Abstract
Freeze tolerance is a critical winter survival strategy for the wood frog, Rana sylvatica. In response to freezing, a number of genes are upregulated to facilitate the survival response. This includes fr10, a novel freeze-responsive gene first identified in R. sylvatica. This study analyzes the transcriptional expression of fr10 in seven tissues in response to freezing, anoxia, and dehydration stress, and throughout the Gosner stages of tadpole development. Transcription of fr10 increased overall in response to 24 h of freezing, with significant increases in expression detected in testes, heart, brain, and lung when compared to control tissues. When exposed to anoxia; heart, lung, and kidney tissues experienced a significant increase, while the transcription of fr10 in response to 40 % dehydration was found to significantly increase in both heart and brain tissues. An analysis of the transcription of fr10 throughout the development of the wood frog showed a relatively constant expression; with slightly lower transcription levels observed in two of the seven Gosner stages. Based on these results, it is predicted that fr10 has multiple roles depending on the needs and stresses experienced by the wood frog. It has conclusively been shown to act as a cryoprotectant, with possible additional roles in anoxia, dehydration, and development. In the future, it is hoped that further knowledge of the mechanism of action of FR10 will allow for increased stress tolerance in human cells and tissues.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The wood frog, or Rana sylvatica, is a woodland frog whose distribution ranges from Alaska and the Maritimes south in the Appalachians to northern Georgia [1]. The climate of this distribution range necessitates that the wood frog have a strategy to deal with sub-zero winter temperatures. As the wood frog has highly water permeable skin, when ice penetrates their hibernacula it will also penetrate the frog. This has resulted in wood frogs utilizing the freezing strategy of freeze tolerance. By freezing and subsequently thawing in a terrestrial habitat, the frog is able to breed very early in the spring using melt-water vernal ponds.
By allowing up to 65 % of their total body water to freeze, the wood frog must deal with problems such as the possibility of physical damage to the subcellular architecture by ice crystals and cessation of all vital functions including heart beat, circulation, breathing, muscle movement, and neural activity [2]. An additional feature of freezing is dehydration, as the formation of extracellular ice results in an increased osmolarity of remaining extracellular fluids, causing an efflux of water from the cells, ultimately resulting in a sharp decrease in cell volume [2]. Other injuries in freezing can result from ischemia; where freezing stops the blood circulation and oxygen delivery. In an attempt to minimize damage caused by these additional stresses, the wood frog has developed adaptations that help to counteract the negative effects of freezing. These include (1) the accumulation of low molecular weight cryoprotectants that work to minimize cellular dehydration and stabilize macromolecular components, (2) the production of ice nucleating proteins that help to control ice growth, and (3) metabolic adaptations, including metabolic arrest mechanisms, which reduce energy consumption during freezing [2]. In a previous study, the use of cDNA libraries prepared from the liver of a frozen wood frog has revealed select genes that are upregulated in response to freezing [3]. While several of these genes were already known to be freeze-responsive, three were discovered to be completely novel clones [3]. They were subsequently named li16, fr47, and fr10 [3–5].
The 10 kDa freeze-responsive protein, or FR10, is a small protein of 90 amino acids. The polypeptide chain consists of 9 strongly basic amino acids and 11 strongly acid amino acids resulting in an overall estimated isoelectric point of 5.25. Computer modeling has predicted the protein structure of FR10 to have a highly hydrophobic core and a secretion signal, which has been suggested to allow FR10 to translocate to the extracellular environment [6]. The cDNA sequence that codes for this protein is composed of 457 bp, and it was found that this sequence shares no significant homology to any known genes [3].
By investigating various environmental stresses (such as freezing, anoxia, and dehydration) in addition to developmental expression, it will be possible to better determine the pattern of expression and possible biological role of FR10. In the future, it is possible that FR10 will be an important tool to improve stress tolerance in human cells and organs.
