Abstract
During the last decade there has been an alarming increase in the appearance of antibiotic-resistant bacteria. The drug-resistant microorganisms dubbed superbugs are projected to kill 10 million people a year by 2050. The annual frequency of deaths from Methicillin-resistant Staphylococcus aureus (MRSA) is rapidly increasing and surpassing those caused by human immunodeficiency virus/acquired immune deficiency syndrome (HIV/AIDS). Muramyl ligase E (MurE), an enzyme involved in the peptidoglycan biosynthesis of the bacterial cell wall, is a highly druggable target in MRSA. Six anti Methicillin-resistant Staphylococcus Aureus peptides were selected for deep bioinformatic analysis. The six anti-Methicillin-resistant Staphylococcus Aureus peptides were modeled and then docked to MurE ligase and their binding interactions were studied. The findings suggested that the interactions of three antimicrobial peptides (Flowlicidin-2, Ostricacin-2 and Ostricacin-3) with MurE could be essential for their inhibitory activity.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
During the last decade there has been an alarming increase in the appearance of antibiotic-resistant bacteria as a result of an increased use of antibiotics combined with the exceptional ability of bacteria to develop resistance (Singh and Mukhopadhyay 2014). The drug-resistant microorganisms dubbed superbugs are projected to kill 10 million people a year by 2050 and will overtake cancer as the leading cause of death worldwide (O’Neill 2016).
Methicillin-resistant Staphylococcus aureus (MRSA) infections are caused by strains of Staphylococcus aureus that have become resistant to the antibiotics (methicillin) commonly used to treat ordinary infections (Ryu et al. 2014). The annual frequency of deaths from MRSA is rapidly increasing and surpassing those caused by human immunodeficiency virus/acquired immune deficiency syndrome (HIV/AIDS) (Bancroft 2007).
Antimicrobial peptides (AMPs) are produced in nature by plants, animals, bacteria and fungi and have been demonstrated to be quite effective in killing a wide selection of bacteria pathogens, including MRSA, and representpromising foundations for developing a new generation of antimicrobial agents (Wang 2010). Additionally, 60 therapeutic peptides have been approved by US Food and Drug Administration (FDA) as antibodies, vaccines, and antimicrobial agents, etc. (Aguilar-Toalá et al. 2019). It has been suggested for a long time that the main mechanism of action of AMPs targets the membrane (Boman 1995) and form pores by oligomerization. However, more recently, an activity on certain intracellular targets has been proven for peptides such as mersadicin (inhibiting the synthesis of peptidoglycan by interaction with the lipid H intermediate) (Brôtz et al. 1998). Such activity could explain a divergence, for some peptides, between the speed of cell death and the speed of membrane destruction (Kim and Brogdeb 2005; Takahashi et al. 2010). Peptidoglycan is one of the main components of the bacterial cell wall, and it represents one of the most frequently used targets for antibacterial agents (Simcic et al. 2012). The mechanism of the peptidoglycan biosynthesis is one the best known and most validated drug targets for antibacterial therapy (Tomašic et al. 2012). The fact that Mur ligases are unique to prokaryotic cells and are vital for the survival of bacteria makes them promising emerging targets for novel antibacterial drug design (Kouidmi et al. 2014). These enzymes successively add l-Ala, d-Glu, meso-A2pm or l-Lys, and d-Ala-d-Ala to the nucleotide precursor UDP-MurNAc, (El Zoeiby et al. 2003; Barreteau et al. 2008). Knowing that compounds that interfere with the biosynthesis and assembly of peptidoglycan have been utilized as antimicrobial agents, Mur enzymes, which catalyze the cytoplasmic steps of the peptidoglycan biosynthesis, are advanced as excellent candidates for antibacterial drug discovery and development (Nikolaidis et al. 2014). Muramyl ligase E (MurE), an enzyme involved in the peptidoglycan biosynthesis of the bacterial cell wall, is a highly druggable target in MRSA (Billones and Bangalan 2019).
