Abstract
Background
The emergence of multidrug-resistant pathogens through excessive and indiscriminate use of antibiotics, together with the lack of highly efficient treatment options for bacterial infections, has raised the development of novel antimicrobial agents to top priority. In this context, cryptic host defense peptides (cHDPs) are being explored as a novel class of antimicrobial agents. In this study, short peptides were designed from the long nonantibacterial protein ToAP2 and analysed for their positive net charge, hydrophobicity, hydrophobic moment, hydrophobic and hydrophilic planes.
Methods
From the designed fragments, five 15 amino acid fragments were synthesised by solid-phase peptide synthesis (SPPS) and analysed for antimicrobial activity against ESKAPE pathogens. All the peptides were subject to cytotoxicity, mode of action, structure and function studies to find the best template for further optimisation.
Results
Among them, two peptides, FKL15 and SKL15, showed better efficiency in killing P. aeruginosa under physiological salt and plasma conditions with no cytotoxicity issues. Further, the peptides destroyed the bacterial membranes and adopted a random coil structure in the presence of the bacteria.
Conclusions
The data indicates that FKL15 and SKL15 are promising antimicrobial peptides against antibiotic-resistant bacteria with great potential to develop as drugs with high economic value.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Antimicrobial agents have made a significant impact on human life, and their success is being threatened due to the emergence of antimicrobial resistance (AMR) in pathogens. Among all, AMR in the ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp) is of great concern for clinicians around the world, as they are part of the normal microflora and opportunistic pathogens (e.g., illness, surgery, transplantation, cancer) (Chang et al. 2022; Serapide et al. 2022). Given the high incidence of AMR in the ESKAPE pathogens around the globe, the world Health Organisation has declared them as critical priority pathogens against whom a new class of antibacterials are needed. The current therapeutic regime, to treat ESKAPE pathogen-associated infections involves the use of antibiotic combinations at high concentrations (Das et al. 2022; Oliveira et al. 2020). However, this excessive or improper use of antibiotics has further led to the development of multidrug-resistant pathogens.
Currently, there is renewed interest in discovering antimicrobial agents, from natural sources, as these molecules are designed and optimised by natural selection (Li et al. 2022a; Rocha and Ferreira 2022; Sarkar et al. 2021). Among the vast repertoire of natural products, toxins and venoms represent a unique group consisting of a variety of bioactive substances ranging from peptides, lipids, enzymes, amines, etc. (Bosso et al. 2020; Luo et al. 2022; Roque-Borda et al. 2022).
Recently, some researchers have started to explore the unique proteins and peptides in the toxins, which have the potential to inhibit and control the menace created by antimicrobial resistance pathogens (Sup Table 1). Interestingly, Scorpion venom’s major bioactive protein components share physicochemical properties, such as cationic nature and structural flexibility, with HDPs (Amorim-Carmo et al. 2022). HDPs are short (~ 50 amino acids), amphipathic, hydrophobic, evolutionary conserved molecules found in all living organisms with a net cationic charge of + 2 to + 9. HDPs are expressed either constitutively or during the inflammatory response to infection in the host. HDPs act by permeabilizing bacterial membranes and simultaneously blocking the over-activation of inflammatory molecules by binding to factors like LPS, and exotoxins (Greve and Cowan 2022; Valenti et al. 2022).
Given this, we decided to further explore the short sequence variant of peptide ToAP2 (FFGTLFKLGSKLIPGVMKLFSKKKER) obtained from the venomous glands of scorpion Tityus obscurus. ToAP2 is a 26 amino acid long peptide, having a + 6 net positive charge, 42% hydrophobicity, and a propensity to form an α-helical structure. ToAP2 and its sequence similar Css54 (FFGSLLSLGSKLLPSVFKLFQRKKE) demonstrated antifungal activity (Guilhelmelli et al. 2016; Park et al. 2020, 2024).
Interestingly, we and others have reported that proteolytic cleavage of the homeostasis-related protein leads to the generation of cryptic host defense peptides (De Oliveira Costa and Franco 2020; Horam et al. 2019; Lata et al. 2023; Papareddy et al. 2010; Pizzo et al. 2018; Raj et al. 2021). Strangely, cryptic HDPs also demonstrate functions similar to those of the natural ones like antimicrobial, antiviral, immunomodulatory etc. (De Oliveira Costa and Franco 2020; Pizzo et al. 2018). Given their bioactive nature, flexibility for modification with natural and unnatural amino acids, cryptic HDPs are regarded as one of the most promising alternative sources for novel agents.
In this work, we have tried to develop a novel 15 amino acids long cryptic peptide from the scorpion Tityus obscurus toxin for potential broad-spectrum antimicrobial activity against the ESKAPE pathogens and assessed the shorter peptides antibacterial activity, toxicity, proteolytic stability, and mechanism of action against 4 bacterial species of the ESKAPE group. Among them, FKL15 and SKL15, shorter than parent ToAP2, exhibited antibacterial activity against Gram-negative bacteria P. aeruginosa without any significant cytotoxicity towards Macrophages (THP-1) and Microglia (BV-2) cells. We here go on to demonstrate the antibacterial potential of the short variant of scorpion venom peptide, ToPA2 for the first time.
Methodology
Materials
All the peptides and LL-37 (LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES) used in this work were synthesized by Fmoc solid-phase synthesis protocol and obtained from Genscript, USA. The purity (> 95%) of the peptides was confirmed by mass spectral analysis (MALDI-TOF) before diluting in H2O (5 mM stock). This stock solution was stored at − 20° C, until use. The microbes used in the present study were K. pneumoniae ATCC 27,736, A. baumannii ATCC 1605, P. aeruginosa ATCC 25,668 and S. aureus ATCC 29,213 were obtained from ATCC, USA.
In Silico Analysis of Peptides
The peptide sequence of ToAP2, a scorpion (Tityus obscurus) toxin was obtained from the Antimicrobial Peptide Database (APD, https://aps.unmc.edu/). The amino acid framework of this peptide was then used to generate 15 amino acid long cryptic analogues, which correspond to the average AMP length. For each designed fragment the positive net charge, hydrophobicity, and hydrophobic moment were calculated using APD’s prediction tools. Further, the distribution pattern of the amino acids in hydrophobic and hydrophilic planes was deduced by preparing helical wheel projections of peptides using HeliQuest (https://heliquest.ipmc.cnrs.fr/cgi-bin/ComputParams.py) and their secondary structures was modelled using the PEP-FOLD3 server (https://bioserv.rpbs.univ-paris-diderot.fr/services/PEP-FOLD3/).