Materials and methods
Animal treatment and tissue preparation
Adult male frogs were collected from the Ottawa region from breeding ponds in early spring. Collected frogs were washed in a tetracycline bath and were kept in damp moss-lined plastic boxes at 5 °C to acclimate for at least 1 week. Control frogs were sampled from this condition. Frogs exposed to freezing conditions were transferred to plastic boxes lined with damp paper towel in a −2.5 °C incubator. Preliminary tests using frogs that have a thermistor secured to their abdomen showed that freezing began within 45–60 min [7]. Therefore, frogs were given 1 h of cooling time before the length of freezing exposure was measured. Frogs were sampled after 24 h freezing for brain, heart, lung, kidney, liver, testes, and hind leg skeletal muscle tissue. These organs/tissues were selected to reflect the movement of ice through the frog (first muscle, moving toward the vital organs, with the liver freezing last) [2], as well as for the importance of those organs in the survival of the frog during/following stress recovery (e.g., testes are essential to ensure future generations, brain controls the functioning of all other organs, etc.). For anoxia exposure, the frogs were placed in 2 L glass bottles that contained 50 ml of dechlorinated tap water which had already been bubbled with N2 gas for 30 min. Frogs were put in the bottles that were immersed in ice while the gassing occurred for another 15 min, after which the bottles were capped, sealed with Parafilm, and placed in the 5 °C incubator. Frogs were sampled after 24 h of anoxia exposure. Frogs destined for dehydration experiments began as 5 °C acclimated frogs. These were individually weighed and ranked from heaviest to lightest and were placed in large plastic buckets (held on ice), until each bucket held 10 frogs of varying masses. The buckets were placed in an incubator set at 5 °C where frogs were allowed to lose water by evaporation. At varying intervals, the frogs were quickly reweighed and ranked, and the amount of body water lost was calculated using the equation \({{\left( {M_{\text{i}} - M_{\text{d}} } \right)} \mathord{\left/ {\vphantom {{\left( {M_{\text{i}} - M_{\text{d}} } \right)} {\left( {M_{\text{i}} \; \times \; \% {\text{H}}_{ 2} {\text{O}}} \right)}}} \right. \kern-0pt} {\left( {M_{\text{i}} \; \times \; \% {\text{H}}_{ 2} {\text{O}}} \right)}}\) where M i is the initial mass of the animal, M d is the mass at each weighing, and %H2O is the percentage of total body mass that is water (for control wood frogs, %H2O is 80.8 ± 1.2 %) [8]. Initial trials using individually marked frogs showed that the frogs retained their ranking at each weighing, proving this method to be an effective way to monitor water loss. The mean rate of water loss during the experiment was about 0.5 % of total body water lost per hour, and animals were sampled after they had lost 40 % of their total body water. Frogs were euthanized by pithing after their desired condition was met. Tissues were rapidly excised, immediately frozen in liquid nitrogen and were stored in a −80 °C freezer until use.
For wood frog development studies, R. sylvatica eggs were collected from ponds around Bishop’s Mill (Ottawa, ON) and were housed in an aquarium containing diluted pond water, bubbled with an air stone. Tadpoles were fed shredded lettuce softened by boiling, as well as a daily supply of goldfish food. Every 2–3 days, all but 1 L of the aquarium water was removed and 3 L of tap water purified with dechlorinated drops was added. The 1 L of aquarium water was retained to maintain bacterial cultures required for the natural flora of the tadpole environment. As the tadpoles developed, they were sacrificed at various stages by submergence into liquid nitrogen. These stages were Gosner stages 14–20, where the embryo has developed a neural fold, Gosner stages 21–25 where it has developed into a recognizable tadpole, Gosner stages 26–30 where a back limb bud has emerged, Gosner stages 31–35 where the back limb bud has developed a “paddle” foot (i.e., no toes), Gosner stages 36–41 where the back limb has developed individual toes, Gosner stages 42–43 where a front limb has developed, and finally Gosner stages 44–45 where the tail has almost completely metabolized and appears as a bud [9].
RNA preparation and cDNA synthesis
All plastic ware, buffers, and solutions used for RNA extraction were treated with 0.1 % (v/v) diethylpyrocarbonate (DEPC) and autoclaved. Frozen tissues were weighed out (100 mg for heart and hind leg skeletal muscle and 50 mg for all others) and ground to a powder under liquid nitrogen using a motor and pestle. To this 1 mL of Trizol™ (Invitrogen) was added, the sample was homogenized using a Polytron PT-10 homogenizer, and 200 µL of chloroform was added and mixed thoroughly. The sample was centrifuged at 10,000×g for 15 min at 4 °C. The aqueous supernatant containing the RNA was removed to a 1.5 mL tube, was precipitated with 500 µL of isopropanol and this was followed by a 20 min incubation at room temperature (RT). The samples were centrifuged at 12,000×g for 15 min at 4 °C; the resulting pellet was washed with 1 mL of 70 % ethanol (in DEPC treated ddH2O) and was centrifuged again at 7,500×g for 5 min at 4 °C. After the resulting pellet was allowed to air dry for 10 min, the pellet was resuspended in 25 µL of DEPC-treated ddH2O. The quality of RNA extracted was judged based on the ratio of absorbances at 260 and 280 nm, while the quantity was determined using a UV spectrophotometer at 260 nm.