In silico analysis, involves computational methods applied to manage, curate, and interpret information related to biological systems (Li-Chan 2015). Bioinformatics approaches may overcome the high cost and the long time consuming by conventional methods, thereby enabling rapid research and study of bioactive peptide (Tu et al. 2018). In the present study, we have focused our attention on MurE which is a UDP-N-acetylmuramoylalanyl-D-glutamate-2, 6-diaminopimelate ligase, that adds m-DAP to UDP-MurNAc-l-Ala-d-Glu (Moraes et al. 2015). The Mur E enzyme of Mur pathway of MRSA is an attractive drug target as it is unique to bacteria and is absent in mammalian cells. MurE adds l-lysine to MRSA and other Gram-positive bacteria (Lto and Strominger 1964). It is therefore imperative to find efficient peptides inhibitors of MurE enzyme, which is essential in the crucial step of prostaglandin biosynthesis (Billones and Bangalan 2019).
Molecular docking, as well as Quantitative structure-activity relationship also have been used to screen and predict the potential peptides (Nongonierma and Fitzgerald 2016). In the current study, six peptides were selected and subjected to further study involving homology modelling and molecular docking against MurE as enzyme target of MRSA strain.
Materials and Methods
Selected Anti-MRSA Peptides
From the 116 anti-MRSA collected peptides (Zouhir et al. 2016), six antiMRSA peptides were selected for further bioinformatic analysis namely: Flowlicidin-2, Pilosulin-1, Pleurocidin, Ostricacin-1, Ostricacin-2 and Ostricacin-3. The choice of these peptides is based on antiMRSA activity and their short sequence (Table 1).
Sequences Analysis
CAMPSign tool (http://www.campsign.bicnirrh.res.in) was used for identification of AMPs belonging to one of the 45 major AMP families present in CAMPR3 (Waghu et al. 2016). The peptides are identified based on family-specific sequence signatures such as patterns and Hidden Markov Models (HMMs) generated for experimentally known AMPs.
ProtParam online tool (http://web.expasy.org/protparam/) was used to determine the physico- chemical parameters of the six selected antiMRSA peptides.
The prediction of peptide antimicrobial activity was performed using the most recent version of: antimicrobial peptide database (APD3) (Wang et al. 2016) and using “Yet Another Database” of AntiMicrobial Peptides (YADAMP) (http://yadamp.unisa.it/) (Stefano et al. 2012).
Molecular Modeling and Models Validation
The MurE enzyme sequence used for homology is that of MRSA 252, was retrieved from UniProt Data Bank (https://www.uniprot.org/. The automated protein structure homology modeling server, Swiss Model (http://www.expasy.org/swissmod/), was used to generate the three-dimensional structural model of MurE enzyme using the crystal structure of MuE enzyme from Staphylococcus aureus MurE (PDB code 4C12) as a template since both enzymes shared about 98.99 % of identity and 100 % of sequence coverage.
Swiss model server was also used to generate the 3D structural model of 4 peptides having a sequence longer than 30 amino acids. For the two short peptides Pleurocidin and Pilosulin 1, PEPstrMOD method (Singh et al. 2015) was used for structure prediction. Peptides models were further refined by the interactive web server 3Drefine publicly available at the URL: http://sysbio.rnet.missouri.edu/3Drefine/ (Bhattacharya et al. 2016). Structures were therefore evaluated and validated by using SAVES v5.0 (Structural Analysis and VErification Server) was used for further checking and validating protein or peptides structures. SAVES (https://services.mbi.ucla.edu/SAVES/) utilizes 6 programs for doing this: Verify3D, ERRAT, Prove, PROCHECK, WHATCHECK and CRYST. Verify3D (Luthy et al. 1992; Bowie et al. 1991) was used to verify the packing quality of each residue in the model. PROCHECK was used to determine the torsion angle restraints for side chains of each amino acid in the predicted models (Laskowski et al. 1993) and thereby to draw Ramachandran plots.
The PyMOL v0.99 program (http://www.pymol.org) was used to visualize and analyze the generated model structure and to construct graphical presentations and illustrative figures.
Protein–Protein Docking Analysis
Protein-protein docking of the antiMRSA peptides with the MurE enzyme was performed using the HDOCK server (http://hdock.phys.hust.edu.cn/). The server automatically predicts the interaction between receptor and ligand molecules through a hybrid algorithm of template-based modeling and ab initio template-free docking. HDOCK incorporates the binding interface information from the PDB and/or user-input biological information like residue restraints and molecular size/shape information obtained from small-angle X-ray scattering, supports both amino acid sequence and structure inputs and uses an intrinsic scoring function for protein-protein interactions. During docking calculation, the top 10 orientations with the best shape complementarity scores in 3D translational space are evaluated by a scoring function (ITScorePP) and the top-scored one is kept (Yan et al. 2020).
