Abstract
Middle East respiratory syndrome coronavirus (MERS-CoV) has caused a high mortality rate since its emergence in 2012 in the Middle East. Currently, no effective drug or vaccine is available for MERS-CoV. Supportive care and prevention are the only ways to manage infection. In this study, we identified an epitope-based vaccine that could be an optimal solution for the prevention of MERS-CoV infection. By deploying an immunoinformatics approach, we predicted a subunit vaccine based on the surface glycoprotein (S protein) of MERS-CoV. For this purpose, the proteome of the MERS-CoV spike protein was obtained from the NCBI GenBank database. Then, it was subjected to a check for allergenicity using the Allergen FP v.1.0 tool. The Vaxijen v.2.0 tool was used to conduct antigenicity tests for binding with major histocompatibility complex class I and II molecules. The solidity of the predicted epitope-allele docked complex was evaluated by a molecular dynamics simulation. After docking a total of eight epitopes from the MERS-CoV S protein, further analyses predicted their non-toxicity and therapeutic immunogenic properties. These epitopes have potential utility as vaccine candidates against MERS-CoV, to be validated by wet-lab testing.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Middle East respiratory syndrome coronavirus (MERS-CoV) has been identified as a novel human coronavirus that poses a major threat to global public health, calling for the urgent development of effective and safe vaccines. MERS-CoV is correlated with an unusually high death rate of almost 35% (Alqahtani et al. 2018; Bermingham et al. 2012; Zaki et al. 2012). The first known infections of MERS-CoV were detected in Saudi Arabia in 2012, and the virus later spread to other countries. Worldwide, 27 countries have reported cases since 2012. In the Western Pacific Region, countries that have experienced imported cases of MERS include China, Malaysia, the Philippines, and the Republic of Korea. The importation of the virus into the Republic of Korea in 2015 led to the largest MERS outbreak outside of the Middle East. This outbreak resulted in 186 laboratory-confirmed cases and 36 deaths (Durai et al. 2015; Ki 2015).
MERS-CoV was determined to be different from all other coronavirus strains that have been found in humans, including the severe acute respiratory syndrome coronavirus (SARS-CoV) and SARS-CoV-2 strains that caused the SARS (LeDuc and Barry 2004) and coronavirus disease 2019 (COVID-19) epidemics, respectively. So far, various types of vaccines targeting SARS-CoV and SARS-CoV-2 have been developed and tested in preclinical models. These include protein subunit vaccines, virus-like particle vaccines, DNA vaccines, RNA vaccines, viral vector vaccines, inactivated whole-virus vaccines, and live-attenuated virus vaccines (Anderson et al. 2020; Folegatti et al. 2020; Jackson et al. 2020; Keech et al. 2020; Logunov et al. 2020; Mulligan et al. 2020; Sahin et al. 2020; Walsh et al. 2020; Zhu, et al. 2020a, b; Zhu, et al. 2020a, b). However, only DNA- and viral vector-based vaccine candidates have been tested in preclinical models for MERS-CoV (Modjarrad et al. 2019; Xia et al. 2020, 2021).
In recent studies, epitope-based vaccine candidates were successfully developed against SARS-CoV-2, human cytomegalovirus, Tropheryma whipplei, nervous necrosis virus, candida fungus, and dengue virus (Akhtar et al. 2021a, b, c; Sunil Krishnan et al. 2020; Jain et al. 2021; Joshi et al. 2020; Joshi and Kaushik 2020; Krishnan et al. 2021). Subunit vaccines have been clinically approved for use against pertussis, influenza, Streptococcus pneumoniae, and Haemophilus influenzae type b (Folegatti et al. 2020; Koch et al. 2020). To our knowledge, such protein subunit vaccines are easy to produce and relatively safe and well-tolerated compared to whole-virus vaccines and viral vector vaccines. They consist of viral antigenic fragments produced by recombinant protein techniques. Thus, our study predicted epitope-based vaccine peptides that have therapeutic properties against MERS-CoV, however, experimental evaluations remain necessary to verify the exact safety and immunogenicity profile of this vaccine.
