Abstract
Double mutation D208Q:K450L was introduced in the beta isoform of human cardiac myosin to remove the salt bridge D208:K450 connecting loop 1 and the seven-stranded beta sheet within the myosin head. Beta isoform-specific salt bridge D208:K450, restricting the flexibility of loop 1, was previously discovered in molecular dynamics simulations. Earlier it was proposed that loop 1 modulates nucleotide affinity to actomyosin and we hypothesized that the electrostatic interactions between loop 1 and myosin head backbone regulate ATP binding to and ADP dissociation from actomyosin, and therefore, the time of the strong actomyosin binding. To examine the hypothesis we expressed the wild type and mutant of the myosin head construct (1–843 amino acid residues) in differentiated C2C12 cells, and the kinetics of ATP-induced actomyosin dissociation and ADP release were characterized using stopped-flow spectrofluorometry. Both constructs exhibit a fast rate of ATP binding to actomyosin and a slow rate of ADP dissociation, showing that ADP release limits the time of the strongly bound state of actomyosin. We observed a faster rate of ATP-induced actomyosin dissociation with the mutant, compared to the wild type actomyosin. The rate of ADP release from actomyosin remains the same for the mutant and the wild type actomyosin. We conclude that the flexibility of loop 1 is a factor affecting the rate of ATP binding to actomyosin and actomyosin dissociation. The flexibility of loop 1 does not affect the rate of ADP release from human cardiac actomyosin.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Myosins are a family of molecular motors, transforming the chemical energy of ATP into mechanical motion of muscles and cells. Myosin head interacts with actin and nucleotide and changes conformation two times in a cycle. The cyclic conformational change of the head results in the production of movement and force. Myosins have different functions in cells and organisms, and therefore, have different kinetic properties. The kinetic differences are determined by the amino acid sequence, which governs protein structure and interactions between structural elements. The sequence analysis suggests that two surface loops of the myosin head, loop 1 and loop 2, likely have regulatory functions, since the sequence variability of these loops significantly exceeds the sequence variability of the core myosin (Murphy and Spudich 2000). Loop 2 is located within the actin binding interface and the chimera studies confirmed that loop 2 regulates myosin interaction with actin (Furch et al. 1998). Loop 1 is located near the active site and hypothesized to regulate interaction with nucleotide, most likely ADP release (Spudich 1994), and therefore, may regulate the velocity of muscle shortening. Loop 1 links two helices of the myosin head, one connecting to loop switch 1 and another to the P-loop of the nucleotide binding site (Fig. 1). P-loop, or Walker A motif, coordinates Mg ion, complexed with ATP within the active site, and stabilizes ATP during hydrolysis. Loop switch 1 coordinates Mg ion and γ phosphate of ATP. This proximity to the loops at the active site makes loop 1 a hypothetical regulatory element, controlling nucleotide retention in actomyosin, such as nucleotide binding and release of products of hydrolysis. To examine the hypothesis of the regulatory role of loop 1 and to determine the mechanism of regulation, multiple chimera studies were performed. The residues in loop 1 were mutated, entire loop 1 was deleted or replaced with loop 1 from other myosins. Obtained results are contradictory. In D. discoideum myosin with loop 1 replaced with the one from Acanthamoeba or rabbit myosin, the rate of ADP release from actomyosin correlated with the same parameter of the source myosin (Murphy and Spudich 1998). No such correlation was observed with smooth myosin (Sweeney et al. 1998), it was concluded that the rate of ADP release correlates with the length of loop 1. It was also found that the electrostatics of loop 1 plays a role in the regulation of ADP release from actomyosin (Sweeney et al. 1998; Clark et al. 2005). Rates of ADP release from actomyosin with β isoforms of the pig and rat cardiac myosin differ about three times, regardless of essentially the same sequence of loop 1 (Pereira et al. 2001), and this study concluded that the rate of ADP release from actomyosin is regulated not by loop 1 but by the other parts of the myosin backbone.