For reverse transcription, 3 µg of RNA was diluted with DEPC-treated ddH2O to a final volume of 10 µL, to which 1 µL of 200 ng/µL oligo dT primer (5′- TTTTTTTTTTTTTTTTT TTTTTV-3′) was added. This was incubated for 5 min at 65 °C, after which the sample was cooled rapidly on ice for 1 min. A total of 4 µL 5 × first strand buffer, 2 µL 100 mM DTT, 1 µL dNTP mixture (25 mM each), and 1 µL reverse transcriptase enzyme M-MLV (all reagents from Invitrogen) were added to the adapted RNA sample. This was incubated for 1 h at 42 °C, and rapidly chilled to 4 °C.
Reverse transcriptase polymerase chain reaction
For RT-PCR, a mixture of 15 µL DEPC-treated ddH2O, 5 µL diluted cDNA, 1.25 µL primer mixture, 0.75 µL 10 × PCR buffer (Invitrogen), 1.5 µL 50 mM MgCl2, 0.5 µL dNTP mixture (25 mM each), and 1 µL of Taq polymerase were combined for a total volume of 25 µL. Primers used were designed using the Primer Design Program v.3 (Scientific and Educational Software) based on the sequence for fr10 (Genbank accession U44831). The primer sequences used in this study are listed in Table 1. The PCR program used was: 95 °C for 7 min, followed by 28 cycles of 94 °C for 1 min, 53 °C for 1 min, and 72 °C for 1.5 min, and finally it was finished with 72 °C for 10 min.
Computer modeling
To determine the structure homology between FR10 and proteins that have been previously identified as either possible homologs, evolutionary precursors, or related proteins we aligned the available crystal and ad initio structures of available proteins. Crystal structures were all downloaded from the RCSB Protein Data Bank (www.rcsb.org). Alignments were carried out using the ‘Sequence and Structural Alignment’ program available in Molecular Operating Environment (MOE) 2011.10 software (Chemical Computing Group, Montreal, QC, Canada). The root mean square deviation (RMSD) was used as the measure of structural similarity between aligned structures.
Although anti-freeze proteins (AFPs) have a common function, they show a high degree of diversity in sequences and structures. Therefore, sequence similarity based search methods often fails to predict AFPs from sequence databases. To provide insight into the possibility that FR10 could function as an anti-freeze protein, we used a random forest approach “AFP-Pred” for the prediction of antifreeze proteins from the FR10 amino acid sequence [10]. AFP-Pred was trained on the dataset containing 181 AFPs and 9193 non-AFPs, achieving an 81.33 % accuracy from training and 83.38 % from testing [10].
Statistics
PCR products were separated on a 1 % agarose gel stained with Sybr Green and visualized using the ChemiGenius imaging system (Syngene, Frederick, MD, USA). To ensure saturation did not occur, the most dilute cDNA samples that produced visible bands were used for quantification by the GeneTools program. Expression of α-tubulin was found to be constantly expressed in all tissues and stresses used in this study and was, therefore, chosen as a suitable housekeeping gene to normalize fr10 transcripts. Data are expressed relative to control values and statistically significant changes in expression were determined carrying out an ANOVA with a post hoc Student–Newman–Keuls (SNK) test using SigmaPlot (v.11). All statistical differences are marked as a result of the post hoc SNK analysis.
Results
Response of fr10 to freezing
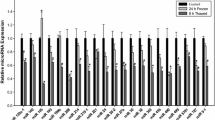
Transcript levels in select frozen wood frog tissues were explored to study the upregulation of fr10 upon freezing. Increases in fr10 expression during freezing were found in 6 of 7 studied tissues when compared to control (Fig. 1). The upregulation upon freezing was greatest in testes tissues where fr10 mRNA levels were 3.72 ± 0.55 fold that of control, and in heart tissue where the freeze-responsive expression increased 3.67 ± 0.32 fold (P < 0.005). Figure 1 shows that after 24 h of freezing, fr10 transcript levels were approximately 1.7 fold higher in brain (1.76 ± 0.24) and lung (1.79 ± 0.17) tissues, compared to their respective control time point (P < 0.05). Only kidney tissue significantly decreased expression of fr10 upon freezing to 55 ± 5 % of control values (P < 0.05).