Radial Diffusion Assay (RDA)
RDA assay was carried out essentially as described elsewhere (Horam et al. 2019; Setty et al. 2017). Briefly, test bacteria were grown to the mid-logarithmic phase in 10 mL of 3% w/v of tryptic soy broth (TSB). Bacteria were centrifuged at 10,000 RPM for 10 min, the supernatant was discarded and the pellet was resuspended in 10 mM Tris-HCl pH 7.4 to get 1-2 × 109 CFU/mL of the bacterial suspension. After washing, bacteria were added to 15 mL of underlay agarose gel (0.03% (w/v) TSB, 1% (w/v) low electroendosmosis (EEO) type agarose and 0.02% (v/v) Tween 20) such that the final concentration of the bacteria reaches 4 × 106 CFU/mL and poured into a 140 mm Petri plate. After solidification, 4 mm diameter holes were punched, followed by the addition of 6 µL peptide solution of different concentrations (25, 50, and 100 µM). Petri plates were incubated for 3 h to allow diffusion of peptides into the gel, followed by pouring of 15 ml of overlay gel. Further petri plate with bacteri and peptide incubation was done for 16–24 h at 37 °C. The antimicrobial activity of peptide was visualized as a zone of inhibition around each well and was measured in mm.
Minimum Inhibitory Concentration (MIC)
Peptides were screened for their in vitro antibacterial activity against reported Gram-positive and Gram-negative pathogenic bacterial strains by broth micro-dilution method as per CLSI guidelines (formerly known as NCLS) as reported elsewhere (Wiegand et al. 2008). Bacteria were grown overnight in 3% TSB to mid-logarithmic phase, centrifuged and washed two times in 10 mM Tris buffer, and re-suspended in 2X of cation adjusted MHB broth (Becton Dickinson, USA) to get 1 × 105 CFU/mL. Serially diluted peptide concentration (160 µM to 0.31 µM) was added to each corresponding well of a 96-well microtiter plate (polypropylene, Costar Corp.) followed by the addition of 50 µL of 2X of cation-adjusted MHB broth containing 1 × 105 CFU/mL bacteria and incubated at 37° C for 16 h. MIC values were determined by mere visual inspection of plates and that value was considered where growth inhibition observed was greater than 90%. MIC determination and the assay were performed in the triplicate. In each assay, a negative control lacking peptide addition is included, alongside LL-37 peptide serving as the positive control.
Viable Count Assay (VCA)
VCA was done as described previously (Horam et al. 2019; Lata et al. 2023). Bacteria of interest were grown in 10 mL of TSB and incubated at 37 °C until the mid-logarithmic phase. After washing with 10mM Tris HCl buffer, the bacteria pellet was re-suspend in 10mM Tris HCl buffer containing 10 mM glucose to get 1 × 109 CFU/mL bacterial suspension. The culture was further diluted to 1000 X in 10mM Tris HCl containing 10 mM glucose, and incubated in the presence of peptides at the indicated concentration for 2 h at 37 °C. After treatment with the peptides, the samples were serially diluted and plated on Todd Hewitt (TH) medium and incubated for 16–18 h at 37 °C. To assess peptide activity under physiological conditions, 1 × 109 CFU/mL bacterial suspension was added to 10 mM Tris HCl + 10 mM glucose + 0.15 M sodium chloride or 10 mM Tris HCl + 10 mM glucose + 20% citrate human plasma. Samples without peptide treatment were considered to represent 100% survival under same test conditions. Mean ± SD values from three biological replicates were used for drawing conclusions.
Hemolysis Assay
For the hemolysis assay, blood was collected from a human volunteer in a vacutainer containing anticoagulant EDTA (BD Vacutainer® EDTA Tubes, 367864) and centrifuged at 800 g for 10 min (Raj et al. 2021). The plasma and buffy coat were removed and erythrocytes were then washed three times in PBS, pH 7.4 and re-suspended in the same (5% suspension) after which they were incubated with end-over-end rotation for 1 h at 37° C in the presence of peptides (50 and 100 µM). The samples were then centrifuged at 800 g for 10 min. The hemoglobin released in the supernatant was measured at 540 nm by transferring 100 µL of supernatant in 96 well plate and expressed as % of Triton X-100 induced hemolysis. 2% Triton X-100 (Sigma-Aldrich, St. Louis, USA) was taken as the positive control.
The use of human blood was approved by the CSIR-CDRI human ethical Committee (CDRI/IEC/2019/A1).
Hemolysis (%) was calculated according to the following equation:
Protease Sensitivity Assay
To investigate the stability of peptides against the protease of both bacterial as well as the human origin, Protease stability studies were done as described elsewhere (Setty et al. 2017). Briefly, test peptides (1 µg) were incubated at 37 °C with V8 protease (0.1 µg 25,000 units/mg), Proteinase K and Human neutrophil elastase (0.4 µg, 29 units/mg), in a total volume of 30 µl for 3 h. After the incubation, the reaction was terminated by heating the sample in 4X lammelie buffer at 100 °C for 5 min, and the degradation of the peptide was analyzed on 16.5% precast SDS-PAGE Tris-Tricine gels after staining with Coomassie Blue R-250, followed by destaining.
MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide) Assay
To assess the effect of peptides on the cells, MTT assay was performed (Gondru et al. 2021; Raj et al. 2021). BV-2 and THP-1, 3000 cells/well were grown in presence of 10% FBS till the 80% confluence stage in 96 well plates, followed by peptide (50 and 100 μm) addition. After 24 h of incubation at 5% CO2 and 37 °C, 20 µL of filter-sterilized MTT (5 mg/mL in PBS) was added to each well and plates were further incubated for 4 h. The MTT-containing medium was removed by aspiration and blue formazan was dissolved in 100 µl of DMSO. Then plates were swirled gently for 10 min at room temperature to completely dissolve formazan followed by measuring the absorbance at 550 nm. The results given here represent Mean ± SD values from triplicate measurements.