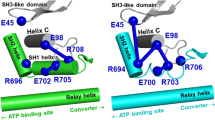
Loop 1 and myosin active site. Permanent salt bridge D208:K450 electrostatically couples loop 1 (cyan) and backbone (gray) of myosin head in β myosin. Red and blue spheres are the positive and negative charged residues. Loop 1 flanks helices, continuing to P-loop (behind ATP) and loop switch 1, coordinating MgATP at the active site
Previously, we ran molecular dynamics simulations and analyzed structural dynamics of two isoforms of human cardiac myosin, α and β (Gargey et al. 2019). The sequence homology of the myosin head of these isoforms is 93%, but the sequences of loop 1 are different (α isoform, 197SIAAIGDRGKKDNANANKGTLE218, and β isoform, 197VIAAIGDRSKKDQSPGKGTLE217). The modeling and computer simulations showed that loop 1 in the β isoform forms a salt bridge D208:K450, which restricts loop 1 motion (Figure S1). Both of these residues are present in the α and β isoforms, but the salt bridge is absent in the α isoform. The rate of ADP release from actomyosin with the α and β isoforms differ significantly, ADP dissociates from actomyosin with the α isoform faster, than from actomyosin with the β isoform (Deacon et al. 2012). We hypothesize that the salt bridge D208:K450, observed in the β isoform, restricts loop 1 motion and this isoform-specific difference of loop 1 flexibility plays a role in the retention of ADP at the active site. We decided to replace both amino acid residues forming the bridge, because of charged residues D203 and R453 nearby, which may interact with K450 or D208 if we design a single residue mutant. We expressed the WT β isoform of the human cardiac myosin and the D208Q:K450L mutant to remove electrostatic interaction between loop 1 and myosin backbone. We studied the rate of ATP-induced actomyosin dissociation and the rate of ADP release from actomyosin in the WT myosin and mutant. We found that the rates of ADP release from the WT and mutant actomyosin are statistically similar and conclude that the flexibility of loop 1 does not contribute to the regulation of ADP release from actomyosin.
Materials and methods
Reagents
N-(1-pyrene)iodoacetamide (pyrene) was from Life Technologies Corporation (Grand Island, NY), phalloidin, ATP, and ADP were from Sigma-Aldrich (Milwaukee, WI). All other chemicals were from Fisher Scientific (Waltham, MA) and VWR (Radnor, PA).
Protein preparation
The β isoform construct of the human cardiac myosin motor domain contains 1–843 residues and a FLAG affinity tag at the C-terminus. Adenoviruses encoded with the wild type and D208Q:K450L myosin mutant were purchased from Vector Biolabs (Malvern, PA), amplified using HEK293 cells (ATCC CRL-1573), and purified using CsCl gradient centrifugation. Recombinant human cardiac myosin construct was expressed in C2C12 (ATCC CRL-1722) mouse myoblast cells. C2C12 cells were grown to a 95% confluence on 15 cm diameter plates and infected with the optimum dosage of virus determined by a viral-titration assay. Cells were allowed to differentiate post-infection and collected 7 days post-infection to extract and purify myosin construct. Collected cells were washed and lysed in the presence of a millimolar concentration of ATP. The cell lysate was incubated with anti-FLAG magnetic beads (Sigma-Aldrich, Milwaukee, WI). Beads were washed and myosin construct was eluted from the beads by 3 × FLAG peptide (ApexBio, Houston, TX). Myosin construct purity was assessed by coomassie-stained SDS–polyacrylamide gels (Figure S2) and protein concentration was determined by measuring the absorbance at 280 nm using extinction coefficient ε280nm = 93,170 M−1 cm−1, calculated using ProtParam tool of ExPASy web server.