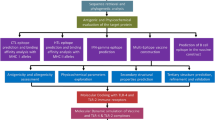
Materials and Methods
Proteomic Data Retrieval
The MERS-CoV surface glycoprotein (S protein) sequence was obtained from the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/) using the accession ID ALW82742.1. The experimentally derived 3D structure of the MERS-CoV S protein was retrieved from the Protein Data Bank (PDB ID: 5X59).
Evaluation of Protein Physicochemical Properties
Using the online program ProtParam, the protein sequence was examined for chemical and physical characteristics such as grand average of hydropathicity (GRAVY), half-life, molecular weight, stability index, and amino acid atomic composition (Gasteiger et al. 2005).
Antigenicity Tests
The Vaxijen v.2.0 server (Doytchinova and Flower 2007; http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html) was used to conduct antigenicity tests. For predicting allergenicity, the protein sequence was expanded for further analysis based on Allergen FP v.1.0. (Dimitrov et al. 2013).
B-Cell Epitope Prediction
B-cell epitopes were predicted using the free web server ABCpred (Saha and Raghava 2006; http://crdd.osdd.net/raghava/abcpred/). The criteria were established at 75% specificity, and 12-residue-long epitopes were deemed sufficient to induce a protective immune response. The top results ranked from 1 to 10 were considered.
T-Cell Epitope Prediction
Epitopes of cytotoxic T lymphocytes (CTLs) are important in vaccine development. Most importantly, it saves money and time as compared to wet-lab studies. Thus, CTL epitopes of target proteins of major histocompatibility complex (MHC) classes I and II were predicted using two different internet tools, the NetMHCpan 4.1 (http://www.cbs.dtu.dk/services/NetMHCpan-4.1/) and NetMHCIIpan 4.0 servers (http://www.cbs.dtu.dk/services/NetMHCIIpan-4.0/; Reynisson et al. 2020). Because these techniques consider a large number of human leukocyte antigen (HLA) alleles during calculation, the results are highly significant. All alleles were chosen for prediction and the sequence was supplied in a simple format.
Toxicity Profiling for Selected Epitopes
After finalizing the epitopes of both MHC class I and class II alleles, the ToxinPred server (Gupta et al. 2013) was used for in silico analysis to differentiate non-toxic and toxic peptides (http://crdd.osdd.net/raghava/toxinpred/). Only non-toxic epitopes were chosen for further investigation.
Molecular Docking
The 3D structures of HLA alleles were retrieved from the Research Collaboratory for Structural Bioinformatics PDB (RCSB PDB) database (Berman 2000), and used for subsequent molecular docking purposes. The PatchDock server (Duhovny et al. 2002; Schneidman-Duhovny et al. 2005a, b) was used for conducting docking experiments (https://bioinfo3d.cs.tau.ac.il/PatchDock/php.php). Thereafter, the FireDock server (Mashiach et al. 2008) was used to screen the best-docked results based on atomic contact energy (ACE; https://bioinfo3d.cs.tau.ac.il/FireDock/php.php).
MD Simulations
The MDWeb tool (Hospital et al. 2012) was deployed for conducting molecular dynamics and simulation studies for obtaining the best-docked complexes (https://mmb.irbbarcelona.org/MDWeb/). Trajectory analysis produced root-mean-square deviation (RMSD) and B-Factor plots that demonstrated the integrity and stability of complexes under simulated environments.
Results
Protein Evaluation
From the protein sequence of the MERS-CoV S protein, various physicochemical properties were computed using ProtParam. AllergenFP v.1.0 results indicated that the protein had non-allergenic properties as it held a higher Tanimoto similarity index of 0.84. Vaxijen v.2.0 analysis with a threshold of 0.4 revealed an antigenicity of 0.4829, indicating that the sequence was a probable antigen. Thus, we confirmed that the MERS-CoV S protein sequence can well be considered for epitope prediction. The abovementioned properties are summarized in Table 1.