Lactate Dehydrogenase (LDH) Assay
To complement the MTT results, an LDH assay was done (Horam et al. 2019; Lata et al. 2023), with minor modification, after the treatment of BV-2 and THP-1 cells with the desired peptide concentration (50 and 100 μm) for 24 h, the supernatant was collected by centrifugation at 2500 RPM and mixed with LDH-based Roche cytotoxicity kit reagents as per the manufacturer’s instructions. The LDH release from 2% Triton X-100 was considered as the positive control (100% LDH release). The results represented here is the mean + SD from three independent experiments.
Liposome Preparation and Leakage Assay
Large unilamellar vesicles (LUV’s) were prepared using the freeze-thaw method. Anionic lipids phosphatidylglycerol (PG), phosphatidylserine (PE) in 3:1 ratio or zwitterionic phosphatidylcholine (PC) (1:4). Subsequently, 10 mM Tris buffer, pH 8 was added together with calcein (8 mg) and left on a shaker overnight. After five freeze-thaw cycles with liquid nitrogen and heating up to 60° C in the water bath, lipid suspensions were extruded 15–20 times through 100 μm polycarbonate membranes mounted in a Lipofast mini extruder (Avestin, Ottawa, Canada) at 25 ° C. Untrapped calcein was removed by passing sample from Sephadex G-25 (Sigma) column three times at 25 ° C with 10 mM Tris buffer, pH 8 as eluent. Calcein released from the Liposomes treated with peptides was measured by monitoring the emitted fluorescence at 520 nm (excitation at 470 nm). An absolute leakage scale was obtained by disrupting liposomes at the end of the experiment through 0.1% Triton X-100. Measurements were performed in triplicate at 25 ° C by using a Tecan Infinite M Nano + spectrometer.
CD Spectroscopy
To find out the secondary structure of peptides, circular dichroism (CD) spectra of peptides were obtained on a Jasco J-1500 Spectropolarimeter as reported else where (Horam et al. 2019; Lata et al. 2023). All measurements were performed in triplicate in a quartz cuvette (1 mm path length) at 37° C with water, buffer and peptide. Liposomes at a concentration of 0.1 mM were monitored in the wavelength range of 190–250 nm. The background value, where no peptide signal is present was substracted, to account for instrumental differences between measurements.
Flow Cytometry
K. pneumoniae ATCC 27,736, A. baumannii ATCC 1605, P. aeruginosa ATCC 25,668 and S. aureus ATCC 29,213 were grown to midlogarithmic phase in 3% TSB medium, centrifuged, and washed with buffer (10 mM Tris buffer, pH 7.4) and resuspended in the same buffer to obtain 1 × 109 CFU/mL of the bacterial suspension. The bacterial suspension was incubated with the peptide (100 µM) at 37° C for 1 h. The bacteria were stained with 200 µl of propidium iodide (10 µM/mL) for 15 min at room temperature in dark under shaking conditions, followed by repeated washing with buffer. The pellet was resuspended in 500 µl of 10 mM Tris buffer, pH 7.4 + 10 mM glucose. For the data acquisition, FACS caliber flow cytometer was used with a gate setting for the intact bacterial cells population and data processing was done in the logarithmic mode as done earlier (Lata et al. 2023).
Scanning Electron Microscopy
Scanning electron microscopy was done to see the membrane damaging effects of the peptides on K. pneumoniae ATCC 27,736, A. baumannii ATCC 1605, P. aeruginosa ATCC 25,668 and S. aureus ATCC 29,213. The processing was done as stated elsewhere (Horam et al. 2019; Raj et al. 2021). Bacteria were grown, washed and treated with the peptide as stated above, After treatment with peptides, bacteria were washed with PBS three times before fixing them, with 2.5% (w/v) glutaraldehyde at 4° C overnight. After the fixing bacteria were washed twice with PBS to remove the fixative and dehydrated through a graded ethanol series (50%, 70%, 90%, and 100%) for 15 min each. The samples were dried using a critical point dryer. The dried bacterial specimens were sputtered with gold using a sputter coater, then visualized under a scanning electron microscope (FEI Quanta 250, Netherlands). The images were obtained at 20 kV using a SE detector.
Statistics
The mean and standard deviation values obtained from the triplicate biological repeat experiments are reported here. The significance was considered at P < 0.05 and the ANOVA or student t-Test was used as indicated in the figure legends. Statistical significance was done using the SigmaStat; SPSS Inc., Chicago, IL Version 14 software.
Results
Synthetic Peptide Design and Characterization
Shorter peptide fragments, with better or broad-spectrum activity than the parent molecule, is of great interest for developing novel and innovative bio-therapies (Ramesh et al. 2016). Usually, peptides of very large size and length are not preferred for development as potential pharmacological candidates due to their high susceptibility to proteolytic degradation, high cytotoxicity, low bioavailability, probable antigenicity, and high cost associated with their synthesis (Fosgerau and Hoffmann 2015; Zane et al. 2021). On the contrary, short peptides with reduced cytotoxicity and higher antimicrobial activity are preferred for designing variants with modifications in charge, hydrophobicity and helicity.
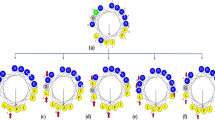
In this work, we aimed to develop 15 mer shorter variants from 26 amino acids long parent peptide that was previously isolated from the venomous gland of the scorpion (Tityus obscurus) (Guilhelmelli et al. 2016). It is demonstrated that continuous stretch of more than 3 hydrophobic amino acids leads to aggregation or gelling of peptides in the solution phase (Matsui et al. 2017; Mueller et al. 2020). Our previous studies have shown that peptides containing less than 12 amino acids demonstrate lower antimicrobial activity (Pasupuleti et al. 2009). To avoid both issues, we decided to keep the size of the peptides to 15 amino acids in length. By using the QSAR guided in silico analysis as reported previously (Lata et al. 2023; Raj et al. 2021), 5 different peptide sequences of length 15 amino acids were selected based on net positive charge (+ 4 to + 6), hydrophobicity (H), and hydrophobic movement (µH) and ease to form alpha-helical structure (Fig. 1A & Sup Table 2). Next, we investigated the evolutionary conservation of the hit molecules in the scorpion toxin and found that peptides FKL15, LGS15 & SKL15 sequence are highly conserved in the scorpion sp. It is interesting to observe that even though the species of scorpions are geographically separated (Sup Table 3), certain amino acids are highly conserved, e.g., the positively charged amino acids like lysine (Fig. 1B).
