Abstract
The present study investigates the effect of the biodegradation time on the properties of condensation segmented nonisocyanate polyurethanes (NIPUs) using the aerobic strain of Gordonia alkanivorans. Polyurethane samples were prepared from phenolsulfonic acid, benzoic acid, polytetramethylene glycol (PTMG 1000) or oligooxypropylene triol (G 1000) and different amounts of formaldehyde. The emulsifying activity, protein concentration, pH, tensile strength and strain at break were evaluated as function of biodegradation time and formaldehyde content. The existence of a segmented structure was confirmed by the existence of two phase transitions in the glass transition temperatures of flexible segments and melting temperatures of hard segments. The tensile strength (TS) of NIPUs based on PTMG 1000 and 50% excess of formaldehyde decreased by about 20%, 60% and 95% after 10, 21 and 90 days of biodegradation. TS decrease was less pronounced for NIPUs prepared from G1000. The 21 days biodegradation caused the most significant changes in the pH, emulsifying activity and protein concentration. The highest degree of cross-linking induced by an excess of formaldehyde led to the highest resistance of NIPU to Gordonia alkanivorans. The DSC, FTIR, SEM and element concentration analyses demonstrated that biodegradation occurred mainly in hydrolysis sensitive hard segments and, to a lesser extent, in flexible oligomerol segments.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
During the last few decades, various research groups around the world have shown great interest in biodegradable conventional polyurethanes (PURs). The investigations concerned mainly the biodegradation of isocyanate based polyaddition PURs [1, 2, 3, 4, 5, 6, 7, 8, 9]. Due to their versatile structure and characteristic performance properties, PURs belong to the most popular group of polymeric materials which are used in applications such as rigid and flexible foams, coatings and biomaterials. Their wide use makes recycling and biodegradability of great importance.
Several conducted studies have shown that the biodegradation process of polymeric materials depends on biological factors, such as bacteria, fungi and enzymes. It is also influenced by environmental factors such as humidity, the presence of oxygen, pH, temperature and the presence of minerals that can affect degradation activity of microorganisms [1, 2, 3, 4, 5 , 6, 7, 8, 9]. Biodegradation of polymers by means of the latter causes the compounds to be broken down into smaller units, which act as a carbon source and lead to the decrease of the mechanical properties. There are many studies on the degradation of polyester polyurethane by microorganisms, especially by fungi. The presence of ester and urethane bonds in the backbone of PURs makes them sensitive to hydrolysis by the enzymes produced by microorganisms, which may act as a carbon source and lead to a decrease in the tensile strength [10, 11, 12].
The first study on the biodegradation of PUR with fungi was carried out more than four decades ago by Darby and Kaplan [5]. Their results showed that polyester based on PUR were very sensitive to biodegradation with Aspergillus niger and Aspergillus flavus, whereas polyether PURs were highly resistant to fungal attack. The biodegradation susceptibility of the polyether PUR was associated with the number of adjacent methylene groups in the main chain. Since then, various fungi and bacteria have been used for the degradation of polyester based PURs. Nakajima-Kambe et al. [6] have shown that the bacterial degradation of polyester PURs was mainly attributed to hydrolysis of the ester bonds by esterases. They isolated and used the bacteria Comamonas acidovorans, which utilizes polyester PUR as its only source of carbon and nitrogen. Obtained results also confirmed that the products of biodegradation would diethylene glycol and adipic acid. Obtained FTIR results by Kay et al. [7] showed that the degradation of PUR was due to the hydrolysis of the ester linkages. Russell et al. [8] investigated the effect of several endophytic fungi on the degradation of polyester PUR. Microorganisms showed the ability to efficiently degrade the polymer in both solid and liquid suspensions. Moreover, it was demonstrated that two Pestalotiopsis microspora utilized PUR as a sole carbon source under both aerobic and anaerobic conditions. The degradation of a polyester PUR by a newly isolated soil bacterium, Bacillus subtilis strain MZA-75 through enrichment technique was confirmed by Shah et al. [9]. Scanning electron microscope (SEM) analysis revealed the appearance of cracks on the surface of samples after biodegradation. However, the Fourier Transform Infrared Spectroscopy (FTIR) spectra showed a decrease in the ester functional groups. Gel permeation chromatography (GPC) data indicated an increase in the polydispersity index, which indicates chain scission as a result of microbial degradation.