T-Cell Epitope Prediction
T-cell epitopes were predicted from the S protein using the NetMHCpan 4.1 server for MHC Class I HLA proteins and the NetMHCIIpan 4.0 server for MHC Class II HLA proteins as both servers work efficiently based on an artificial neural network schematic framework. Tables S1 and S2 present the screened epitopes based on the binding affinity and rank generated by the NetMHC servers. Supplementary Excel Sheets 1 and 2 present the full T-cell epitopes for both MHC I and MHC II HLA allelic determinants.
B-Cell Epitope Prediction
The ABCpred tool was deployed for B-cell epitope prediction. B-cell epitopes bind to B-cell receptors (BCRs) and induce immune responses against MERS-CoV. Along with T-cell epitopes, B-cell epitopes are highly useful in generating immunogenicity (Table S3).
Toxicity and Antigenicity Tests
Allergenicity prediction using AllergenFP v.1.0 revealed that the complete S protein sequence represented a probable non-allergen. Vaxijen v.2.0 analysis of T-cell and B-cell epitopes from the MERS-CoV S protein using a threshold of 1.1 facilitated the screening of epitopes of high antigenicity, indicating that the selected protein had probable antigenic and immunogenic properties. Thus, we confirmed that the protein sequence can be considered for epitope prediction. After this analysis, the screened epitopes were analyzed using the ToxinPred tool that further confirmed their non-toxic features (Table 2).
MHC Proteins and BCR Structure Retrieval
All receptor structures were retrieved from the RCSB PDB database. The structural files for MHC class II HLA DRB1:0101 (PDB ID: 1AQD), MHC Class I HLA-A*0101 (PDB ID: 1W72), and BCR (PDB ID: 5IFH) were downloaded successfully.
Molecular Docking Studies
Molecular docking studies were conducted using the PatchDock and FireDock tools (Dina Schneidman-Duhovny et al. 2005a, b). Figure 1 presents all 11 docked complexes, of which eight complexes had suitable ACE values from − 9 to − 5 kcal/mol (Krishnan et al. 2021). The eight selected complexes are presented in Table 3.
The obtained results also indicated perfect binding between the ligand and receptor as presented in Fig. 2. Two B-cell epitopes, LEPRSGNHCPAG and QNCTAVGVRQQR, bound to the BCR_FAB domain during molecular docking and showed chemical interactions in visualizations with PyMOL.
Similarly, the T-cell epitopes GRGVFQNCTAVGVRQ and EGGGWLVASGSTVAM interacted with MHC class II HLA determinants (Fig. 3A and B). Additionally, the T-cell epitopes YSNITITYQGLF, YIDLKELGNYTY, SYIDLKELGNYT, and PTNFSFGVTQEY were observed to interact with MHC class I HLA determinants (Fig. 3C–F). All such T-cell epitope interactions were observed in the binding pocket of core protein receptors using the PyMOL visualization tool.
MD Simulations
For epitopes interacting with the HLA allele structures, the RMSD values and atomic fluctuation per amino acid residue were acquired, allowing for ideal pair selection and confirmation. The MDWeb tool was deployed for this purpose, and RMSD and B-factor values in the appropriate range were successfully obtained (Sunil Krishnan et al. 2020). The RMSD and B-factor plots of the B-cell epitope with the lowest ACE value interacting with the BCR_FAB domain are indicated in Fig. 4.
Figures 5 and 6 present RMSD and B-factor plots, respectively, for all T-cell epitopes interacting with MHC class I and class II HLA determinants. The obtained results indicated perfect molecular stability in the docked complex in short-run simulations using the MDWeb tool.
Discussion
MERS-CoV and SARS-CoV-2 are emerging infectious viruses that are extremely harmful to humans. Effective immunization and protective measures against these infections are still largely undiscovered. Gaps in our understanding of these pathogens’ protective immunity pose challenges to vaccine development (Sunil Krishnan et al. 2020; Park et al. 2019).