Helical Wheel Projection Of The Peptides. (a) In helical wheel projections hydrophobic residues are represented as yellow, negatively charged residues are red, positively charged residues are blue, and the particular polar residue is violet. A molecular model of the peptide is present in the center of the wheel. (b) Alignment of ToAP2 peptide sequence: conserved amino acids are represented with * and identical amino acids are represented with #
Our initial antimicrobial studies of the peptides against, S. aureus, K. pneumoniae, A. baumannii, and P. aeruginosa, showed that all the shortlisted peptides have antimicrobial activity (Fig. 2A, ). Among the peptides investigated, all of them exhibited broad-spectrum activity, in viable count assay at > 6 µM (Fig. 2B, upper panel). To understand further the antimicrobial activity, MIC assay was done as prescribed by the NCCLS guidelines, the peptides show activity at higher concentrations (Sup Table 2) indicating that salts might antagonize the antimicrobial activity.
Antibacterial activity of ToAP2 peptide analogs. (a) The activity of peptides as assessed by radial diffusion assay (RDA) against S. aureus ATCC 29,213, P. aeruginosa ATCC 25,668 A. baumannii ATCC 1605, and K. pneumoniae ATCC 27,736 (bottom to top). The clear zones corresponding to the inhibitory effect of each peptide were measured after incubation at 37 °C for 16–24 h. The color of the cell indicates the zone of inhibition (in mm) observed after the peptide treatment. The values (25, 50, 100 µM ) indicate the tested concentration. The values in the cell indicate the exact zone of inhibition observed. (b) For the viable count assay, bacteria were subjected to different concentrations of peptides as mentioned above and incubated for 2 h at 37 °C in 10 mM Tris buffer pH 7.4 alone, Tris buffer + 0.15 M NaCl and Tris buffer + 20% citrate human plasma. The peptide-treated samples were diluted and plated on MH agar plates and survival (CFU/mL) of the bacteria was counted
It is well reported in the literature that the HDPs antimicrobial activity is dependent on the microenvironment and tend to lose their activity partially or completely in the presence of high salt or plasma (Chen et al. 2021; Maharjan et al. 2022; Roshanak et al. 2021; Wang et al. 2021; Zhao et al. 2021). We, therefore, examined the influence of physiological conditions on the antimicrobial activity of the peptides. As shown in Fig. 2, peptides displayed 100% killing at 10 times higher concentrations (30 µM) in the presence of salt and plasma (Fig. 2B, middle and lower panel). As expected, we observed variability in the sensitivities towards the bacteria by different peptides and our results are consistent with the previous results obtained with other peptides in the presence of plasma or salt (Horam et al. 2019; Raj et al. 2021). Interestingly, LL-37 also showed the same phenomenon, i.e., an increase in the concentration. Nevertheless, more convincing data are emerging that show that the AMPs contribute to enhanced bacterial killing under in vivo conditions, likely through synergism or immune modifiers (Li et al. 2022b; Sandhu et al. 2022).
Cytotoxic Activities of Peptide
Armed with this information, cytotoxicity of the peptides against THP-1 (human macrophage cell line), human erythrocytes, and BV-2 (brain microglia) were performed at 50 and 100 µM. Results indicated that cells are healthy with a high percent (> 80%) of survival and no significant (< 10%) cytotoxicity was detected for our peptides in MTT assay (Fig. 3A, B and C). Based on the antimicrobial activity results in the presence of plasma, it can be argued that the observed lack of peptide cytotoxicity could be due to binding to the components or degradation by proteases present in the plasma. To shed light on this aspect, protease stability studies were performed using protease of bacterial origin with broad and narrow specificity. Results showed that the tested peptides were stable up to 3 h in the presence of the protease, except proteinase K which has a broad specificity (Fig. 3D). This contrasted with the benchmark peptide LL37 which was cleaved by all the bacterial proteases.
Cytotoxicity assay of peptides. (a) C57/BL6 murine-derived microglial cell, BV2 and human-derived monocyte, THP-1 were incubated with peptides at 50 and 100 µM and then LDH enzyme released was measured (b) hemolytic effects of the peptides on human erythrocytes was analysed at 50 and 100 µM. Hemoglobin released was measured at 540 nm and is expressed as % of Triton X-100 induced hemolysis. (c) On the contrary, the survival of BV2 and THP-1 cells in the presence of peptides was measured by using the live-cell ability to convert MTT to blue formazan as an indicator. All the values (mean + SD) represented here are from three biological repeats. (d) Protease sensitivity assay. Peptides (200 µg) were incubated with V8 protease, Proteinase K, and Human neutrophil elastase at 37 °C for 3 h and analyzed on 16.5% precast Tris-tricine SDS PAGE, faint or no staining indicates partial or complete cleavage
Mode of Action Studies
Scanning electron microscopy, flow cytometry and membrane permeabilization investigations were carried out to gain deeper insights into the mode of action. Our EM studies clearly showed that the tested peptides could damage the membrane and induce ghost-like structures on the membranes, with the release of intracellular contents (Fig. 4). Notably, shorter cryptic antimicrobial peptides from various proteins also showed ghost-like appearances on the membranes with extracellular extrusions. Although all the bacteria showed signs of membrane damage, the degree of the damage induced varied among the peptides probably due to architectural differences between the bacteria membrane components. e.g. capsule of K. pneumoniae, outer cell walls in A. baumannii. Never the less flow cytometry analysis on the test bacteria confirmed the EM results that membranes were damaged, thus accessible to the propidium iodide (Fig S1).
Scanning electron microscopy (SEM) images of bacteria treated with peptides. Peptides (100 µM) were treated with (a) K. pneumoniae ATCC 27,736, (b) A. baumannii ATCC 1605, (c) P. aeruginosa ATCC 25,668 and (d) S. aureus ATCC 29,213 and analyzed by scanning electron microscopy (FEI Quanta 250, Netherlands). Peptide-treated bacterial cells showed bursting and leakage of intracellular contents. LL37 was represented as positive (+) control and while cells untreated with peptide served as the negative control
Our VCA assay in the presence of the physiological conditions showed that the peptides partially inhibition antimicrobial activity at low concentration probably due to binding to salts or plasma (Fig. 2, lower row) (Maharjan et al. 2022; Roshanak et al. 2021). To rule out the role of plasma in the absence of cytotoxicity, liposome leakage assay was performed to demonstrate the ability of peptides to differentiate between the prokaryotic and eukaryotic membrane mimics. Results of calcein leakage from liposome mimicking the bacterial membrane, i.e., PE/PG demonstrated that at 1 µM concentration 80% of release occurred within 10 min of peptide addition (Fig. 5A). The sole purpose of using mimic membranes was to quantitatively monitor the membrane damage effects of the peptides. On contrary, except FKL15, all the other peptides failed to induce membrane damage in the liposome mimicking the eukaryotic membranes, i.e., PC (Fig. 5B). These results confirm our observation that the peptides are highly specific for bacterial membranes.