While most studies have been carried out with bacteria and fungi extracted from the soil, few investigations have concerned the degradation of PURs by microorganisms isolated from plants [13, 14]. Nakkabi et al. [13] studied the biodegradation of a commercial poly(ester-urethane) by using isolated microorganisms from cedar wood. FTIR data showed the disappearance of the characteristic peak of urethane of PUR samples after biodegradation. The loss of the peak was associated with the hydrolysis of the ester bond in the urethane linkage, confirming the effective degradation of PUR by microorganisms. However, Aldila et al. [14] investigated the degradation PUR by bacteria isolated from decayed teak wood. They analysed the biodegradation of the structure of PUR by FTIR spectroscopy. The results obtained confirmed the disappearance of the peak characteristic of the urethane bond, thus explaining the biodegradation process.
In order to reduce the use of petroleum-based ingredients as well as increase environmental protection, the utilization of renewable resources for the preparation of biodegradable polymeric materials has been the subject of increasing interest during these last decades. Due to their minimal effect on the environment, their versatile structures and biodegradability, bio-based PURs have been widely used in recent decades in several fields [15, 16, 17, 18, 19, 20, 21, 22]. The biodegradation of a PUR foam prepared from castor oil was explored by Cangemi et al. [15]. The FTIR results showed changes in absorption band at 1042 cm−1 of ester groups, confirming the biodegradation of the sample. However, Oprea [17] studied the fungal biodegradation of PUR elastomers based on polyethylene glycol and castor oil. The effects of the hard segment of the PUR elastomers on the fungal biodegradation behaviour was investigated. The biodegradation of PUR elastomer samples was confirmed by the decrease of the tensile strength and strain at break. The strain at break decreased from 670–1180% to 500–680% and the tensile strength decreased from 11.5–27.5 MPa to 4–11.5 MPa after 130 days of fungal biodegradation. Yeganeh and Hojati-Talemi [18] prepared and studied the physical, mechanical and viscoelastic properties of a biodegradable PUR networks based on castor oil (CO) and polyethylene glycol (PEG) as potential candidates in biomedical applications. Obtained results showed that the biodegradation process and the mechanical properties of PURs were affected by the content of PEG or CO in PUR samples. The biodegradability of PUR based on polyols from renewable resources such as soya-based vegetable oils was also studied by Orgilés-Calpena et al. [19]. The PURs were synthesised from an aromatic, 4,4′-diphenylmethane diisocyanate and an aliphatic, 1,6-hexamethylene diisocyanate, and 1,4-butanediol as a chain extender. The authors defined the optimal amount of soybean oil to give samples with maximum adhesion strength. Gogoi and Karak [20] have synthesized a sustainable hyperbranched PUR using polyphenolic tannic acid instead of vegetable oil as the bio-based component. The prepared PUR showed improved thermal stability and mechanical properties such as tensile strength and strain at break. It also underwent biodegradation by Pseudomonas aeruginosa.
An interesting investigation concerned the biodegradation of PUR prepared from polyethylene glycol (PEG) which was mixed with plant components such as molasses, lignin, woodmeal, or coffee grounds [23]. The study showed that the tensile and compressive strengths as well as glass transition temperature of the PUR increased with increasing plant content. It was also shown that the PURs prepared from plants were biodegradable in soil.
It should be noted that although nonisocyanate polyurethanes (NIPUs) have been widely investigated during the last few decades, only few studies have focused on their biodegradation [24, 25, 26, 27, 28]. PURs with potential application as biodegradable scaffold were prepared from carbonated soybean oil (CSBO) and selected amine curing agents through a nonisocyanate route by Jalilian and Yeganeh [24]. CSBO was prepared from epoxidized soybean oil and carbon dioxide gas using a catalyst. The chemical structure of obtained NIPUs was attested by FTIR spectroscopy and their hydrolytic degradation was confirmed. Farhadian et al. [25] synthesized nonisocyanate poly(ester amide/urethane) by reacting a synthesized carbonated sunflower oil (CSFO) without solvent with amines obtained from castor oil and oleic acid. Obtained NIPU networks which showed an excellent thermal stability, a low water absorption and a capacity for biodegradation by the presence of aliphatic ester groups are suitable for biomedical applications.
Recently we have investigated the biodegradation of nonisocyanate condensation polyurethanes synthesized from phenol sulphonic acid, various oligoetherol, urea and formaldehyde [27, 28].