RT-PCR analysis showing the effects of 24 h freezing on fr10 mRNA transcript levels in select tissues of the wood frog. a Histogram show mean values (±SEM, n = 4–5 independent determinations) for fr10. b Representative fr10 and tubulin bands. Data were analyzed using analysis of variance with a post hoc Student–Newman–Keuls test where the letters ‘a,’ ‘b,’ and ‘c’ represent significant differences between indicated stress and control, where ‘a’ indicates P < 0.05, ‘b’ indicates P < 0.01 and ‘c’ indicates P < 0.005
Response of fr10 to anoxia and dehydration
Transcription of fr10 was further analyzed after 24 h anoxic and 40 % total body water dehydration, as these two stresses are critical components of freezing. After 24 h of anoxia, three tissues displayed significant increases in transcript levels compared to control. These tissues included heart with increases of 1.82 ± 0.08 fold, 1.68 ± 0.09 for lung, and 1.71 ± 0.24 for kidney tissue when compared to control values (Fig. 2; P < 0.05). Conversely, four tissues showed no change in transcription. In response to 40 % dehydration, both heart and brain increased in fr10 mRNA levels to 1.57 ± 0.15 and 1.54 ± 0.08 fold from control values, while lung and muscle decreased fr10 transcription to 65 ± 4 % and 50 ± 7 % of control values, respectively (Fig. 3; P < 0.05).
RT-PCR analysis showing the effects of 24 h of anoxia on fr10 mRNA transcript levels in select tissue of the wood frog. a Histogram show mean values (±SEM, n = 4–5 independent determinations) for fr10. b Representative fr10 and tubulin bands. All other information as mentioned in Fig. 1
RT-PCR analysis showing the effects of 40 % dehydration on fr10 mRNA transcript levels in select tissues of the wood frog. a Histogram show mean values (±SEM, n = 4–5 independent determinations) for fr10. b Representative fr10 and tubulin bands. All other information as mentioned in Fig. 1
Expression of fr10 during development
Transcript levels of fr10 were also examined during the stages of development of the wood frog, from embryo to tadpole to juvenile frog (Fig. 4). While fr10 was present in all stages of development, expression was found to be relatively constant throughout development. Almost all stages of development had expression levels of fr10 significantly higher than Gosner Stages 14–20, which was used as our control value. However, Gosner stages 31–35 and 42–43 (stages 4 and 6, respectively) have decreased transcription compared to stages 14–20 (stage 1) to 72 ± 7 and 61 ± 6 % that of control, respectively (P < 0.05). These stages are characterized by the development of a foot that has yet to grow toes (called a “paddle foot”) during Stages 31–35, while stages 42–43 represent frogs that appear to be fully formed but have yet to metabolize their long tail [9].
Changes in fr10 mRNA transcript levels throughout tadpole development of the wood frog. a Histogram show mean values (±SEM, n = 4–5 independent determinations) for fr10. b Representative fr10 and tubulin bands. Data were analyzed using analysis of variance with a post hoc Student–Newman–Keuls test where the letters ‘a,’ ‘b,’ and ‘c’ represent significant differences between Gosner stages 14–20 (used as a control) and the indicated stage whereas, ‘d’ indicates significant differences between the indicated stage and the stage immediately preceding it; ‘a’ indicates P < 0.05, ‘b’ indicate P < 0.01, while ‘c’ and ‘d’ indicate P < 0.005
Computational analysis of FR10 protein
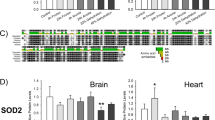
Structural alignment of available apolipoproteins and AFPs to FR10 were used in an attempt to identify; (1) a possible function, or (2) evolutionary precursors based on structural homology [6]. The structural comparisons to apolipoproteins were included given that previous studies have documented possible sequence and structural similarities between FR10 and apolipoprotein A2 [6]. Results suggest that FR10 protein is more structurally similar to apolipoproteins E and A, compared to type 2 and type 3 AFPs (Fig. 5). When compared to FR10, the RMSD was found to 0.45 Å for ApoE3 and ApoE2, 0.31 Å for the type IV AFP LS-12 and 0.27 Å for an unknown Xenopus leavis protein. These results are especially interesting given the low degree of primary protein sequence conservation of these proteins with FR10.
Structural homology of FR10 to apolipoproteins and anti-freeze proteins. Structural homology between FR10, apolipoproteins, anti-freeze proteins, and select proteins was determined by the root-mean-squared deviation (RMSD) between overlaid structures. The RMSD is the measure of the average distance between the atoms of the aligned structures. Analysis was carried out using MOE
Given the high degree of structural homology of FR10 to the type IV AFP LS-12, we used a computation approach to predict whether FR10 is a candidate AFP. Importantly, AFP-Pred first builds a positive prediction model using 119 distinct structural features from known AFPs proteins and a negative model from all other non-AFPs. The analysis decides what features are significantly present in AFPs and uses them to predict new ice binding proteins. Our analysis used 181 AFPs and 9193 non-AFPs to build the models and has been previously shown to have ~85 % accuracy in predictions. Results of AFP-Pred indicate that FR10 is a candidate type IV AFP.
