastic cell loading models. Bone 30:347–351
Rubin J, Rubin C, Jacobs CR (2006) Molecular pathways mediating mechanical signaling in bone. Gene 367:1–16
You J, Reilly GC, Zhen X, Yellowley CE, Chen Q, Donahue HJ, Jacobs CR (2001) Osteopontin gene regulation by oscillatory fluid flow via intracellular calcium mobilization and activation of mitogen-activated protein kinase in MC3T3-E1 osteoblasts. J Biol Chem 276:13365–13371
Wadhwa S, Godwin SL, Peterson DR, Epstein MA, Raisz LG, Pilbeam CC (2002) CC Fluid flow induction of cyclo-oxygenase 2 gene expression in osteoblasts is dependent on an extracellular signal-regulated kinase signaling pathway. J Bone Miner Res 17:266–274
Wang B, Du T, Wang Y, Yang C, Zhang S, Cao X (2011) Focal adhesion kinase signaling pathway is involved in mechanotransduction in MG-63 cells. Biochem Biophys Res Commun 410:671–676
Lee DY, Yeh CR, Chang SF, Lee PL, Chien S, Cheng CK, Chiu JJ (2008) Integrin-mediated expression of bone formation-related genes in osteoblast-like cells in response to fluid shear stress: roles of extracellular matrix, Shc, and mitogen-activated protein kinase. J Bone Miner Res 23:1140–1149
Nishimoto S, Nishida E (2006) MAPK signalling: ERK5 versus ERK1/2. EMBO Rep 7:782–786
Pi X, Yan C, Berk BC (2004) Big mitogen-activated protein kinase (BMK1)/ERK5 protects endothelial cells from apoptosis. Circ Res 94:362–369
Mehta PB, Jenkins BL, McCarthy L, Thilak L, Robson CN, Neal DE, Leung HY (2003) MEK5 overexpression is associated with metastatic prostate cancer, and stimulates proliferation, MMP-9 expression and invasion. Oncogene 22:1381–1389
Dinev D, Jordan BW, Neufeld B, Lee JD, Lindemann D, Rapp UR, Ludwig S (2001) Extracellular signal regulated kinase 5 (ERK5) is required for the differentiation of muscle cells. EMBO Rep 2:829–834
Haversath M, Catelas I, Li X, Tassemeier T, Jager M (2012) PGE(2) and BMP-2 in bone and cartilage metabolism: 2 intertwining pathways. Can J Physiol Pharmacol 90:1434–1445
McAllister TN, Du T, Frangos JA (2000) Fluid shear stress stimulates prostaglandin and nitric oxide release in bone marrow-derived preosteoclast-like cells. Biochem Biophys Res Commun 270:643–648
Kujubu DA, Fletcher BS, Varnum BC, Lim RW, Herschman HR (1991) TIS10, a phorbol ester tumor promoter-inducible mRNA from Swiss 3T3 cells, encodes a novel prostaglandin synthase/cyclooxygenase homologue. J Biol Chem 266:12866–12872
Forwood MR (1996) Inducible cyclo-oxygenase (COX-2) mediates the induction of bone formation by mechanical loading in vivo. J Bone Miner Res 11:1688–1693
Rawlinson SC, El-Haj AJ, Minter SL, Tavares IA, Bennett A, Lanyon LE (1991) Loading-related increases in prostaglandin production in cores of adult canine cancellous bone in vitro: a role for prostacyclin in adaptive bone remodeling? J Bone Miner Res 6:1345–1351
Ajubi NE, Klein-Nulend J, Nijweide PJ, Vrijheid-Lammers T, Alblas MJ, Burger EH (1996) Pulsating fluid flow increases prostaglandin production by cultured chicken osteocytes–a cytoskeleton-dependent process. Biochem Biophys Res Commun 225:62–68
Ogasawara A, Arakawa T, Kaneda T, Takuma T, Sato T, Kaneko H, Kumegawa M, Hakeda Y (2001) Fluid shear stress-induced cyclooxygenase-2 expression is mediated by C/EBP beta, cAMP-response element-binding protein, and AP-1 in osteoblastic MC3T3-E1 cells. J Biol Chem 276:7048–7054
Chen NX, Geist DJ, Genetos DC, Pavalko FM, Duncan RL (2003) Fluid shear-induced NFkappaB translocation in osteoblasts is mediated by intracellular calcium release. Bone 33:399–410
Li P, Ma YC, Shen HL, Han H, Wang J, Cheng HJ, Wang CF, Xia YY (2012) Cytoskeletal reorganization mediates fluid shear stress-induced ERK5 activation in osteoblastic cells. Cell Biol Int 36:229–236
Frangos JA, McIntire LV, Eskin SG (1988) Shear stress induced stimulation of mammalian cell metabolism. Biotechnol Bioeng 32:1053–1060
Klein T, Shephard P, Kleinert H, Komhoff M (2007) Regulation of cyclooxygenase-2 expression by cyclic AMP. Biochim Biophys Acta 1773:1605–1618
English JM, Vanderbilt CA, Xu S, Marcus S, Cobb MH (1995) Isolation of MEK5 and differential expression of alternatively spliced forms. J Biol Chem 270:28897–28902
Regan CP, Li W, Boucher DM, Spatz S, Su MS, Kuida K (2002) Erk5 null mice display multiple extraembryonic vascular and embryonic cardiovascular defects. Proc Natl Acad Sci USA 99:9248–9253
Cavanaugh JE (2004) Role of extracellular signal regulated kinase, 5 in neuronal survival. Eur J Biochem 271:2056–2059
Nishimoto S, Kusakabe M, Nishida E (2005) Requirement of the MEK5-ERK5 pathway for neural differentiation in Xenopus embryonic development. EMBO Rep 6:1064–1069
Song H, Jin X, Lin J (2004) Stat3 upregulates MEK5 expression in human breast cancer cells. Oncogene 23:8301–8309
Kim SM, Lee H, Park YS, Lee Y, Seo SW (2012) ERK5 regulates invasiveness of osteosarcoma by inducing MMP-9. J Orthop Res 30:1040–1044
Zhao LG, Chen SL, Teng YJ, An LP, Wang J, Ma JL, Xia YY (2014) The MEK5/ERK5 pathway mediates fluid shear stress promoted osteoblast differentiation. Connect Tissue Res 55:96–102
Pavalko FM, Chen NX, Turner CH, Burr DB, Atkinson S, Hsieh YF, Qiu J, Duncan RL (1998) Fluid shear-induced mechanical signaling in MC3T3-E1 osteoblasts requires cytoskeleton-integrin interactions. Am J Physiol 275:C1591–C1601
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant Nos. 81071478 and 81450042).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jiang, J., Zhao, Lg., Teng, Yj. et al. ERK5 signalling pathway is essential for fluid shear stress-induced COX-2 gene expression in MC3T3-E1 osteoblast. Mol Cell Biochem 406, 237–243 (2015). https://doi.org/10.1007/s11010-015-2441-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-015-2441-z