The purpose of the present work was the study of the structure effect on the properties and biodegradation of linear and branched nonisocyanate condensation PURs based on benzoic acid.
Materials and methods
Materials
The raw materials used for the preparation of NIPUs:
-
Phenol sulfonic acid in form of 65% aqueous solution from Sigma Aldreich Chemie GmbH Riedest, Steinheim, Germany
-
Benzoic acid, from Schuchardt, Munchen, Germany;
-
Polytetramethylene glycol with molecular weight of 1000 g/mol from Du Pont Co., Germany;
-
Oligooxypropylene triol (trade name Rokopol G1000) from Rokita (Brzeg Dolny, Poland), with an average molecular weight of 1000 g/mol and hydroxyl number of 118 mg KOH/g;
-
Formalin with concentration 34–37wt. %, from “Zakłady Azotowe” in Tarnow, Poland;
-
Tetrabutyl titanate, from Schuchardt, Munchen, Germany;
-
Urea and ethyl urethane, from POCh Co., Gliwice, Poland.
Preparation of condensation nonisocyanate polyurethanes
Nonisocyanate condensation polyurethanes (NIPUs) were synthesized from urea, phenol sulfonic acid, benzoic acid, formaldehyde and PTMG 1000 or G 1000.
The prepared NIPUs had a segmental structure with flexible and hard segments.
In the first step, compounds containing the flexible segments were prepared by the amidation reaction of the hydroxyl groups of PTMG 1000 or G 1000 using ethyl urethane to give carbamates according to the procedure described in our previous work [29, 30].
In the next step, the compounds containing the hard segments were synthesized from urea, formaldehyde, phenolsulfonic acid and benzoic acid.
Finally, one mole of oligomer with hard segments was mixed with one mole of carbamate based on PTMG 1000 or G 1000 and formaldehyde (in the form of formalin). The stoichiometric amount of formaldehyde was used to carry out the polycondensation reaction of the mixtures containing hard and soft segments while, the excess of formaldehyde was added to crosslink the prepared polyurethane samples. The formed thin layers were heated at 50 °C for 2 h. As a result of the heating, thin transparent and elastomeric films were obtained. The compositions of synthesized NIPUs is presented in the Table 1. The structure of nonisocyanate polyurethanes based on PTMG and G 1000 is shown in Scheme 1.
The obtained polymers were subjected to 90 days biodegradation under the influence of aerobic bacterial strain—Gordonia alkanivorans.
Biodegradation
The bacterial strain was isolated from soil contaminated with petroleum oil and deposited in the culture collection. The degradation factor was selected for its high degradation activity and ability to produce biosurfactants.
The biodegradation process of NIPUs was carried out in mineral medium (with the following composition: glucose 1.0 g, yeast extract 1.0 g, ammonium chloride 1.5 g and sodium hydrogene phosphate NaH2PO4 0.75 g) under immersion culture. Initially, the pH of the medium was adjusted to 6.7 before autoclaving. Then the polymer pieces were previously subjected to 1-h surface sterilization by UV radiation. The prepared samples were then introduced into the culture medium (24-h inoculum of the Gordonia alkanivorans strain). The biodegradation process was carried out on a rotary shaker at 160 rpm at 30 °C for 90 days. At the same time, so-called control tests were carried out that did not contain a bacterial strain.
Evaluation of NIPU properties after biodegradation process
Evaluation of physical properties
The biodegradability of the NIPU samples with Gordonia alkanivorans was determined by changes of pH, emulsifying activity (Pearce and Kinsella’s method, 1978) protein concentration (Bradford, Bradford’s method, 1976) as well as weight loss.
The variations of tensile strength and strain at break of NIPU samples were determined at room temperature according to DIN EN ISO 257 using an Instron 5566 tensile machine at a deformation rate of 100 mm/min. The samples used for testing had 10 cm in length and 1 cm in width.
Characterization
The PUR samples were filtered under vacuum and then washed with 2 M sodium hydroxide (to remove bacterial cells) and distilled water. Then the samples before and after biodegradation were dried for 24 h at room temperature and analyzed by the following methods:
-
FTIR was performed by using Nicolet iN10 (Thermo Scientific, US) spectrometer recording the IR spectra from 4000 cm−1 to 700 cm−1 with a resolution of 4 cm−1. The test was carried out with ATR mode with Germanium ATR crystal.