Discussion
To survive the winter in a terrestrial habitat, the wood frog has developed a variety of behavioral and biochemical adaptations that allow the wood frog to freeze and thaw with minimal consequence. One such biochemical adaption is the ability to upregulate certain genes in response to freezing. The use of cDNA libraries prepared from the livers of frozen wood frogs revealed the novel clone fr10 was an example of such a gene. As a result, extensive research has been performed on this gene, both in the past [3] and the present study. Both studies have concluded that fr10 is expressed universally in all tissues tested, and that most often the gene is upregulated in response to freezing. Figure 2 shows that of the 7 tissues examined, only kidney tissue failed to increase their transcription after 24 h of freezing. These results are similar to those found by Cai and Storey, with the exception of skeletal muscle which showed no change in their study [3].
To obtain additional information about the pattern of fr10 transcription, mRNA levels were observed in wood frogs that had been under anoxic stress. The freezing of the lungs, blood, and heart during winter hibernation prevents the delivery of oxygen to the tissues, resulting in a state of anoxic stress in the wood frog. When fr10 mRNA levels were observed in tissues of wood frogs that had undergone 24 h of anoxia, it was found that all organs experienced either no change or a significant increase in their fr10 transcript levels when compared to controls (Fig. 2). Interestingly, the three organs that showed a significant increase (kidney, lung, and heart tissues) are arguably those organs most significantly affected by anoxic conditions. Comparatively, previous studies have documented that the initial onset of anoxia results in an increase in ventilation frequency, heart rate, pulmonary blood flow, and systemic blood flow in the red-eared slider turtle (Trachemys scripta elegans) [11]. Like the wood frog, the red-eared slider turtle can tolerate anoxia during the winter at low temperatures. In the turtle, the initial physiological responses to lack of oxygen are intended to increase oxygen consumption and delivery to tissues, and they last up to 1 h [11]. After this point, the turtle switches to long-term solutions for the anoxic conditions it faces by reducing its heart rate by 80 % of control values, from 5 min−1 to <1 min−1 [11, 12]. Additionally, during prolonged anoxia, the blood of the turtle will progressively become hyperkalemic and acidic as blood lactate levels are greatly elevated during anoxia [13–15]. It is possible that similar conditions exist in the anoxic frog, where the combination of acidosis, hyperkalemia, and oxygen deprivation may negatively affect the inotropic ability of the heart [16–25]. The kidney and lungs are also affected by these changes in the blood, as they are responsible for the maintenance of acid base balance. Specifically, the lung and kidney work in unison to maintain acid base homeostasis, with the lungs regulating bicarbonate (HCO3 −) concentration and the kidneys reabsorbing bicarbonate from the urine and excreting hydrogen ions into urine. In addition, the kidney regulates the changing blood pressure seen during anoxia by maintaining ion and water balance and it produces hormones, such as erythropoietin, which is known to be released in response to anoxia in the turtle [26]. Interestingly, fr10 did not experience increased transcription in the brain, a tissue that is known to be sensitive to low oxygen conditions. It is possible that fr10 is not necessary in this organ, as other genes/proteins provide protection from anoxia. For example, phosphoglycerate kinase 1 experiences an increase in both transcription and translation in the brain in response to anoxia, with an 11-fold increase in PGK1 protein. Transcript levels also increased in the liver upon short anoxia exposure, which is another tissue that did not show an anoxia dependent increase in fr10 transcription [27]. It is believed that elevated amounts of PGK1 would enhance the glycolytic capacity of the brain and liver, and perhaps this would render fr10 non-essential. Based on lung, heart, and kidney tissues that did experience on increased transcription of fr10, it is hypothesized that fr10 may play a role in the protection of those organs most affected by anoxic conditions. The mechanism of this protection, however, is not yet fully known.