Interface covalent bonds, hydrogen bonds and salt bridges between antiMRSA peptide and MurE enzyme within the predicted docking models were evaluated using the online tool PDBePISA (http://www.ebi.ac.uk/pdbe/pisa/).
The PyMOL program was ultimately used for the visualization of structures showing the interactions between toxin and receptor.
Results and Discussion
Analysis of Selected AntiMRSA Peptides
Through CAMPR3 database, the three Ostricacin peptides were identified as Defensin family members, Flowlicidin-2 as Cathelicidin family member, Pleurocidin as Pleurocidin member while Pilosulin-1 was not identified to belong to any AMP family.
The physico-chemical analysis of the six selected anti-MRSA was performed using ProtParam tool (Table 2). The inspection showed that the number of amino acids vary from 20 to 41 residues with a molecular weight ranging from 2109 to 4689 Da. All the pHi are in the basic zone and the highest value (12.85) in given by Flowlicidin-2. All peptides have high estimated half-life expect Flowlicidin-2 and Ostricacin-1 with exhibit the lowest estimated half-life when expressed in Yeast (3 min) or E. coli (2 min). Among the six peptides, only Ostricacin-2 was predicted to be unstable.
The antimicrobial peptide database (APD3) (Wang et al. 2016) and yet another database of antimicrobial peptides (YADAMP) (http://yadamp.unisa.it/) (Stefano et al. 2012) databases were used for prediction of peptide antimicrobial activity. As shown by Table 3, Pleurocidin, Ostricacin-1, Ostricacin-2 and Ostricacin-3 exhibit antiMRSA activity and have different animal sources. However, Flowlicidin-2 and Pilosulin-1 have not been recognized by this database.
Homology Modeling and Structural Inspection
To gain further insights into the structure of the 6 selected peptides, a search for homology modeling. According to the peptide length, Swiss-model server was used to predict the models of Flowlicidin-2, Ostricacin-1, Ostricacin-2 and Ostricacin-3 based on available RMN structures, while models of Pleurocidin and Pilosulin-1 were predicted using PEPstrMOD method (Singh et al. 2015) recommended for short peptides. Related Data to modeling method, PDB template, model oligomerization, sequence identity between peptide and template as well as sequence coverage are summarized in Table 4.
To validate the predicted models, their stereochemical quality was assessed by Ramachandran plots (Fig. 1). All models showed a high percent of residues (ranging from 95 to 100%) in the most favored regions and allowed regions. A very low percent of residues in the disallowed region supports high geometry of models. In addition, the packing quality of each residue in the model was evaluated using Verify 3D. For all models, the majority of residues have a score indicating acceptable side chain environment and reasonably good structures. Accordingly, and based on all obtained data, predicted models were validated and retained for further investigation.
The 3D structure inspection revealed huge structural divergence between the 6 selected antiMRSA peptides (Fig. 2). For Ostricacin peptides, the tertiary structure is principally marked by the occurrence of anti-parallel β-strands. However, Flowlicidin-2 is especially marked by a helical organization at the C-terminal extremity. Pilosulin-1 and Pleurocidin models indicated the presence of a noticeable helix at the N-terminal end.
Structure Inspection of MurE Enzyme
MurE enzyme of MRSA 252 is a 494 amino acids protein with about 54.153 kDa and a theoretical pHi of 5.32 as revealed by ProtParam analysis. The 3D-structural model of MurE was predicted using the crystal structure of MuE enzyme from Staphylococcus aureus MurE (PDB code 4C12) (Ruane et al. 2013) as a template since both enzymes shared about 98.99% of identity and 100% of sequence coverage. The Ramachandran plot analysis of the generated model revealed that 960 residues (98%) of the model were in the favored, 16 residues (1.6%) in the allowed regions and only 4 residues (0.4%) in the disallowed regions. As more than 99 % of the residues were in the favored and allowed regions, the model was consequently validated. The model was also checked and validated using the structural analysis and verification server (SAVES) and accordingly it was retained for analysis and further investigation.
The examination of the MurE enzyme model showed a homodimer organization (Fig. 3) and the presence of 8 ligands (4 Mg2+ ions, 2 UML and 2 UDP) establishing several interactions with the protein residues as illustrated in Table 5.