-
Differential scanning calorimetry (DSC) measurements were carried out on sample size ~ 10 mg by using Mettler Toledo apparatus in a nitrogen atmosphere. The samples were heated from -70 to 250 °C at a rate of 10 °C/min, then cooled to -70 °C at a rate of 10 °C/min and finally heated to 250 °C at the same rate of 10 °C/min.
-
The morphology of samples was investigated with scanning electron microscope Nova Nano SEM 450 (FEI company, Thermo Fischer Scientific (the Czech Republic) operating in high vacuum mode and accelerating voltage of 5 kV.
Results and discussion
pH analysis
The change in pH is used to assess the degree of polymer biodegradation carried out using specific types of microorganisms. Figure 1 shows the effect of the biodegradation time using Gordonia alkanivorans on the pH of culture medium in the form of NIPU obtained by using polytetramethylene glycol with an average molecular weight of 1000 (designated PTMG 1000) as the oligomerol and different amount of formaldehyde. Significant changes in pH values were observed for all tested NIPU samples, confirming thus the occurrence of the biodegradation process. The most significant changes in pH values of culture medium were observed in all samples after 21 days of experiment, which may suggest the intensification of the biodegradation process at this stage. Moreover, it should be pointed out that the ΔpH of samples submitted to biodegradation during 21 days decreased with increasing amount of formaldehyde. On the opposite, it was found that the formaldehyde content did not affect the ΔpH after 10 and 90 days of NIPU samples biodegradation.
The pH of the culture medium in the form of polyurethanes (NIPU) obtained using oligooxypropylene triol with an average molecular weight of 1000 (designated G 1000) is presented in Fig. 2 as function of biodegradation time using Gordonia alkanivorans and excess of formaldehyde. The bacterial activity was clearly marked after 21 days of biodegradation of G1000 based NIPU samples, analogically to previously described results for PTMG 1000 based NIPU as shown in Fig. 1. The use of excess formaldehyde led to a lower susceptibility of NIPU samples to biodegradation in comparison to NIPUs prepared from stoichiometric amount of formaldehyde. This was confirmed by smaller pH changes for the samples which were submitted to 21 and 90 days of biodegradation and prepared with 10% and 50% excess of formaldehyde. The increase in ΔpH values during biodegradation may be connected not only to the polymer structure but also to the presence of biosurfactants produced by Gordonia alkanivorans strain in the culture medium. The lower the ΔpH, the more resistant the NIPU sample is to Gordonia alkanivorans. At the onset of biodegradation (not exceeding seven days), alkaline metabolites may be released, which does not allow a clear interpretation of the biodegradation process on the basis of changes in pH [5, 29]. Thus, the changes in pH measured after biodegradation times greater than 7 days (i.e. 10, 21, 90 days, Fig. 1 and Fig. 2) already explain the studied phenomenon.
The summary of above pH results suggested that NIPU samples based on PTMG 1000 and G1000 with excess of formaldehyde were the most resistant to bacteria after 21 and 90 days of biodegradation. Moreover, it can be noted that the higher the amount of formaldehyde used, the greater the resistance of the tested samples to microorganisms. Gautam et al. [31] found that, the pH of polyester PUR waste increased steadily over a period of 12 days of biodegradation using Pseudomonas chlororaphis. They confirmed that polyester PUR can be successfully biodegraded by Pseudomonas chlororaphis under defined laboratory conditions. The analysis of breakdown products during the biodegradation of polyester PUR confirmed the formation of adipic acid as confirmed by the results of Nakajima-Kambe et al. [32].
Emulsifying activity
The effect of biodegradation time on the emulsifying activity of NIPUs prepared from different amounts of formaldehyde and types of polyols is shown in Fig. 3 and Fig. 4. Emulsifying activity is defined as the ability of bacteria to produce biosurfactants. A higher value of emulsifying activity indicates that a greater amount of biosurfactants is produced, i.e. the greater bacterial growth, and therefore the greater degree of the sample biodegradation.
It can be noted that maximum values of emulsifying activity have been reached for all tested NIPU samples after 21 days of biodegradation. High emulsifying activity values (2.0 ÷ 2.5 for NIPU samples based on PTMG 1000 and 1.7 ÷ 1.8 for samples prepared from G 1000 were observed, confirming the maximum rate of biodegradation of the polymer samples.