The formation of extracellular ice crystals in the frog during winter hibernation causes water to leave the cells, dramatically decreasing their overall volume and making dehydration a component stress of freezing. Figure 3 shows that dehydration results in a very organ-specific response of fr10 expression, with brain and heart significantly increasing its expression compared to control. This pattern of expression allows for several interpretations of what the role of fr10 might be in dehydration. For example, dehydration tolerant R. sylvatica have been shown to defend water content of the core organs during dehydration of the whole organism [28]. The results of this previous study indicated that there is no significant difference in water content for liver, kidney, and heart at 50 % dehydration [28]. Given that at 40 % dehydration in heart tissue displayed a significant increase in fr10 mRNA, while kidney and liver saw no change compared to control, it is conceivable that fr10 acts to defend water content of these organs. Supporting this speculation, in organs where water loss may not be as critical, such as skeletal muscle, fr10 levels decreases allowing free-water release into extracellular spaces. Meanwhile organs with critical survival functions, such as heart, could experience an increase in fr10, which helps to strongly defend against water loss. Those core organs with important roles in dehydration, such as kidney which is responsible for ion balance, and liver which releases glucose in response to dehydration, show no change in fr10 mRNA levels. Finally, fr10 levels seem to reflect the workload on that organ for the duration of the dehydration stress. For example, both muscle and lung have decreased fr10 mRNA after 40 % dehydration and both experienced a decreased workload during dehydration. While overwintering, the skeletal muscle of the frog is primarily inactive as the frog assumes a posture that reduces exposed surface area and therefore evaporative loss [29], while oxygen consumption decreases by about 25 % as animals dehydrates at low temperature [30]. Meanwhile the heart, brain, kidney, and liver all arguably experience an increased work load as the heart must pump harder against viscous blood, the brain must coordinate dehydration behavior, the kidney must maintain ion balance, and the liver must produce glucose. Correspondingly, fr10 levels either maintain what was experienced during control conditions (liver and kidney) or significantly increase (brain and heart). It should be noted that the proposed function of fr10 is purely speculative at this time and require future experiments to confirm. However, it appears that within dehydration stress fr10 is involved in the protection of organs most important to the wood frog, namely the heart and brain tissues.
To further develop the potential role of fr10, expression was observed throughout the metamorphosis and development of the wood frog, from Gosner stages 14–20 to stages 44–45 [9]. As seen in Fig. 4, fr10 expression appears to be Gosner stage specific, with no clear overall trend emerging. The majority of developmental time points expressed fr10 at a transcription level of approximately 1.4 (where fr10 expression at Gosner stages 14–20 was assigned a transcription level of 1), with the exception of a significant decrease in Gosner stages 31–35 and 42–43. Interestingly, both of these stages are involved with the development of appendages, as stages 31–35 are marked by the development of a “paddle” foot (i.e., no digits), and stages 42–43 are distinguished by the forelimbs appearing. Additionally, a recent study has found that water restriction (volume) in early Gosner stage wood frog tadpoles (earlier than stage 38) reduced tadpole size and increased glucocorticoid levels during critical developmental periods [31]. As both in nature and in the lab environment these tadpoles would have never experienced freezing temperatures, fr10 must play another role in addition to that of a cryoprotectant. It is interesting to speculate that FR10 expression may help defend water content in developing tadpoles in this manner. It is possible that this protein has a generic water-regulatory role depending on the need of the wood frog or the life stage of the frog.
The overall characteristics of its wide spread distribution and its presence during freezing, argues for a critical function of fr10 in the cryoprotection of organs. A study done by Biggar et al. confirmed the cryoprotective action of FR10 by transfecting freeze-intolerant silkworm cell line with a FR10 fusion protein [6]. Those cells overexpressing transfected FR10 showed a 1.7 ± 0.2 fold greater survival after freezing at −6 °C for 1 h, and recovering for 24 h. This survival advantage persisted in the 2 h freezing (and 24 h recovery) stress group as well. As for the role of FR10, Biggar et al. also predicted the protein to have an extracellular action due to the high abundance of FR10 protein released into the extracellular medium [6]. This was further supported by computer modeling which predicted the structure of FR10 to have a highly hydrophobic core (Fig. 6), which would perhaps allow the protein to translocate to the extracellular environment, as well as the presence of a secretion signal present in the FR10 amino acid sequence, which predicted an extracellular localization. Finally, similar to the present study, FR10 was found to have a similar structure to the longhorn sculpin antifreeze protein 12 (LS-12) type IV AFP, despite having virtually no amino acid similarity to each other (Fig. 5) [6]. It has been previously suggested that both FR10 and LS-12 proteins may have evolved independently from pre-existing parental apolipoprotein genes [6]. Structural homology results from this study seem to support this hypothesis, with both FR10 and LS-12 showing very high structural similarity to Apolipoprotein types E2 and E3 (Fig. 5).