Liposome leakage assay and CD studies. (a, b) The membrane disruption effect of the peptides was measured by monitoring the emitted fluorescence of calcein released from anionic bacterial membrane (PE/PG) or zwitterionic eukaryotic membrane (PC). (c, d, e, f) For CD studies, CD spectra of peptides were obtained in Milli Q, 10 mM Tris buffer pH 7.4, PE/PG, and PC liposomes. Readings were taken in triplicate under stirring conditions at 100 µM at 37 °C. The background value (where peptide was not present) was subtracted to nullify the instrumental differences between measurements
We next investigated the peptide’s ability to form helical structures in the presence of membranes. As exemplified in Fig. 5D, E and F peptides showed the random structure in the presence of 10 mM Tris buffer, PE/PG, and PC except for LL-3, which adopts a perfect α-helical structure in all conditions. The CD-structures results indicate that peptides assume random structure in the presence of membranes. Based on our and other results, we are of the opinion that the peptides might have used electrostatic attraction and hydrophobicity for antimicrobial activity, rather than having a helical structure for the antimicrobial activity (Duan et al. 2021; Horam et al. 2019; Krishnamoorthy et al. 2023; Lata et al. 2023; Theansungnoen et al. 2022).
Discussion
The central conceptually novel finding in this study is the identification of short cryptic epitopes from the toxic protein with antimicrobial activity. ESKAPE pathogens, which account for 70% of bacterial infections in human, have an unprecedented ability to inactivate the small molecule based antimicrobial agents by hydrolysis or modification; alteration or bypassing of drug targets, permeation barriers, and removal of antibiotics out of the cell via efflux pump (Oliveira et al. 2020; Denissen et al. 2022). The discovery and development of a novel class, broad-spectrum antimicrobial agents that act on the evolutionary conserved, unique, and hard-to-mutate targets should be the utmost priority. In this regard, host defense peptides (HDPs) have captured the attention of researchers in recent years because of their efficiency as the first line of defense against multi-drug-resistant pathogens (Heselpoth et al. 2022; Pasupuleti et al. 2012; Sarkar et al. 2021).
In the present investigation, five new synthetic peptides were designed TFL15, FKL15, SKL15, LIP15, LGS15, and analyzed for their antimicrobial and hemolytic activities. The peptide sequences were deduced from the parent ToAP2. Even though the investigated peptides have different amino acid sequences, they share similar biophysical parameters which allow them to interact with the membranes (Sup Table 2). All the peptides were 15 amino acids long with a net positive charge in the range of + 4 to + 6 and rich in basic amino acids. The peptides showed activity at higher concentrations illustrating the fact that a blend of cationicity, amphipathicity, and hydrophobicity are essential for the activity, especially at physiological concentrations. Our data is consistent with the previous reports from our lab and others (Apostolopoulos et al. 2021; Horam et al. 2019; Ko et al. 2020; Raj et al. 2021).
It is well reported that metal cations like Na+, Mg2+, and Fe3+ decrease the antimicrobial activity of the HDPs by masking the electrostatic charges on the peptides as well as the membranes leading to lower peptide-membrane interactions (Ko et al. 2020). As reported, peptides showed varying degrees of antibacterial activity in physiological buffers and plasma conditions. We believe that the loss of activity at lower concentrations in the plasma could be due to the loss of initial electrostatic interactions or binding to the plasma components. Our data show that the inhibition of peptide’s antimicrobial activity in the plasma is more than in the presence of physiological salt alone. Studies by Su Jin Ko et al. with melectin peptide as a template showed that divalent cations inhibit the peptide more in comparison to mono or trivalent ions (Ko et al. 2020). At high cations concentrations, the membrane rigidity also increases due to the decrease in the electrostatic interactions between the negatively charged phospholipids. The lower antimicrobial activity in the plasma could be due to the high concentration of Na+ and Mg2+ as they have been shown to hamper the initial electrostatic interaction of the peptides and binding with membrane. Probably, peptides LIP15 and LGS15 activity are more affected by divalent cation than the FKL15 and SKL15, as those lost their activity in the salt and plasma significantly. On the contrary, FKL15 higher hydrophobicity might have helped in deeper penetration into membranes and subsequent cell damage (Edwards et al. 2016; Tan et al. 2021). Nevertheless, there is now enough evidence that demonstrates that the HDPs do have antibacterial activity under physiological conditions, and work in synergism with other natural HDPs present in the host as well as by their immunomodulatory activities (De San Nicolas et al. 2022; Ridyard et al. 2023; Sharma and Bisht 2023; Xia et al. 2023).
Our toxicity as well as the liposome leakage data demonstrate that the peptides are selective towards the prokaryotic membranes. Bacterial membranes are rich in acidic phospholipids like phosphatidylglycerol (PG), phosphatidylserine (PE) and cardiolipin, which are negatively charged surfaces. On the contrary, eukaryotic membranes are rich in mostly zwitterionic lipids (e.g., phosphatidylcholine), and sterols which are mostly neutral in charge. This electrostatic difference might play a role in the interaction of the peptide with the membranes (Miyazaki and Shinoda 2022; Utterstrom et al. 2022). Furthermore, it is very clear that in addition to binding to the membranes, the peptides have to undergo oligomerization and assembly on the surface before inducing the membrane rupture. Likely, all these mechanisms, together with the high electronegativity of P. aeruginosa LPS might explain the herein noted higher activity of the SKL15 and FKL15 peptides. Probably, the capsular nature of K. pneumoniae may explain why there is a loss of activity or higher MIC values. However, further in depth investigations are needed to clearly understand the lack of high activity.