Actin was prepared from New Zealand white junior doe (3–4 months of age) rabbit leg and back muscles (Strzelecka-Golaszewska et al. 1980; Margossian and Lowey 1982; Waller et al. 1995). The sex of the animal does not produce any variation in prepared protein (Lexell et al. 1994). F-actin was labeled with pyrene iodoacetamide (Life Technologies Corporation, Grand Island, NY) with the molar ratio 6:1, label:actin. After labeling, actin was cleaned from the excess of label, re-polymerized, stabilized with phalloidin at the molar ratio of 1:1, and dialyzed for 2 days at T = 4 °C against the experimental buffer. Concentration of unlabeled G-actin was determined spectrophotometrically assuming the extinction coefficient ε290nm = 0.63 (mg ml−1)−1 cm−1 (Houk and Ue 1974). Concentration of labeled G-actin and labeling efficiency were determined spectroscopically using the following expressions: [G-actin] = [A290nm–(A344nm·0.127)]/26,600 M−1 and [pyrene] = A344nm/22,000 M−1 (Takagi et al. 2008). Actin labeling efficiency was 60–80%. The experimental buffer contained 20 mM MOPS (3-[N-morpholino]propanesulfonic acid) pH 7.3, 50 mM KCl, 3 mM MgCl2. All reported concentrations are final concentrations.
Acquisition of fluorescent transients
In the ATP-induced actomyosin dissociation experiment, 0.25–1 µM actomyosin was rapidly mixed with ATP solution of variable concentrations. In the ADP inhibition of the ATP-induced actomyosin dissociation experiment, 0.25–1 µM actomyosin was rapidly mixed with the premixed ATP and ADP solution. The concentration of ATP in solution was 0.9 mM and the concentration of ADP varied from 10 to 200 µM. Transient fluorescence of pyrene-labeled actin was measured with a Bio-Logic SFM-300 stopped flow transient fluorimeter (Bio-Logic Science Instruments SAS, Claix, France), equipped with an FC-15 cuvette. The pyrene fluorescence was excited at 365 nm and detected using a 420 nm cutoff filter. Multiple transients were acquired and averaged to improve the signal-to-noise ratio. 8000 points were acquired in each experiment. All experiments were performed at T = 20 °C.
Analysis of fluorescence transients
Obtained transients were fitted with the numerical solution of the system of ordinary differential equations, corresponding to the reaction scheme of ATP-induced actomyosin dissociation (Fig. 2; Eq. S1) and ADP release from actomyosin (Fig. 3; Eq.S2). In both cases, the traces obtained at different nucleotide concentrations for the same protein preparation were fitted simultaneously. Rate constants were obtained in experiments with human cardiac myosin constructs from at least three independent preparations. The transients of the ATP induced actomyosin dissociation were also fitted with the one-exponential function S(t) = So + A·exp[− kobs·(t − t0)], where S(t) is the observed signal at the time t, A is the signal amplitude, t0 is the time before the flow stops, and kobs is the observed rate constant. Transients, obtained for the same actomyosin preparation at different concentrations of the nucleotide were fitted together, assuming the known value of t0, measured in a separate experiment, and the constant value of S0, which depends on the concentration and labeling efficiency of pyrene-labeled actin in the actomyosin preparation. The dependence of the observed rates kobs on the ATP concentration was fitted by a hyperbola, kobs = Vmax·[ATP]/(Kapp + [ATP]), allowing the determination of the maximum rate, Vmax (the horizontal asymptote). The rate constant k+2T is the Vmax, and the equilibrium constant of the collision complex formation K1T is 1/Kapp. To determine the bimolecular rate (K1Tk+2T), the