MurE-antiMRSA Docking Analysis
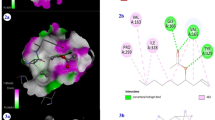
The protein-protein docking models of antiMRSA peptides and MurE enzyme were predicted by the updated version of HDOCK server (Yan et al. 2020). As shown by Fig. 4, the binding of antiMRSA peptides to MurE enzyme is occurred in different manner. Subsequently, the interface interactions between the enzyme and the coupled peptide were evaluated using PDBePISA to more elucidate the binding mode and involved interactions and residues (Table 6). Interestingly, among the six peptides, only Flowlicidin-2 established interactions with UML9 via covalent bonds. These covalent bonds could be responsible of irreversible inhibition which could explain the low minimum inhibitory concentration (MIC) and then the high observed antimicrobial activity of Flowlicidin-2 (Table 7).
On the other hand, Ostricacin-2 exhibited interactions with only two amino acid in the chain B namely Glu246 and Glu264 (Fig. 5) and has an antimicrobial activity very closed to Flowlicidin-2 (1.59 versus 1.58 µg/ml). In addition, Pilosulin-1, Pleurocidin, Ostricacin-1 and Ostricacin-3 interacted with both chain A and B of MurE enzyme. Ostricacin-3 which had a good antimicrobial activity (Table 7) exhibited the highest number of interactions between peptide-MurE when compared with other studied AMPs. In order to verify whether the number or the type (covalent or uncovalent) of interactions peptide-MurE is essential for the efficiency of the anti-microbial peptides, The peptides should be generated, tested in the same conditions and even site-directed mutated to alter binding mode and confirm hypothesis.
Conclusions
Analysis of the bioactive peptides by conventional methods is expensive and time-consuming. Emerging bioinformatics approaches may overcome these drawbacks to enable rapid research and study of bioactive peptide (Tu et al. 2018). In the current work, diverse methods in silico were applied in order to analyze the properties and the structure of antiMRSA peptides as well as their interactions with MurE enzyme. The six anti-MRSA peptides were then docked to the enzyme target and their binding energies were calculated. Three AMPs (Flowlicidin-2, Ostricacin-2 and Ostricacin-3) displayed greater inhibitory potential against MurE ligase compared to the others. These three peptides can be chemically synthesized and their activities against the MRSA strain can be tested in further study.
This study is an essential step to understand the mode of action of antiMRSA peptides on Mur enzymes such as MurE of MRSA.
References
Aguilar-Toalá JE, Hernández-Mendoza A, González-Córdova AF, Vallejo Cordoba B, Liceaga AM (2019) Potential role of natural bioactive peptides for development of cosmeceutical skin products. Peptides 122:170170
Bancroft EA (2007) Antimicrobial resistance: it’s not just for hospitals. JAMA 298:1803–1804
Barreteau H, Kovac A, Boniface A, Sova M, Gobec S, Blanot D (2008) Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol 32:168–207
Bhattacharya D, Nowotny J, Cao R, Cheng J (2016) 3D refine: an interactive web server for efficient protein structure refinement. Nucl Acids Res 44(W1):W406–W409
Billones JB, Bangalan MAT (2019) Structure-based discovery of inhibitors against MurE in methicillin-resistant Staphylococcus aureus. Orient J Chem 35:618–625
Boman HG (1995) Peptide antibiotics and their role in innate immunity. Annu Rev Immunol 13:61–92
Bowie JU, Luthy R, Eisenberg D (1991) A method to identify protein sequences that fold into a known three-dimensional structure. Science 253:164–170
Brôtz H, Bierbaum G, Leopold K, Reynolds PE, Sahl HG (1998) The lantibiotic mersacidin inhibits peptidoglycan synthesis by targeting lipid ii. Antimicrob Agents Chemother 42:154–160
El Zoeiby A, Sanschagrin F, Levesque RC (2003) Structure and function of the Mur enzymes: development of novel inhibitors. Mol Microbiol 47:1–12
Kim A, Brogden (2005) Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol 3:238–250
Kouidmi I, Levesque RC, Paradis-Bleau C (2014) The biology of Mur ligases as an antibacterial target. Mol Microbiol 94:242–253
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 26:283–291