Helical wheel projections of peptides showed both hydrophilic and hydrophobic faces and the in silico modeling indicated that the peptides assume a helical structure ( Fig. 1A). On the contrary, CD analysis of the peptides suggests that peptides assume random structure in the presence of membranes and buffer. Given this, we are opinion of that our lead peptides resemble other classical linear HDPs with regularly interspersed hydrophobic residues like those derived from GST, pneumolysin, and growth factors (Horam et al. 2019; Malmsten et al. 2007; Raj et al. 2021; Svensson et al. 2010). Our previous works have demonstrated that a combination of helicity and net charge is required for cytotoxicity (Pasupuleti et al. 2008). Thus it is likely that lack of helicity might contribute to a lack of cytotoxicity against BV-2, THP-1, and erythrocytes.
One of the drawbacks of this study is that we did not try to increase the peptide’s activity by increasing the charge or hydrophobicity or helicity other parameters which influence the activity. Even though, there are many reported methods of enhancing the activity in the literature (Han et al. 2021; Kang et al. 2022; Mwangi et al. 2023). For valid reasons, we refrained from proceeding in this line. For example, the replacement of each position in tripeptide would generate around 35 billion individual peptide sequences. Given the practical feasibility issues, it cannot be explored in this work. The other method is to develop a novel HDP sequence by de novo method, based upon our years of experience and literature reports. However, we believe that each peptide sequence is a unique and simple replacement of amino acids at any position will not work as positioning charged or hydrophobic amino acids at the strategic position is very crucial for activity. We in future studies wish to explore this option as carry out such kind of studies is beyond the scope of this paper.
It is now very clear that the HDPs are widely distributed in life forms, and they are not just antimicrobial molecules but have diverse functions such as cell signaling, wound healing, immunomodulatory, antiviral, angiogenesis, and controlling both the adaptive and innate immune systems. The results obtained in this study will have implications in our understanding of toxic proteins and multifunctionality of the HDPs. It will be logical to use naturally occurring peptides as a test sequence and apply the acquired rational drug design knowledge (structural prerequisites, helix stabilization, QSAR modelling, etc.) for further refinements to obtain a better bioactive and protease resistant molecule.
Conclusion
In light of this, our findings indicate that a strategy based on the selection of peptides from toxic proteins may prove to be a starting point for the generation of new variants with enhanced activities or discovering additional novel sequences as lead candidates for the development of AMPs. Further short antimicrobial peptide templates with extended broad-spectrum antimicrobial activities can be identified.
References
Amorim-Carmo B, Parente AMS, Souza ES, Silva-Junior AA, Araújo RM, Fernandes-Pedrosa MF (2022) Antimicrobial peptide analogs from scorpions: modifications and structure-activity. Front Mol Biosci 9:887763
Apostolopoulos V, Bojarska J, Chai TT, Elnagdy S, Kaczmarek K, Matsoukas J, New R, Parang K, Lopez OP, Parhiz H, Perera CO, Pickholz M, Remko M, Saviano M, Skwarczynski M, Tang Y, Wolf WM, Yoshiya T, Zabrocki J, Zielenkiewicz P, AlKhazindar M, Barriga V, Kelaidonis K, Sarasia EM, Toth I (2021) A Global Review on short peptides: frontiers and perspectives. Molecules 26(2):430
Bosso A, Di Maro A, Cafaro V, Di Donato A, Notomista E, Pizzo E (2020) Enzymes as a reservoir of host defence peptides. Curr Top Med Chem 20(14):1310–1323
Chang RYK, Nang SC, Chan HK, Li J (2022) Novel antimicrobial agents for combating antibiotic-resistant bacteria. Adv Drug Deliv Rev 187:114378
Chen SP, Chen EH, Yang SY, Kuo PS, Jan HM, Yang TC, Hsieh MY, Lee KT, Lin CH, Chen RP (2021) A systematic study of the stability, safety, and efficacy of the de novo designed antimicrobial peptide pepd2 and its modified derivatives against Acinetobacter baumannii. Front Microbiol 12:678330
Das S, Bombaywala S, Srivastava S, Kapley A, Dhodapkar R, Dafale NA (2022) Genome plasticity as a paradigm of antibiotic resistance spread in ESKAPE pathogens. Environ Sci Pollut Res Int 29(27):40507–40519
De Oliveira DMP, Forde BM, Kidd TJ, Harris PNA, Schembri MA, Beatson SA, Paterson DL, Walker MJ (2020) Antimicrobial resistance in ESKAPE pathogens. Clin Microbiol Rev 33(3):e00181–19
de Oliveira Costa B, Franco OL (2020) Cryptic host defense peptides: multifaceted activity and prospects for medicinal chemistry. Curr Top Med Chem 20(14):1274–1290
De San Nicolas N, Asokan A, Rosa RD, Voisin SN, Travers MA, Rocha G, Dantan L, Dorant Y, Mitta G, Petton B, Charriere GM, Escoubas JM, Boulo V, Pouzadoux J, Meudal H, Loth K, Aucagne V, Delmas AF, Bulet P, Montagnani C, Destoumieux-Garzon D (2022) Functional Diversification of Oyster Big Defensins Generates Antimicrobial Specificity and Synergy against Members of the Microbiota. Mar Drugs 20(12):745
Denissen J, Reyneke B, Waso-Reyneke M, Havenga B, Barnard T, Khan S, Khan W (2022) Prevalence of ESKAPE pathogens in the environment: antibiotic resistance status, community-acquired infection and risk to human health. Int J Hyg Environ Health 244:114006
Duan X, Zhang M, Chen F (2021) Prediction and analysis of antimicrobial peptides from rapeseed protein using in silico approach. J Food Biochem 45(4):e13598