This study aimed to use immunoinformatics approach to screen of vaccine epitopes to identify the most antigenic protein of MERS-CoV as well as the B- and T-cell epitopes that map onto this protein. The immunoinformatics method, which is based on bioinformatics breakthroughs, is a viable and necessary tool for developing vaccines against new, highly dangerous microorganisms. In this investigation, critical dominant immunogens were screened against MERS-CoV using an immunoinformatics-driven strategy.
The results revealed that the S protein was a superior antigenic protein. Indeed, all current MERS vaccination trials focus on the S protein of MERS-CoV, which facilitated the identification of the host cell DPP4 receptor and causes a strong immune response (Wang et al. 2013). After docking a total of eight epitopes from the MERS-CoV S protein, analyses predicted that these epitopes are non-toxic and have therapeutic immunogenic properties. Two B-cell epitopes, LEPRSGNHCPAG and QNCTAVGVRQQR, bound to the BCR_FAB domain during molecular docking. Additionally, the T-cell epitopes GRGVFQNCTAVGVRQ and EGGGWLVASGSTVAM interacted with MHC class II HLA determinants. Similarly, the T-cell epitopes YSNITITYQGLF, YIDLKELGNYTY, SYIDLKELGNYT, and PTNFSFGVTQEY were observed to interact with MHC class I HLA determinants. Recent studies using a similar approach successfully identified epitope-based vaccine candidates against SARS-CoV-2, human cytomegalovirus, Tropheryma whipplei, nervous necrosis virus, candida fungus, and dengue virus (Akhtar et al. 2021a, b, c; Sunil Krishnan et al. 2020; Jain et al. 2021; Joshi et al. 2020; Joshi and Kaushik 2020; Krishnan et al. 2021). Thus, the identified T-cell and B-cell epitopes have therapeutic potential against the MERS virion.
Conclusions
MERS-CoV causes a severe respiratory disorder, and better vaccines need to be developed to control this pathogen. The main target of this study was to identify peptide epitopes from MERS-CoV for both T cells and B cells that have therapeutic potential. Our modern in silico study demonstrates a fast strategy for vaccine development against MERS-CoV that nevertheless requires wet-lab testing for final validation.
References
Akhtar N, Joshi A, Kaushik V, Kumar M, Mannan MA-U (2021a) In-silico design of a multivalent epitope-based vaccine against Candida auris. Microb Pathog 155:104879. https://doi.org/10.1016/j.micpath.2021.104879
Akhtar N, Joshi A, Singh B, Kaushik V (2021b) Immuno-informatics quest against COVID-19/SARS-COV-2: determining putative T-cell epitopes for vaccine prediction. Infect Disord Drug Targets 21(4):541–552. https://doi.org/10.2174/1871526520666200921154149
Akhtar N, Joshi A, Singh J, Kaushik V (2021c) Design of a novel and potent multivalent epitope based human cytomegalovirus peptide vaccine: an immunoinformatics approach. J Mol Liq 335:116586. https://doi.org/10.1016/j.molliq.2021.116586
Alqahtani FY, Aleanizy FS, El Hadi A, Mohamed R, Alanazi MS, Mohamed N, Alrasheed MM, Abanmy N, Alhawassi T (2018) Prevalence of comorbidities in cases of Middle East respiratory syndrome coronavirus: a retrospective study. Epidemiol Infect 147:e35–e35. https://doi.org/10.1017/S0950268818002923
Anderson EJ, Rouphael NG, Widge AT, Jackson LA, Roberts PC, Makhene M, Chappell JD, Denison MR, Stevens LJ, Pruijssers AJ, McDermott AB, Flach B, Lin BC, Doria-Rose NA, O’Dell S, Schmidt SD, Corbett KS, Swanson PA II, Padilla M et al (2020) Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N Engl J Med 383(25):2427–2438. https://doi.org/10.1056/NEJMoa2028436