dependence of the observed rates on the ATP concentration was fitted by a straight line at small concentrations of ATP. Transients of the ADP inhibition experiment can be fitted only with a two-exponential function, where the rate constant of the fast process presumably reflects the ATP-induced actomyosin dissociation, and the slower process is governed by ADP release from actomyosin. The fit to the two-exponential function confirms that ADP is not in fast equilibrium with actomyosin, and the reaction (Fig. 3; Eq. S2) must be treated as a parallel reaction. Therefore, obtained transients were fitted with the numerical solution of the differential equations, corresponding to the reaction scheme in Fig. 3, assuming no fast equilibrium of actomyosin and ADP (Fig. 4). All exponential data fits were performed with Origin 8 (OriginLab Corp, Northampton MA). Differential equations were solved numerically using Wolfram Mathematica built-in symbol NDSolve. The numerical solution was fitted to the experimental data using the built-in symbol NMinimize, which searches for a global minimum. We first fitted the data of ATP-induced actomyosin dissociation to obtain fitted parameters k+1T, k−1T, k+2T, k−2T (Fig. 2). All transients from the same protein preparation were fitted simultaneously (globally) since they all have the same intensity of pyrene-actin fluorescence at the origin (the mixing point) and the end of the transient (due to complete actomyosin dissociation). Obtained rate constants were averaged, and mean values were used in the global fits of the transients of the ADP inhibition experiments to determine fitted parameters k+1D and k+1D. Obtained kinetic rates (fitted parameters) were averaged and reported in Table 1 as mean values and standard deviations. Wolfram Mathematica scripts are provided in the Supplementary Information file.
Typical transients obtained in the study. Pyrene labeled actomyosin is rapidly mixed with ATP (60 μM, black dots), or the mixture of ATP and ADP ([ATP] = 900 μM, [ADP] = 100 μM, blue dots). Normalized pyrene actin fluorescence reflects actomyosin dissociation. The transients are fitted to the numerical solution of differential equations (Eqs. S1 and S2), red traces. Fitted parameters are k+1T, k−1T, k+2T, k−2T for the ATP-induced actomyosin dissociation experiment, and k+1D, k+1D for the ADP inhibition experiment. All transients obtained for the same myosin construct preparation are fitted globally with the same set of kinetic constants. Different signal to noise ratio reflects different labeling efficiency of actin. A WT myosin construct, B D208Q:K450L myosin mutant. The rate of actomyosin dissociation is faster in the mutant and a larger portion of transient is hidden in the dead time of the spectrofluorometer
Statistical analysis of the obtained results
The independent two-sample t-test analysis was used to examine whether the mean values of obtained kinetic constants of the WT and the mutant are statistically similar or different. Kinetic constants obtained in fits of different datasets were assumed independent, normally distributed with unequal variances. The null hypothesis was that the mean values of kinetic constants for the WT and mutant are equal. The alternative hypothesis was that the mean values of kinetic constants are not equal. The p value was calculated for determining whether to reject the null hypothesis. A p value less than the significance level of 0.05 indicates that the null hypothesis can be rejected. The statistical significance of results was examined with the statistics package integrated into Origin (OriginLab Corp, Northampton, MA) software.