Edwards IA, Elliott AG, Kavanagh AM, Zuegg J, Blaskovich MA, Cooper MA (2016) Contribution of amphipathicity and hydrophobicity to the antimicrobial activity and cytotoxicity of beta-hairpin peptides. ACS Infect Dis 2(6):442–450
Fosgerau K, Hoffmann T (2015) Peptide therapeutics: current status and future directions. Drug Discov Today 20(1):122–128
Gondru R, Kanugala S, Raj S, Ganesh Kumar C, Pasupuleti M, Banothu J, Bavantula R (2021) 1,2,3-triazole-thiazole hybrids: synthesis, in vitro antimicrobial activity and antibiofilm studies. Bioorg Med Chem Lett 33:127746
Greve JM, Cowan JA (2022) Tackling antimicrobial stewardship through synergy and antimicrobial peptides. RSC Med Chem 13(5):511–521
Guilhelmelli F, Vilela N, Smidt KS, de Oliveira MA, da Cunha Morales A Álvares, Rigonatto MC, da Silva Costa PH, Tavares AH, de Freitas SM, Nicola AM, Franco OL, Derengowski LD, Schwartz EF, Mortari MR, Bocca AL, Albuquerque P, Silva-Pereira I (2016) Activity of Scorpion Venom-derived antifungal peptides against Planktonic Cells of Candida spp. and Cryptococcus neoformans and Candida albicans Biofilms. Front Microbiol 7:1844
Han Y, Zhang M, Lai R, Zhang Z (2021) Chemical modifications to increase the therapeutic potential of antimicrobial peptides. Peptides 146:170666
Heselpoth RD, Euler CW, Fischetti VA (2022) PaP1, a broad-spectrum lysin-derived cationic peptide to treat polymicrobial skin infections. Front Microbiol 13:817228
Horam S, Raj S, Tripathi VC, Pant G, Kalyan M, Reddy TJ, Arockiaraj J, Pasupuleti M (2019) Xenobiotic binding domain of glutathione s-transferase has cryptic antimicrobial peptides. Int J Pept Res Ther 25(4):1477–1489
Kang SJ, Nam SH, Lee BJ (2022) Engineering approaches for the development of Antimicrobial peptide-based antibiotics. Antibiot (Basel) 11(10):1338
Ko SJ, Park E, Asandei A, Choi JY, Lee SC, Seo CH, Luchian T, Park Y (2020) Bee venom-derived antimicrobial peptide melectin has broad-spectrum potency, cell selectivity, and salt-resistant properties. Sci Rep 10(1):10145
Krishnamoorthy R, Adhikari P, Anaikutti P (2023) Design, synthesis, and characterization of non-hemolytic antimicrobial peptides related to human cathelicidin LL-37. RSC Adv 13(23):15594–15605
Lata M, Telang V, Gupta P, Pant G, Kalyan M, Arockiaraj J, Pasupuleti M (2023) Evolutionary and in silico guided development of novel peptide analogues for antibacterial activity against ESKAPE pathogens. Curr Res Microb Sci 4:100183
Li X, Zuo S, Wang B, Zhang K, Wang Y (2022a) Antimicrobial mechanisms and clinical application prospects of antimicrobial peptides. Molecules 27(9):2675
Li Z, Jing X, Yuan Y, Shui Y, Li S, Zhao Z, Deng B, Zhang W (2022b) In vitro and in vivo activity of phibilin against Candida albicans. Front Microbiol 13:862834
Luo L, Kamau PM, Lai R (2022) Bioactive peptides and proteins from Wasp venoms. Biomolecules 12(4):527
Maharjan R, Khan AI, Nadeem-Ul-Haque M, Maresca M, Choudhary MI, Shaheen F, Simjee SU (2022) Serum stable and low hemolytic temporin-sha peptide analogs disrupt cell membrane of methicillin-resistant Staphylococcus aureus (MRSA). Probiotics Antimicrob Proteins 14(2):391–405
Malmsten M, Davoudi M, Walse B, Rydengard V, Pasupuleti M, Morgelin M, Schmidtchen A (2007) Antimicrobial peptides derived from growth factors. Growth Factors 25(1):60–70
Matsui D, Nakano S, Dadashipour M, Asano Y (2017) Rational identification of aggregation hotspots based on secondary structure and amino acid hydrophobicity. Sci Rep 7(1):9558
Miyazaki Y, Shinoda W (2022) Cooperative antimicrobial action of melittin on lipid membranes: a coarse-grained molecular dynamics study. Biochim Biophys Acta Biomembr 1864(9):183955
Mueller LK, Baumruck AC, Zhdanova H, Tietze AA (2020) Challenges and perspectives in chemical synthesis of highly hydrophobic peptides. Front Bioeng Biotechnol 8:162
Mwangi J, Kamau PM, Thuku RC, Lai R (2023) Design methods for antimicrobial peptides with improved performance. Zool Res 44(6):1095–1114
Papareddy P, Rydengård V, Pasupuleti M, Walse B, Mörgelin M, Chalupka A, Malmsten M, Schmidtchen A (2010) Proteolysis of human thrombin generates novel host defense peptides. PLoS Pathog 6(4):e1000857
Park J, Oh JH, Kang HK, Choi MC, Seo CH, Park Y (2020) Scorpion-venom-derived antimicrobial peptide Css54 exerts potent antimicrobial activity by disrupting bacterial membrane of zoonotic Bacteria. Antibiot (Basel) 9(11):831
Park J, Kim H, Kang DD, Park Y (2024) Exploring the therapeutic potential of scorpion-derived Css54 peptide against Candida albicans. J Microbiol 62(2):101–112
Pasupuleti M, Walse B, Svensson B, Malmsten M, Schmidtchen A (2008) Rational design of antimicrobial C3a analogues with enhanced effects against Staphylococci using an integrated structure and function-based approach. Biochemistry 47(35):9057–9070
Pasupuleti M, Chalupka A, Morgelin M, Schmidtchen A, Malmsten M (2009) Tryptophan end-tagging of antimicrobial peptides for increased potency against Pseudomonas aeruginosa. Biochim Biophys Acta 1790(8):800–808
Pasupuleti M, Schmidtchen A, Malmsten M (2012) Antimicrobial peptides: key components of the innate immune system. Crit Rev Biotechnol 32(2):143–171
Pizzo E, Cafaro V, Di Donato A, Notomista E (2018) Cryptic antimicrobial peptides: identification methods and current knowledge of their immunomodulatory properties. Curr Pharm Des 24(10):1054–1066
Raj S, Venugopal U, Pant G, Kalyan M, Arockiaraj J, Krishnan MY, Pasupuleti M (2021) Anti-mycobacterial activity evaluation of designed peptides: cryptic and database filtering based approach. Arch Microbiol 203(8):4891–4899