Moreover, it is seen that the excess of formaldehyde influenced the value of the emulsifying activity after 21 days of biodegradation. The higher the amount of formaldehyde, the lower the emulsifying activity. The two types of NIPU with the highest degree of crosslinking (using a 50% excess of formaldehyde) were the most resistant to the Gordonia alkanivorans.
The values of emulsifying activity of the tested NIPU samples were comparable after 10 and 90 days of biodegradation and significantly lower after 21 days of testing. This clearly indicated a lower activity of the strain after 10 and 90 days of testing, regardless of the amount of formaldehyde involved during the preparation of NIPU samples.
Microbial growth analysis
The Bradford method is the one of the methods used for the assessment of microbial growth during the biodegradation process. This growth was estimated indirectly by measuring protein concentration [33]. During the analysis, the complex between the Coomassie Brillant Blue dye (Brandford reagent) and the protein is created. The ionic bonds are formed between the dye and amino groups of the protein. The intensity of the color is proportional to the amount of protein, and the absorbance value is proportional to the protein concentration. Hence, this method has been chosen to the mark of the biodegradation kinetic of PURs.
Figure 5 shows the protein absorbance of NIPU samples prepared from PTMG 1000. The highest values of measured values were recorded after 21 days of biodegradation. This may suggest the highest rate of polymer biodegradation. The addition of formaldehyde slightly affected the protein concentration values (except for the NIPU samples containing 50% excess formaldehyde and subjected to 21 days of biodegradation process), regardless of the biodegradation time. It should be remembered that the tested NIPUs constitute the medium for the bacteria used for the biodegradation process. Furthermore, it can be noted that the higher the formaldehyde content, the lower the protein absorbance value. This is probably due to the increasing degree of polymer cross-linking induced by increased amount of added formaldehyde.
The variation of protein concentration for NIPUs prepared from G 1000 and various amounts of formaldehyde is presented in Fig. 6. Obtained results are analogous to those of NIPUs prepared from PTM G 1000 (Fig. 5). It should be noted that the fastest biodegradation process took place after 21 days of testing, which confirmed the highest values of protein concentration for all based G1000 based NIPUs. However, a direct relationship can be found between the degree of cross-linking of PURs and their biodegradability. The higher formaldehyde content, the slightly lower the protein concentration, and hence the better sample sensitivity to the used bacteria.
Weight loss
The weight loss was carried on PTMG 1000 and G1000 based NIPU samples as function of biodegradation time and formaldehyde content. As shown in Fig. 7, the tested samples showed a relatively small weight loss caused by the action of microorganisms during the first 14 days of biodegradation followed by a significant increase for the period from 21 to 90 days. Samples of the two polymers showed comparable trends in weight loss changes during biodegradation. Moreover, one can observe that, the amount of formaldehyde used in the synthesis of NIPU affected the degradation tendency of NIPUs. The resistance of NIPU prepared from PTMG 1000 to Gordonia alkanivorans increased with increasing amount of formaldehyde), thus confirming the positive effect of polymer cross-linking on its resistance to microorganisms. In the case of G 1000, the highest resistance to Gordonia alkanivorans was exhibited by the sample containing 10% excess of formaldehyde. On the other hand, polyurethanes synthesized from a stoichiometric amount and 50% excess of formaldehyde showed a similar resistance to bacteria and slightly lower than the NIPUs with 10% excess of formaldehyde. The tendency for weight loss appeared to be consistent with the variation in protein concentration and emulsifying activity during the biodegradation process.
Tensile properties
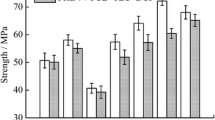
The mechanical properties were evaluated aiming at confirming the biodegradation of the samples. Figure 8 shows the effect of biodegradation time and excess of formaldehyde on the tensile strength (TS) of NIPUs prepared from PTMG 1000. It can be observed, that the strength of all tested NIPU samples decreased with increasing biodegradation time, regardless of formaldehyde content. However, the most significant decrease was obtained with NIPU samples based on excess of formaldehyde. It decreased by approximately 20%, 60% and 95% after 10, 21 and 90 days of biodegradation under conditions of maximum excess of formaldehyde (i.e. 50%). Similar results were obtained for NIPUs prepared from G1000 as shown in Fig. 9. The noticeable decrease was more pronounced and reached 25%, 60% and 90% after 10 days, 3 weeks and 90 days of biodegradation, respectively, compared to undegraded sample. The degradation effect was most probably related to the base compounds that were used for the preparation of NIPUs.