Results
Mutation D208Q:K450L increases the rate of actomyosin dissociation
ATP binding and actomyosin dissociation were monitored using fluorescence of pyrene labeled actin. Pyrene fluorescence is quenched when actin strongly binds myosin and increases upon actomyosin dissociation. In the case of the WT myosin construct the maximum rate of actomyosin dissociation was achieved at 450–900 μM ATP. In the case of the mutant the rate of actomyosin dissociation is higher. At ATP concentration of 150 μM and higher obtained transients were flat, indicating that the actomyosin dissociation occurs during the dead time of the stopped flow fluorometer. Therefore, for the mutant we opted for a lower concentration range of ATP (15–100 μM). In the assumption of the rapid equilibrium of the ATP-actomyosin collision complex formation (step 1 in Fig. 2) we fitted obtained transients with single-exponential function to determine reaction rates at different concentrations of ATP, and the rates were fitted with a hyperbola to determine Vmax and Kapp (Fig. 5). The rate constant K1Tk+2T was determined directly from the slope of the linear fit of the dependence of reaction rates on ATP concentration (Fig. 5), K1Tk+2T = 2.1 ± 0.2 μM−1 s−1 (WT) and 3.7 ± 0.8 μM−1 s−1 (mutant), p = 0.003, Table 1. We also fitted obtained transients with the numerical solution of the system of differential equations corresponding to the reaction scheme of ATP-induced actomyosin dissociation (Fig. 2). For the WT actomyosin, both fits produced similar values for the rate constant k+2T, 491.5 ± 74.1 s−1 for the fit with a hyperbola, and 406.2 ± 14.1 s−1 for the fit with differential equations. The values for K1Tk+2T were about two times different in the fits (2.1 ± 0.2 μM−1 s−1 and 4.6 ± 1.0 μM−1 s−1 for the fit with a hyperbola and with differential equations respectively), the fit with differential equations suggests tighter binding of ATP and actomyosin in the collision complex than the fit of transients with an exponential function and a hyperbola. For the mutant both fits produced similar results, k+2T = 866.4 ± 317.3 s−1 and 961.0 ± 97.1 s−1, Kapp = 200.9 ± 94.0 μM and 238.9 ± 106.5 μM, K1Tk+2T = 3.7 ± 0.8 μM−1 s−1 and 4.3 ± 2.0 μM−1 s−1 for the fit with a hyperbola and with differential equations respectively. Data for the WT actomyosin were reported in our previous study (Gargey et al. 2021). Obtained rate constants k+1T and k−2T are statistically similar for the WT and mutant (k+1T = 7.4 ± 1.4 µM−1 s−1 (WT) and 9.9 ± 1.0 µM−1 s−1 (mutant), p = 0.184, k−2T = 3.6 ± 2.5 s−1 (WT) and 0.9 ± 0.9 s−1 (mutant), p = 0.366, Table 1). Due to the high standard deviation of the rate constant k−1T for the mutant, it is difficult to conclude if the rate constant k−1T is different in the WT and mutant actomyosin. The high value of the standard deviation of the rate constant k−1T reflects shallow χ-squared surface for this parameter in our fits. Interestingly, the rate constant k+1T is well defined in our fits, which gives equilibrium constant Kapp = 238.9 ± 106.5 µM, corresponding well to the data in Fig. 5. We expected k−1T to be of the same order of magnitude as k+2T because if k−1T ≫ k+2T, that leads to the reduced probability of actomyosin dissociation, and the case of k−1T ≪ k+2T makes the reaction a one-step process with a linear dependence on ATP concentration, which is not observed in our experiments. The rate constants obtained from our experiments clearly show an increased rate of ATP-induced actomyosin dissociation in the mutant. The rate constants k+2T for the WT and mutant are statistically different (k+2T = 406.2 ± 14.1 s−1 (WT) and 961.0 ± 97.1 s−1 (mutant), p = 0.001, Table 1). Therefore, our data show that D208Q:K450L mutation leads to an increased rate of ATP-induced actomyosin dissociation.