Berman HM (2000) The protein data bank. Nucleic Acids Res 28(1):235–242. https://doi.org/10.1093/nar/28.1.235
Bermingham A, Chand MA, Brown CS, Aarons E, Tong C, Langrish C, Hoschler K, Brown K, Galiano M, Myers R, Pebody RG, Green HK, Boddington NL, Gopal R, Price N, Newsholme W, Drosten C, Fouchier RA, Zambon M (2012) Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Eurosurveillance. https://doi.org/10.2807/ese.17.40.20290-en
Dimitrov I, Naneva L, Doytchinova I, Bangov I (2013) AllergenFP: allergenicity prediction by descriptor fingerprints. Bioinformatics 30(6):846–851. https://doi.org/10.1093/bioinformatics/btt619
Doytchinova IA, Flower DR (2007) VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics 8:4–4. https://doi.org/10.1186/1471-2105-8-4
Duhovny D, Nussinov R, Wolfson HJ (2002) Efficient unbound docking of rigid molecules. Lecture notes in computer science. Springer, Berlin, Heidelberg, pp 185–200. https://doi.org/10.1007/3-540-45784-4_14
Durai P, Batool M, Shah M, Choi S (2015) Middle East respiratory syndrome coronavirus: transmission, virology and therapeutic targeting to aid in outbreak control. Exp Mol Med 47(8):e181–e181. https://doi.org/10.1038/emm.2015.76
Folegatti PM, Bittaye M, Flaxman A, Lopez FR, Bellamy D, Kupke A, Mair C, Makinson R, Sheridan J, Rohde C, Halwe S, Jeong Y, Park Y-S, Kim J-O, Song M, Boyd A, Tran N, Silman D, Poulton I et al (2020) Safety and immunogenicity of a candidate Middle East respiratory syndrome coronavirus viral-vectored vaccine: a dose-escalation, open-label, non-randomised, uncontrolled, phase 1 trial. Lancet Infect Dis 20(7):816–826. https://doi.org/10.1016/S1473-3099(20)30160-2
Gasteiger E, Hoogland C, Gattiker A, Duvaud SE, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. The proteomics protocols handbook. Humana Press, Totowa, pp 571–607
Gupta S, Kapoor P, Chaudhary K, Gautam A, Kumar R, Raghava GPS (2013) In silico approach for predicting toxicity of peptides and proteins. PLoS ONE 8(9):e73957. https://doi.org/10.1371/journal.pone.0073957
Hospital A, Andrio P, Fenollosa C, Cicin-Sain D, Orozco M, Gelpí JL (2012) MDWeb and MDMoby: an integrated web-based platform for molecular dynamics simulations. Bioinformatics 28(9):1278–1279. https://doi.org/10.1093/bioinformatics/bts139
Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, McCullough MP, Chappell JD, Denison MR, Stevens LJ, Pruijssers AJ, McDermott A, Flach B, Doria-Rose NA, Corbett KS, Morabito KM, O’Dell S, Schmidt SD, Swanson PA II et al (2020) An mRNA vaccine against SARS-CoV-2—preliminary report. N Engl J Med 383(20):1920–1931. https://doi.org/10.1056/NEJMoa2022483
Jain P, Joshi A, Akhtar N, Krishnan S, Kaushik V (2021) An immunoinformatics study: designing multivalent T-cell epitope vaccine against canine circovirus. J Genet Eng Biotechnol 19(1):121–121. https://doi.org/10.1186/s43141-021-00220-4
Joshi A, Kaushik V (2020) In-silico proteomic exploratory quest: crafting T-cell epitope vaccine against Whipple’s disease. Int J Pept Res Ther. https://doi.org/10.1007/s10989-020-10077-9
Joshi A, Joshi BC, Mannan MA-U, Kaushik V (2020) Epitope based vaccine prediction for SARS-COV-2 by deploying immuno-informatics approach. Inform Med Unlocked 19:100338–100338. https://doi.org/10.1016/j.imu.2020.100338