The reduction in the tensile strength of polyester polyurethane has been explained differently by other researchers depending on the ingredients used [10, 12, 17]. According to Barratt et al. [10], the decrease in tensile strength of polyester PUR was caused by microbial activity and cracks formation and not by chemical hydrolysis of the polymer. However, Oprea [17] attributed the significant decrease in tensile strength of polyurethane elastomers caused by 130 days of fungal biodegradation to the chain scission of macromolecules and the formation of pores in the films.
The strain at break of NIPUs prepared from PTMG 1000 and G1000 is presented in Figs. 10 and 11 respectively, as a function of biodegradation time and formaldehyde content. In Fig. 10, it can be seen that the strain at break of the NIPU based on PTMG 1000 decreased with the increase in the biodegradation time and that the excess of formaldehyde had no significance. It appears that a 50% excess of formaldehyde led to the highest degree of NIPU cross-linking, which corresponded to the smallest strain at break. The greater the excess of formaldehyde, the smaller the strain at break and hence the higher the degree of crosslinking of the NIPUs. As expected, the most significant decrease in strain at break associated with the degradation of the tested samples was observed after 21 and 90 days of biodegradation. Similar results on a substantial decrease in the tensile strength and the strain at break of NIPUs upon exposure to microorganisms have been reported by other researchers [6, 7, 34, 35]. In the G1000 based NIPUs (Fig. 11), the amount of incorporated formaldehyde clearly affected the strain at break of the samples. The greater the amount of formaldehyde added, the smaller the strain at break, and hence the greater the degree of cross-linking of the NIPU samples. The decrease of the strain at break of polyester polyurethanes was due to the hydrolysis of the ester linkages [7, 9].
Analogically to the previous data shown in Fig. 10, the fastest biodegradation occurred after 21 and 90 days, resulting in the greatest decrease in the estimated property. However, after 90 days of biodegradation, the strain at break reached insignificant values and confirmed the complete degradation of all tested NIPU samples and no influence of the amount of formaldehyde used. A high degree of polymer chain cross-linking logically leads to high strength and low elongation at break due to restrictions in chain movement. The significant decrease in strain at break may also be associated with significant sample weight loss after 90 days of biodegradation (Fig. 7).
The NIPU synthesized from G 1000 and stoichiometric amount of formaldehyde having the best biodegradability and tensile strength as well as satisfactory strain at break, could be considered as the best composition.
Differential scanning calorimetry
The Differential Scanning Calorimetry (DSC) thermograms of NIPU samples before and after 90 days of biodegradation are shown in Fig. 12. The samples were prepared from G 1000 (symbol I), PTMG 1000 (symbol II), stoichiometric amount of formaldehyde (symbol a) and 50% excess of formaldehyde (symbol c). Thermograms of samples before biodegradation and subjected to sterilization (control sample) were designed with 0, while samples after 90 days of biodegradation are designed with letter B.
The presence of two phase transitions in the glass transition temperatures of the flexible segments (Tg) and the melting temperatures of the hard segments (Tm in the range of 230 − 250 °C) observed during the first heating, confirmed the segmented structure of the tested NIPUs. However, due to the action of Gordonia alkanivorans, the melting temperatures were reduced to approx. 180 °C in most of the tested samples (Ia, Ic and IIc), indicating the degradation process of the NIPU hard segments.
However, we can observe less visible change in the hard segments of NIPU based on G 1000 and stoichiometric amount of formaldehyde (sample IIc), which explains the greater resistance of these samples to bacteria. We did not notice significant changes in the Tg values of NIPU based on PTMG 1000 and G 1000 and the stoichiometric amount of formaldehyde (samples a). The results suggested the resistance of the flexible segments to bacteria. However, a 50% excess of formaldehyde led to a marked increase in the Tg. The greatest increase in Tg was exhibited by samples based on branched G 1000 (sample IIc). A shift in Tg to higher temperatures has been observed during the biodegradation of an oil based polyurethane and has been attributed with the plasticization of the samples as the result of the attack on free or dangling-pendant chains [36].
DSC analysis demonstrated that biodegradation occurred mainly in hard domains sensitive to hydrolysis and to a lesser extent, in flexible domains. The same results were obtained with thermoplastic biodegradable polyurethanese prepared from polyester polyols, aliphatic diisocyanates and chain extenders [37]. The weight loss was directly proportional to hard segment content and soft segment underwent little or no degradation under the test conditions.