Predicted ab initio structure of novel freeze responsive protein, FR10. Structures were predicted using Quark and optimized with MOE as previously described [6]
In addition to the high degree of structural homology between FR10 and LS-12, this study also successfully predicted the FR10 as a candidate type IV AFP with the use of AFP-Pred software [10]. Although the presence of a type IV AFP in a freeze-tolerant frog might seem contradictory, numerous recent studies have suggested that some type IV AFPs may not have biologically relevant roles in thermal hysteresis [32]. Interestingly, LS-12 only exhibits AFP ability in a concentration dependant manner. However, at protein concentrations naturally found in the plasma of longhorn sculpin fish, the anti-freeze activity of LS-12 ranges from low to undetectable [32]. It is possible that both FR10 and LS-12 function in a manner related to anti-freeze, and could provide a role of FR10 during anoxic and dehydration stresses. In the beginning, thermal hysteresis in insects was first discovered in association with the Malpighian tubules and postulated to be a way of helping to recover water from feces, so that insects did not dehydrate as a consequence [33]. Although the function is not clear at this time, it is possible that FR10 might have had its origins as a peptide that aided water conservation and could be acting to help protect the wood frog from dehydration-related injury during freezing.
The overall function of fr10 within the wood frog still remains unknown, however, by examining its pattern of expression several characteristics have become evident. Expression of fr10 is universal in wood frog organs and the majority of organs tested experienced early and sustained transcription over the course of freezing. Further investigation of expression revealed that fr10 is transcribed under anoxic and dehydration conditions, both of which are component stresses of freezing. Finally, fr10 appears to be important throughout development of the wood frog from embryo to fully developed frog, and therefore fr10 must have a freeze-independent role in cryoprotection. In the future, it is possible that FR10 will be an important tool to improve stress tolerance in human cells and organs. The most significant application of this would be to improve tissue survival for the purpose hypothermic or cryo-preservation of tissues and organs for transplantation.
References
Wilbur HM (1977) Interactions of food level and population density in Rana sylvatica. Ecology 58(1):206–209
Storey KB, Storey J (1996) Natural freezing survival in animals. Ann Rev Ecol Evol Syst 27:365–386
Cai Q, Storey K (1997) Upregulation of a novel gene by freezing exposure in the freeze-tolerant wood frog (Rana sylvatica). Gene 198:305–312
McNally J, Sturgeon C, Storey K (2003) Freeze-induced expression of a novel gene, fr47, in the liver of the freeze-tolerant wood frog, Rana sylvatica. Biochim Biophys Acta 1625:183–191
McNally J, Wu S, Sturgeon C, Storey K (2002) Identification and characterization of a novel freeze-inducible gene, li16, in the wood frog Rana sylvatica. FASEB J 16:902–904
Biggar KK, Kotani E, Furusawa T, Storey K (2013) Expression of freeze-responsive proteins, FR10 and Li16, from freeze-tolerant frogs enhances freezing survival of BmN insect cells. FASEB J 27(8):3376–3383
Storey JM, Storey KB (1985) Triggering of cryoprotectant synthesis by the initiation of ice nucleation in the freeze tolerant frog, Rana sylvatica. J Comp Physiol B 156:191–195
Churchill TA, Storey KB (1994) Effects of dehydration on organ metabolism in the frog Pseudacris crucifer: hyperglycaemic responses to dehydration mimic freezing induced cryoprotectants production. J Comp Physiol B 164:492–498
Gosner K (1960) A simplified table for staging anuran embryos and larvae with notes on identification. Herpetologica 16(3):183–190
Kandaswamy KK, Chou KC, Martinetz T, Moller S, Suganthan PN, Sridharan S, Pugalenthi G (2011) AFP-Pred: a random forest approach for predicting antifreeze proteins from sequence-derived properties. J Theor Biol 270:56–62
Hicks JW, Wang T (1998) Cardiovascular regulation during anoxia in the turtle: an in vivo study. Physiol Zool 71(1):1–14
Stecyk JA, Farrell AP (2007) Effects of extracellular changes on spontaneous heart rate of normoxia and anoxia acclimated turtles (Trachemys scripta). J Exp Biol 210:421–431