ATP-induced actomyosin dissociation. Squares, WT, data from (Gargey et al. 2021), N = 3. Circles, mutant, N = 5. A Observed reaction rate constants at low [ATP], fitted with a straight line, the second-order reaction rate constant is determined from the slope of the line. K1Tk+2T = 2.1 ± 0.2 μM−1 s−1 and 3.7 ± 0.8 μM−1 s−1 for the WT and mutant, respectively. B Reaction rate constants fitted with a hyperbola. For the mutant, the reaction rate is too fast for accurate measure above [ATP] = 100 μM. Rate k+2 T = 491.5 ± 74.1 s−1 and 866.4 ± 317.3 s−1 for the WT and mutant, respectively. Equilibrium dissociation constant Kapp = 214.2 ± 24.9 μM and 200.9 ± 94.0 μM. Data points are mean ± SD. N is the number of biological replicates
Mutation D208Q:K450L does not affect the rate of ADP release from actomyosin
ADP release from actomyosin was monitored via the change of pyrene fluorescence, assuming that ADP release is a slower process than the ATP-induced actomyosin dissociation. In the experiment, actomyosin was rapidly mixed with the premixed ATP and ADP. We kept the concentration of ATP constant (900 µM) and varied ADP concentration (10–200 µM). Previously we found that ADP has a high affinity to actomyosin with human cardiac myosin construct (Gargey et al. 2021), and obtained transients are best fitted with a double exponential function, confirming that ADP is not in fast equilibrium with actomyosin. Therefore, we fitted obtained transients to the solution of the differential equations, corresponding to the reaction scheme of parallel reaction, without the assumption of the fast equilibrium of actomyosin and ADP upon rapid mixing. Traces, obtained at different concentrations of ADP were fitted simultaneously to determine the reaction rate constants (Table 1). We found that for the WT and mutant, the rate constants k+D and k−D are statistically similar [k+D = 26.6 ± 8.1 µM−1 s−1 (WT) and 36.9 ± 19.5 µM−1 s−1 (mutant), p = 0.247, k−D = 199.6 ± 51.8 s−1 (WT) and 144.1 ± 16.8 s−1 (mutant), p = 0.918, (Table 1)]. Our data show that D208Q:K450L mutation does not affect the rate of ADP dissociation from actomyosin.
Discussion
The results of our study do not support our initial hypothesis that the restricted motion of loop 1 affects the rate of ADP release from actomyosin. Instead, we found that the rate of ATP-induced actomyosin dissociation is affected by the removal of the restriction of loop 1 motion. Human cardiac myosin has a high affinity to ADP, and the rate of ADP release is significantly slower than the rate of ATP-induced actomyosin dissociation. Observed in the mutant the faster rate of actomyosin dissociation likely does not change the timing of the actomyosin cycle and the force production in muscle. Obtained our results add information on the role of loop 1 in myosin kinetics, which was of interest recently (Spudich 1994; Murphy and Spudich 1998, 2000; Pereira et al. 2001). We understand that the role of loop 1 might be myosin-specific and our findings may apply solely to human cardiac myosin. By introducing D208Q:K450L mutation we introduced a perturbation in protein function, and we believe that the obtained results may help to clarify the microscopic mechanism of the myosin motor.
ATP binding to actomyosin is a two-step process, formation of the weak collision complex, and then the tight binding, which results in actomyosin dissociation. Two loops at the active site play a role in ATP binding to actomyosin, P-loop and loop switch 1. According to the current view, ATP first binds P-loop and then loop switch 1 closes to stabilize and coordinate MgATP in the active site. It is believed that this motion of loop switch 1 is linked to actomyosin dissociation. Another loop of the active site, loop switch 2, subsequently closes and interacts with loop switch 1 to ensure successful ATP hydrolysis. After ATP hydrolysis, abstracted phosphate is released via the “trapdoor” of destabilized and opened loop switch 1 (Reubold et al. 2003). The fluorescence of pyrene actin reflects a transition from the strong to weak actomyosin binding, therefore the fluorescence change due to actomyosin dissociation can be interpreted as an indicator of the transition of loop switch 1 from the open to the closed state.