Ramesh S, Govender T, Kruger HG, de la Torre BG, Albericio F (2016) Short AntiMicrobial peptides (SAMPs) as a class of extraordinary promising therapeutic agents. J Pept Sci 22(7):438–451
Ridyard KE, Elsawy M, Mattrasingh D, Klein D, Strehmel J, Beaulieu C, Wong A, Overhage J (2023) Synergy between human peptide LL-37 and Polymyxin B against Planktonic and Biofilm cells of Escherichia coli and Pseudomonas aeruginosa. Antibiot (Basel) 12(2):389
Rocha GA, Ferreira RB (2022) Antimicrobial polysaccharides obtained from natural sources. Future Microbiol 17:701–716
Roque-Borda CA, Gualque MWL, da Fonseca FH, Pavan FR, Santos-Filho NA (2022) Nanobiotechnology with therapeutically relevant macromolecules from animal venoms: venoms, toxins, and antimicrobial peptides. Pharmaceutics 14(5):891
Roshanak S, Shahidi F, Tabatabaei Yazdi F, Javadmanesh A, Movaffagh J (2021) Buforin I an alternative to conventional antibiotics: evaluation of the antimicrobial properties, stability, and safety. Microb Pathog 161(Pt B):105301
Sandhu AK, Yang Y, Li WW (2022) Vivo antibacterial efficacy of antimicrobial peptides modified metallic implants horizontal line systematic review and meta-analysis. ACS Biomater Sci Eng 8(5):1749–1762
Sarkar T, Chetia M, Chatterjee S (2021) Antimicrobial peptides and proteins: from nature’s reservoir to the laboratory and beyond. Front Chem 9:691532
Serapide F, Quirino A, Scaglione V, Morrone HL, Longhini F, Bruni A, Garofalo E, Matera G, Marascio N, Scarlata GGM, Cicino C, Russo A, Trecarichi EM, Torti C (2022) Is the pendulum of antimicrobial drug resistance swinging back after. COVID-19? Microorganisms 10(5):957
Setty SC, Horam S, Pasupuleti M, Haq W (2017) Modulating the antimicrobial activity of Temporin l through introduction of fluorinated phenylalanine. Int J Pept Res Ther 23(2):213–225
Sharma L, Bisht GS (2023) Synergistic effects of short peptides and antibiotics against bacterial and fungal strains. J Pept Sci 29(1):e3446
Svensson SL, Pasupuleti M, Walse B, Malmsten M, Morgelin M, Sjogren C, Olin AI, Collin M, Schmidtchen A, Palmer R, Egesten A (2010) Midkine and pleiotrophin have bactericidal properties: preserved antibacterial activity in a family of heparin-binding growth factors during evolution. J Biol Chem 285(21):16105–16115
Tan R, Wang M, Xu H, Qin L, Wang J, Cui P, Ru S (2021) Improving the activity of antimicrobial peptides against aquatic pathogen bacteria by amino acid substitutions and changing the ratio of hydrophobic residues. Front Microbiol 12:773076
Theansungnoen T, Phosri S, Bumrungthai S, Daduang J, Klaynongsruang S, Daduang S (2022) Novel non-cytotoxic antimicrobial peptides WSKK11 and WSRR11 with potent activity against Cutibacterium acnes. J Antimicrob Chemother 77(4):1012–1019
Utterstrom J, Barriga HMG, Holme MN, Selegard R, Stevens MM, Aili D (2022) Peptide-folding triggered phase separation and lipid membrane destabilization in cholesterol-rich lipid vesicles. Bioconjug Chem 33(4):736–746
Valenti GE, Alfei S, Caviglia D, Domenicotti C, Marengo B (2022) Antimicrobial peptides and Cationic nanoparticles: a broad-spectrum weapon to fight multi-drug resistance not only in bacteria. Int J Mol Sci 23(11):6108
Wang T, Zou C, Wen N, Liu X, Meng Z, Feng S, Zheng Z, Meng Q, Wang C (2021) The effect of structural modification of antimicrobial peptides on their antimicrobial activity, hemolytic activity, and plasma stability. J Pept Sci 27(5):e3306
Wiegand I, Hilpert K, Hancock RE (2008) Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3(2):163–175
Xia X, Song S, Zhang S, Wang W, Zhou J, Fan B, Li L, Dong H, Luo C, Li B, Zhang X (2023) The synergy of thanatin and cathelicidin-BF-15a3 combats Escherichia coli O157:H7. Int J Food Microbiol 386:110018
Zane D, Feldman PL, Sawyer T, Sobol Z, Hawes J (2021) Development and regulatory challenges for peptide therapeutics. Int J Toxicol 40(2):108–124
Zhao X, Zhang M, Muhammad I, Cui Q, Zhang H, Jia Y, Xu Q, Kong L, Ma H (2021) An antibacterial peptide with high resistance to trypsin obtained by substituting d-amino acids for trypsin cleavage sites. Antibiot (Basel) 10(12):1465
Acknowledgements
MP is grateful to the Ex-Director(S), CDRI, Lucknow, India, CSIR New Delhi [HCP0039, HCP0035, MLP0107], MOES (MoES-2/DS/6/2007 PC-IV) and DBT [BT/PR10484/MED/29/281/2013] for providing the financial assistance. The authors, ML are grateful to CSIR (31/004(1282)/2014-EMR-I) New Delhi, India. PG is grateful to CSIR (31/004(1365)/2019-EMR-I), New Delhi, India. VT is thankful to the DBT fellowship (DBT/2019/CDRI/1195). The authors thank Mr. Angney Lal and Mr. Atul Krishna for their technical assistance when and where needed during the study. CSIR-CDRI communication number: 123/2022/MP.
Author information
Authors and Affiliations
Contributions
ML, VT, PG and GP planned, carried out the experiments and analyzed the results. MP, MK and JA conceived planned, supervised and interpreted the results. ML devised the proof outline and wrote the manuscript. MP and JA contributed to the final version of the manuscript. All authors discussed the results and contributed to the final manuscript.
Corresponding authors
Ethics declarations
Ethical Approval
The study was approved by the Human ethics committee as per CSIR-CDRI human ethical permission (CDRI/IEC/2019/A1).
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lata, M., Telang, V., Gupta, P. et al. Synthetic Short Cryptic Antimicrobial Peptides as Templates for the Development of Novel Biotherapeutics Against WHO Priority Pathogen. Int J Pept Res Ther 30, 57 (2024). https://doi.org/10.1007/s10989-024-10632-8
Accepted:
Published:
DOI: https://doi.org/10.1007/s10989-024-10632-8