Li-Chan EC (2015) Bioactive peptides and protein hydrolysates: research trends and challenges for application as nutraceuticals and functional food ingredients. Curr Opin Food Sci 1:28e37
Lto E, Strominger JL (1964) Enzymatic synthesis of the peptide in bacterial uridine nucleotides, iii. purification and properties of 1-lysin-adding enzyme. J Biol Chem 239:210–214
Luthy R, Bowie JU, Eisenberg D (1992) Assessment of protein models with three-dimensional profiles. Nature 356:83–85
Moraes GL, Gomes GC, Monteiro de Sousa PR, Alves CN, Govender T, Kruger HG, Lameira J (2015) Structural and functional features of enzymes of Mycobacterium tuberculosis peptidoglycan biosynthesis as targets for drug development. Tuberculosis 95:95–111
Nikolaidis I, Favini-Stabile S, Dessen A (2014) Resistance to antibiotics targeted to the bacterial cell wall . Protein Sci 23:243–259
Nongonierma A, Fitzgerald D (2016) Learnings from quantitative structure activity relationship (QSAR) studies with respect to food protein-derived bioactive peptides. RSC Adv 6:75400e75413
O’Neill J (2016) Tackling drug-resistant infections globally: final report and recommendations. Rev Antimicrobial Resist. Available from https://amr-review.org
Ruane KM et al (2013) Specificity determinants for lysine incorporation in Staphylococcus aureus peptidoglycan as revealed by the structure of a MurE enzyme ternary complex. JBC 288:33439–33448
Ryu S, Song PI, Seo CH, Cheong H et al (2014) Colonization and infection of the skin by S. aureus: immune system evasion and the response to cationic antimicrobial peptides. Int J Mol Sci 15:8753–8772
Simcic M, Sosic I, Hodoscek M, Barreteau H, Blanot D, Gobec S, Grdadolnik SG (2012) The binding mode of second-generation sulfonamide inhibitors of MurD: Clues for rational design of potent MurD inhibitors. PLoS ONE 7(12):e52817
Singh S, Singh H, Tuknait A, Chaudhary K, Singh B, Kumaran S, Raghava GP (2015) PEPstrMOD: structure prediction of peptides containing natural, non-natural and modified residues. Biol Direct 10:73
Singh M, Mukhopadhyay K (2014) Alpha-melanocyte stimulating hormone: an emerging anti-inflammatory antimicrobial peptide. Biomed Res Int 2014:874610
Stefano PP, Sessa L, Concilio S, Iannelli P (2012) YADAMP: yet another database of antimicrobial peptides. Int J Antimicrob Agents 39:346–351
Takahashi D, Shukla SK, Prakash O, Zhang G (2010) Structural determinants of host defense peptides for antimicrobial activity and target cell selectivity. Biochimie 92:1236–1241
Tomašic T, Šink R, Zidar N, Fic A, Contreras-Martel C, Dessen A, Mašic LP (2012) Dual inhibitor of MurD and MurE ligases from Escherichia coli and Staphylococcus aureus. ACS Med Chem Lett 3:626–630
Tu M, Cheng S, Lu W, Du M (2018) Advancement and prospects of bioinformatics analysis for studying bioactive peptides from food-derived protein: sequence, structure, and functions. TrAC Trends Anal Chem 105:7–17
Waghu FH, Barai RS, Gurung P, Idicula-Thomas S (2016) CAMPR3: a database on sequences, structures and signatures of antimicrobial peptides. Nucl Acids Res 44:1094–1097
Wang G, Li X, Wang Z (2016) APD3: the antimicrobial peptide database as a tool for research and education. Nucl Acids Res 44:1087–1093
Wang G (2010) Antimicrobial peptides: discovery, design and novel therapeutic strategies. CABI Wallingford
Yan Y, Tao H, He J, Huang SY (2020) The HDOCK server for integrated protein–protein docking. Nat Protoc 15:1829–1852
Zouhir A, Jridi T, Nefzi A, Ben Hamida J, Sebei K (2016) Inhibition of methicillin-resistant Staphylococcus aureus (MRSA) by antimicrobial peptides (AMPs) and plant essential oils. Pharm Biol 54:3136–3150
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Informed Consent
All authors gave informed consent to the submission of this manuscript.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zouhir, A., Jemli, S., Omrani, R. et al. In Silico Molecular Analysis and Docking of Potent Antimicrobial Peptides Against MurE Enzyme of Methicillin Resistant Staphylococcus Aureus. Int J Pept Res Ther 27, 1253–1263 (2021). https://doi.org/10.1007/s10989-021-10165-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10989-021-10165-4