Our data show that only actomyosin dissociation step K1Tk+2T is affected while ADP release is unchanged. The data are consistent with an increase in the rate constant of actin dissociation, k+2T but due to the high error in the determination of the rate constant k−1T we cannot be confident that K1T is unchanged. According to the crystal structures of myosin with ADP and ATP analogs, loop switch 1 is closed when the nucleotide is bound to the active site (Fisher et al. 1995; Gulick et al. 1997). The difference between ADP and ATP analogs is in the coordination of the loops and the nucleotide. When ATP analog is bound to the active site, P-loop coordinates γ and β phosphates and Mg ion, and loop switch 1 coordinates Mg ion and γ phosphate. When ADP is bound, all γ phosphate coordinations are lost, and loop switch 1 coordinates only Mg ion whereas P-loop coordinates Mg ion and β phosphate of ADP. Observed faster rate k+2T indicates faster closure of loop switch 1 in the mutant. Since the only difference between the WT and the mutant is the removed electrostatic interaction between loop 1 and myosin backbone, we suggest that increased flexibility of loop 1 in the mutant results in increased dynamics of relative positions of loop switch 1 and P-loop in the mutant. Such increased dynamics of the loops decrease the time required to loop switch 1 close and position MgATP in the active site for successful hydrolysis, assuring a faster rate k+2T. The rate k−D is similar in the WT and mutant, and this similarity indicates that only coordination of ADP by P-loop governs kinetics of interaction of ADP and actomyosin. There is no direct interaction between ADP and loop switch 1, and therefore dynamics of loop switch 1 do not affect the rate of ADP dissociation. According to the “trapdoor” mechanism, loop switch 1 opens upon ATP hydrolysis, therefore, the interaction of loop switch 1 with Mg ion is weak, this interaction does not stabilize loop switch 1 in the closed state and does not affect the retention of ADP in the active site. Thus, our data show that the electrostatic interaction of loop 1 with the backbone of the myosin head affects the relative dynamics of loop switch 1 and P-loop at the active site. Increased dynamics of loop switch 1 and P-loop at the active site affects the rate of loop switch 1 closure and thus the rate of actomyosin dissociation, but it does not affect the rate of ADP dissociation from human cardiac actomyosin.
Obtained rate constants of nucleotide binding to actomyosin k+1T and k+D are lower than the theoretical rate of the diffusion-limited nucleotide binding [255 µM−1 s−1, (Gargey et al. 2021)]. Both MgATP and MgADP complexes possess electric charge [(MgATP)2− and (MgADP)−, (Cowan 1991)], therefore, we conclude that electrostatic repulsion governs the rate of nucleotide binding, and the difference of the reaction rates k+1T and k+D reflects the charge difference of MgATP and MgADP. In the WT actomyosin, the rate of ATP dissociation from the collision complex k−1T is higher than the rate of ADP dissociation k−D. Assuming that the formation of the collision complex reflects only nucleotide binding to P-loop, one can suggest that the faster rate k−1T is the consequence of the charge difference of these nucleotides.
Data availability
Raw data generated during the current study are available from the corresponding author on reasonable request. All analyzed data are included in this published article and its supplementary information file.
Code availability
Origin (OriginLab Corp, Northampton, MA) and Wolfram Mathematica (Champaign, IL) were used in data analysis. Wolfram Mathematica scripts are available in the supplementary information file.
References
Clark R, Ansari MA, Dash S, Geeves MA, Coluccio LM (2005) Loop 1 of transducer region in mammalian class I myosin, Myo1b, modulates actin affinity, ATPase activity, and nucleotide access. J Biol Chem 280(35):30935–30942
Cowan JA (1991) Metallobiochemistry of magnesium. Coordination complexes with biological substrates: site specificity, kinetics and thermodynamics of binding, and implications for activity. Inorg Chem 2740–2747:2740–2747