Ultsch GR, Jackson DC (1982) Long-term submergence at 3°C of the turtle Chrysemys picta bellii, in normoxic and severely hypoxic water. I. Survival, gas exchange and acid–base status. J Exp Biol 96:11–28
Jackson DC, Ultsch GR (1982) Long-term submergence at 3°C of the turtle Chrysemys picta bellii, in normoxic and severely hypoxic water: II. Extracellular ionic responses to extreme lactic acidosis. J Exp Biol 96:29–43
Herbert CV, Jackson DC (1985) Temperature effects on the responses to prolonged submergence in the turtle Chrysemys picta bellii: I. Blood acid–base and ionic changes during and following anoxic submergence. Physiol Zool 58:665–669
Yee HP, Jackson DC (1984) The effects of different types of acidosis and extracellular calcium on the mechanical activity of turtle atria. J Comp Physiol B 154:385–391
Wasser JS, Freund EV, Gonzalez LA, Jackson DC (1990) Force and acid–base state of turtle cardiac tissue exposed to combined anoxia and acidosis. Am J Physiol 259:R15–R20
Wasser JS, Inman KC, Arendt EA, Lawler RG, Jackson DC (1990) 31P-NMR measurements of pHi and high-energy phosphates in isolated turtle hearts during anoxia and acidosis. Am J Physiol 259:R521–R530
Farrell AP, Franklin CE, Arthur PG, Thorarensen H, Cousins KL (1994) Mechanical performance of an in situ perfused heart from the turtle Chrysemys scripta during normoxia and anoxia at 5 and 15°C. J Exp Biol 191:207–229
Jackson DC, Shi H, Singer JH, Hamm PH, Lawler RG (1995) Effects of input pressure on in vitro turtle heart during anoxia and acidosis: a 31P-NMR study. Am J Physiol 268:R683–R689
Shi H, Hamm PH, Lawler RG, Jackson DC (1999) Different effects of simple anoxic lactic acidosis and simulated in vivo anoxic acidosis on turtle heart. Comp Biochem Physiol 122A:173–180
Shi H, Jackson DC (1997) Effects of anoxia, acidosis and temperature on the contractile properties of turtle cardiac muscle strips. J Exp Biol 200:1965–1973
Kalinin A, Gesser H (2002) Oxygen consumption and force development in turtle and trout cardiac muscle during acidosis and high extracellular potassium. J Comp Physiol B 172:145–151
Overgaard J, Gesser H (2004) Force development, energy state and ATP production of cardiac muscle form turtles and trout during normoxia and severe hypoxia. J Exp Zool 207:1915–1924
Overgaard J, Wang T, Nielsen OB, Gesser H (2005) Extracellular determinants of cardiac contractility in the cold anoxic turtle. Physiol Biochem Zool 78:976–995
Milton SL, Nayak G, Lutz PL, Prentice HM (2006) Gene transcription of neuroglobin is upregulated by hypoxia and anoxia in the brain of the anoxia-tolerant turtle Trachemys scripta. J Biomed Sci 13:509–514
Wu S, Storey JM, Storey KB (2009) Phosphoglycerate kinase 1 expression responds to freezing, anoxia, and dehydration stresses in the freeze tolerant wood frog, Rana sylvatica. J Exp Zool 311A:57–67
Churchill TA, Storey KB (1993) Dehydration tolerance in wood frogs: a new perspective on development of amphibian freeze tolerance. Am Physiol Soc 265:R1324–R1332
Heatwole H, Torres F, Blasini de Austin S, Heatwole A (1969) Studies on anuran water balance—Dynamics of evaporative water loss by the Coquí, Eleutherodactylus portoricensis. Comp Biochem Physiol 28:245–269
Wu S, De Croos JNA, Storey KB (2008) Cold acclimation-induced up-regulation of the ribosomal protein L7 gene in the freeze tolerant wood frog, Rana sylvatica. Gene 424:48–55
Crespi EJ, Warne RW (2013) Environmental conditions experienced during the tadpole stage alter post-metamorphic glucocorticoid response to stress in an amphibian. Integr Comp Biol 53(6):989–1001
Gauthier SY, Scotter AJ, Lin FH, Baardsnes J, Fletcher GL, Davies PL (2008) A re-evaluation of the role of type IV antifreeze protein. Cryobiology 57:292–296
Ramsay JA (1964) The rectal complex of the Mealworm Tenebrio molitor L. (Coleoptera tenebrionidae). Phil Trans R Soc Lond B 248(748):279–314
Acknowledgments
Thanks to J. M. Storey for editorial review of the manuscript. Research was supported by a discovery grant from the Natural Sciences and Engineering Research Council (NSERC) of Canada to KBS and the Canada Research Chairs program. Both KS and KKB held a NSERC CGS scholarship.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sullivan, K.J., Biggar, K.K. & Storey, K.B. Transcript expression of the freeze responsive gene fr10 in Rana sylvatica during freezing, anoxia, dehydration, and development. Mol Cell Biochem 399, 17–25 (2015). https://doi.org/10.1007/s11010-014-2226-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-014-2226-9