Keech C, Albert G, Cho I, Robertson A, Reed P, Neal S, Plested JS, Zhu M, Cloney-Clark S, Zhou H, Smith G, Patel N, Frieman MB, Haupt RE, Logue J, McGrath M, Weston S, Piedra PA, Desai C et al (2020) Phase 1–2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N Engl J Med 383(24):2320–2332. https://doi.org/10.1056/NEJMoa2026920
Ki M (2015) 2015 MERS outbreak in Korea: hospital-to-hospital transmission. Epidemiol Health 37:e2015033. https://doi.org/10.4178/epih/e2015033
Koch T, Dahlke C, Fathi A, Kupke A, Krähling V, Okba NMA, Halwe S, Rohde C, Eickmann M, Volz A, Hesterkamp T, Jambrecina A, Borregaard S, Ly ML, Zinser ME, Bartels E, Poetsch JSH, Neumann R, Fux R et al (2020) Safety and immunogenicity of a modified vaccinia virus Ankara vector vaccine candidate for Middle East respiratory syndrome: an open-label, phase 1 trial. Lancet Infect Dis 20(7):827–838. https://doi.org/10.1016/S1473-3099(20)30248-6
Krishnan GS, Joshi A, Akhtar N, Kaushik V (2021) Immunoinformatics designed T cell multi epitope dengue peptide vaccine derived from non structural proteome. Microbial Pathogenesis 150:104728. https://doi.org/10.1016/j.micpath.2020.104728
LeDuc JW, Barry MA (2004) SARS, the first pandemic of the 21st century. Emerg Infect Dis 10(11):e26. https://doi.org/10.3201/eid1011.040797_02
Logunov DY, Dolzhikova IV, Zubkova OV, Tukhvatulin AI, Shcheblyakov DV, Dzharullaeva AS, Grousova DM, Erokhova AS, Kovyrshina AV, Botikov AG, Izhaeva FM, Popova O, Ozharovskaya TA, Esmagambetov IB, Favorskaya IA, Zrelkin DI, Voronina DV, Shcherbinin DN, Semikhin AS et al (2020) Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: two open, non-randomised phase 1/2 studies from Russia. Lancet 396(10255):887–897. https://doi.org/10.1016/s0140-6736(20)31866-3
Mashiach E, Schneidman-Duhovny D, Andrusier N, Nussinov R, Wolfson HJ (2008) FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res 36(Web Server):W229–W232. https://doi.org/10.1093/nar/gkn186
Modjarrad K, Roberts CC, Mills KT, Castellano AR, Paolino K, Muthumani K, Reuschel EL, Robb ML, Racine T, Oh M-D, Lamarre C, Zaidi FI, Boyer J, Kudchodkar SB, Jeong M, Darden JM, Park YK, Scott PT, Remigio C et al (2019) Safety and immunogenicity of an anti-Middle East respiratory syndrome coronavirus DNA vaccine: a phase 1, open-label, single-arm, dose-escalation trial. Lancet Infect Dis 19(9):1013–1022. https://doi.org/10.1016/S1473-3099(19)30266-X
Mulligan MJ, Lyke KE, Kitchin N, Absalon J, Gurtman A, Lockhart S, Neuzil K, Raabe V, Bailey R, Swanson KA, Li P, Koury K, Kalina W, Cooper D, Fontes-Garfias C, Shi P-Y, Türeci Ö, Tompkins KR, Walsh EE et al (2020) Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 586(7830):589–593. https://doi.org/10.1038/s41586-020-2639-4
Park BK, Maharjan S, Lee SI, Kim J, Bae J-Y, Park M-S, Kwon H-J (2019) Generation and characterization of a monoclonal antibody against MERS-CoV targeting the spike protein using a synthetic peptide epitope-CpG-DNA-liposome complex. BMB Rep 52(6):397–402. https://doi.org/10.5483/BMBRep.2019.52.6.185
Reynisson B, Alvarez B, Paul S, Peters B, Nielsen M (2020) NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res 48(W1):W449–W454. https://doi.org/10.1093/nar/gkaa379
Saha S, Raghava GPS (2006) Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins Struct Funct Bioinform 65(1):40–48
Sahin U, Muik A, Derhovanessian E, Vogler I, Kranz LM, Vormehr M, Baum A, Pascal K, Quandt J, Maurus D, Brachtendorf S, Lörks V, Sikorski J, Hilker R, Becker D, Eller A-K, Grützner J, Boesler C, Rosenbaum C et al (2020) COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature 586(7830):594–599. https://doi.org/10.1038/s41586-020-2814-7
Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ (2005) PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res 33(Web Server):W363–W367. https://doi.org/10.1093/nar/gki481
Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ (2005) PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res 33(Web Server issue):W363–W367. https://doi.org/10.1093/nar/gki481
Sunil Krishnan G, Joshi A, Kaushik V (2020) T cell epitope designing for dengue peptide vaccine using docking and molecular simulation studies. Mol Simul 46(10):787–795. https://doi.org/10.1080/08927022.2020.1772970
Walsh EE, Frenck RW Jr, Falsey AR, Kitchin N, Absalon J, Gurtman A, Lockhart S, Neuzil K, Mulligan MJ, Bailey R, Swanson KA, Li P, Koury K, Kalina W, Cooper D, Fontes-Garfias C, Shi P-Y, Türeci Ö, Tompkins KR et al (2020) Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N Engl J Med 383(25):2439–2450. https://doi.org/10.1056/NEJMoa2027906
Wang N, Shi X, Jiang L, Zhang S, Wang D, Tong P, Guo D, Fu L, Cui Y, Liu X, Arledge KC, Chen Y-H, Zhang L, Wang X (2013) Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res 23(8):986–993. https://doi.org/10.1038/cr.2013.92
Xia S, Duan K, Zhang Y, Zhao D, Zhang H, Xie Z, Li X, Peng C, Zhang Y, Zhang W, Yang Y, Chen W, Gao X, You W, Wang X, Wang Z, Shi Z, Wang Y, Yang X et al (2020) Effect of an inactivated vaccine against SARS-CoV-2 on safety and immunogenicity outcomes: interim analysis of 2 randomized clinical trials. JAMA 324(10):951–960. https://doi.org/10.1001/jama.2020.15543
Xia S, Zhang Y, Wang Y, Wang H, Yang Y, Gao GF, Tan W, Wu G, Xu M, Lou Z, Huang W, Xu W, Huang B, Wang H, Wang W, Zhang W, Li N, Xie Z, Ding L et al (2021) Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: a randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect Dis 21(1):39–51. https://doi.org/10.1016/S1473-3099(20)30831-8
Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus ADME, Fouchier RAM (2012) Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367(19):1814–1820. https://doi.org/10.1056/nejmoa1211721
Zhu F-C, Guan X-H, Li Y-H, Huang J-Y, Jiang T, Hou L-H, Li J-X, Yang B-F, Wang L, Wang W-J, Wu S-P, Wang Z, Wu X-H, Xu J-J, Zhang Z, Jia S-Y, Wang B-S, Hu Y, Liu J-J et al (2020a) Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet (London, England) 396(10249):479–488. https://doi.org/10.1016/S0140-6736(20)31605-6
Zhu F-C, Li Y-H, Guan X-H, Hou L-H, Wang W-J, Li J-X, Wu S-P, Wang B-S, Wang Z, Wang L, Jia S-Y, Jiang H-D, Wang L, Jiang T, Hu Y, Gou J-B, Xu S-B, Xu J-J, Wang X-W et al (2020b) Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet (London, England) 395(10240):1845–1854. https://doi.org/10.1016/S0140-6736(20)31208-3
Funding
No funding was received to assist with the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization: TLN; Methodology: TLN; Formal analysis and investigation: TLN; Writing—original draft preparation: TLN; Writing—review and editing: TLN and YL; Supervision: HK.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Research Involving Human and/or Animal Participants
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nguyen, T.L., Lee, Y. & Kim, H. Immunogenic Epitope-Based Vaccine Prediction from Surface Glycoprotein of MERS-CoV by Deploying Immunoinformatics Approach. Int J Pept Res Ther 28, 77 (2022). https://doi.org/10.1007/s10989-022-10382-5
Accepted:
Published:
DOI: https://doi.org/10.1007/s10989-022-10382-5