Deacon JC, Bloemink MJ, Rezavandi H, Geeves MA, Leinwand LA (2012) Identification of functional differences between recombinant human alpha and beta cardiac myosin motors. Cell Mol Life Sci 69(13):2261–2277
Fisher AJ, Smith CA, Thoden JB, Smith R, Sutoh K, Holden HM, Rayment I (1995) X-ray structures of the myosin motor domain of Dictyostelium discoideum complexed with MgADP.BeFx and MgADP.AlF4. Biochemistry 34(28):8960–8972
Furch M, Geeves MA, Manstein DJ (1998) Modulation of actin affinity and actomyosin adenosine triphosphatase by charge changes in the myosin motor domain. Biochemistry 37(18):6317–6326
Gargey A, Ge J, Tkachev YV, Nesmelov YE (2019) Electrostatic interactions in the force-generating region of the human cardiac myosin modulate ADP dissociation from actomyosin. Biochem Biophys Res Commun 509(4):978–982
Gargey A, Iragavarapu SB, Grdzelishvili AV, Nesmelov YE (2021) Electrostatic interactions in the SH1-SH2 helix of human cardiac myosin modulate the time of strong actomyosin binding. J Muscle Res Cell Motil 42(2):137–147
Gulick AM, Bauer CB, Thoden JB, Rayment I (1997) X-ray structures of the MgADP, MgATPgammaS, and MgAMPPNP complexes of the Dictyostelium discoideum myosin motor domain. Biochemistry 36(39):11619–11628
Houk TW Jr., Ue K (1974) The measurement of actin concentration in solution: a comparison of methods. Anal Biochem 62(1):66–74
Lexell J, Jarvis JC, Currie J, Downham DY, Salmons S (1994) Fibre type composition of rabbit tibialis anterior and extensor digitorum longus muscles. J Anat 185(Pt 1):95–101
Margossian SS, Lowey S (1982) Preparation of myosin and its subfragments from rabbit skeletal muscle. Methods Enzymol 85:55–71
Murphy CT, Spudich JA (1998) Dictyostelium myosin 25–50K loop substitutions specifically affect ADP release rates. Biochemistry 37(19):6738–6744
Murphy CT, Spudich JA (2000) Variable surface loops and myosin activity: accessories to a motor. J Muscle Res Cell Motil 21(2):139–151
Pereira JS, Pavlov D, Nili M, Greaser M, Homsher E, Moss RL (2001) Kinetic differences in cardiac myosins with identical loop 1 sequences. J Biol Chem 276(6):4409–4415
Reubold TF, Eschenburg S, Becker A, Kull FJ, Manstein DJ (2003) A structural model for actin-induced nucleotide release in myosin. Nat Struct Biol 10(10):826–830
Spudich JA (1994) How molecular motors work. Nature 372(6506):515–518
Strzelecka-Golaszewska H, Prochniewicz E, Nowak E, Zmorzynski S, Drabikowski W (1980) Chicken-gizzard actin: polymerization and stability. Eur J Biochem 104(1):41–52
Sweeney HL, Rosenfeld SS, Brown F, Faust L, Smith J, Xing J, Stein LA, Sellers JR (1998) Kinetic tuning of myosin via a flexible loop adjacent to the nucleotide binding pocket. J Biol Chem 273(11):6262–6270
Takagi Y, Yang Y, Fujiwara I, Jacobs D, Cheney RE, Sellers JR, Kovacs M (2008) Human myosin Vc is a low duty ratio, nonprocessive molecular motor. J Biol Chem 283(13):8527–8537
Waller GS, Ouyang G, Swafford J, Vibert P, Lowey S (1995) A minimal motor domain from chicken skeletal muscle myosin. J Biol Chem 270(25):15348–15352
Acknowledgements
We thank the anonymous reviewers for critical reviews of an earlier version of the manuscript.
Funding
This work was supported by the National Institutes of Health, Grant No. HL132315.
Author information
Authors and Affiliations
Contributions
YN designed research, AG and YN performed research and wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Authors declare no conflict of interest.
Ethical approval
Actin was produced from rabbit skeletal tissue. All experimental protocols were approved by the Institutional Animal Care and Use Committee of UNC Charlotte and all experiments were performed in accordance with relevant guidelines and regulations.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gargey, A., Nesmelov, Y.E. Electrostatic interaction of loop 1 and backbone of human cardiac myosin regulates the rate of ATP induced actomyosin dissociation. J Muscle Res Cell Motil 43, 1–8 (2022). https://doi.org/10.1007/s10974-021-09611-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10974-021-09611-z