Fourier transform infrared spectroscopy
The biodegradation mechanism of the NIPU samples was assessed by the Fourier Transform Infrared Spectroscopy (FTIR method). The FTIR spectra of NIPU samples based PTMG 1000 and G 1000 are shown in Fig. 13 and Fig. 14, respectively.
The most typical bands of synthesized NIPUs were identified in FTIR spectra, confirming the chemical structure of undegraded sterilized samples (control sample). The spectra of unsterilized and undegraded samples designated “before biodegradation” were obtained and compared to control samples to verify the effect of sterilization on the structure of the NIPU samples. All main groups of the flexible segment such as C − O (1100 cm−1), C − O − C (1100 cm−1), and C − H (2850 cm−1) in the aliphatic chain of oligomerols have been identified. The presence of characteristic bands such as C = C (1500 cm−1) and C − H (3000 cm−1) in the aromatic rings of the acids used, O − H (3300 cm−1) from phenol sulfonic acid, N − H groups (3300 cm−1) in urethanes and C = O (1710 cm−1) in urea, N − C (2200 cm−1) in urethane grups and S = O (950 cm−1) in SO3H groups were also present and are associated with the hard segment.
However, it can be seen that some of the bands disappeared, decreased in height or/and were shifted after the biodegradation process. Distinct changes can be observed in the intensity of the characteristic bands of hard segments groups in NIPUs. The peak intensity of O − H and N − H groups in urethanes and urea, N − C in urethane groups decreased after biodegradation of NIPUs and the bands from O − H and N − H in urea groups were slightly shifted. As shown in Fig. 13, similar decreases were observed for sulfonic groups primarily in branched G1000 based NIPUs, which are an integral part of the hard segments of the polymer. It is shown that, the biodegradation in polyurethanes occurred mainly in the crystalline urea regions. Various studies have attributed the biodegradation of polyurethanes by microorganisms of different origin to the reduction or disappearance of the peak characteristic of urethane groups [6, 11, 14, 38, 39, 40].
The decrease in the intensity of these bands or their disappearance indicates that the hydrolytically unstable groups constitutes a breeding ground for the bacteria. It appears that the biodegradation in condensation NIPUs occurred in the urea groups, urethane groups and in sulfonic groups. Similar results were obtained for other NIPUs prepared from 2-hydroxynaphthalene-6-sulfonic acid and phenol sulfonic acid [28].
Surface morphology and element composition
Kay’s research group found a clear relationship with the type of functional groups in the polymer and its tendency to biodegrade [7]. The hydrophilic nature of these groups promotes bacterial activity. In addition, the presence of N, S, C and O atoms in such hydrophilic systems can generate polymer biodegradability. The above assumptions were supported by elemental analysis of biodegradable PUR compositions.
The results of the elemental analysis of the NIPUs prepared from PTM1000 are presented in Fig. 15 as a function of the biodegradation time. After the 90 day of the test, no traces of nitrogen and sulfur were identified in the samples and no reduction in the content of other elements, i.e. C, O. Similar results were obtained for G1000 based NIPUs. It is assumed that, Gordonia alkanivorans attacked hydrophilic groups containing nitrogen and sulfur and to a lesser extent carbon and oxygen. Nitrogen and sulfur in very small quantities constituted the main media for the bacteria used.
Figure 16 shows the Scanning Electron Microscope (SEM) micrographs of NIPU samples obtained from linear PTMG 1000 before and after 90 days of biodegradation. Micrographs of NIPU samples based on branched G 1000 are presented in Fig. 17. As can be seen, the morphologies of both types of NIPUs based on PTMG 1000 and G1000 as well as the stoichiometric amount of formaldehyde were slightly different. As indicated previously, NIPUs obtained from linear oligomerol had a crystalline structure. As can be seen in Fig. 16(a), the PTMG 1000 based NIPU has shown before the biodegradation process a uniformly distributed crystalline phase in the amorphous phase of the flexible segments. Moreover, G 1000 based NIPU samples were also characterized with a similar structure before biodegradation (Fig. 17). However, after 90 days of bacterial action, the structure of both polymers changed with numerous visible voids and micro cracks in the samples. The obtained results may suggest the sensitivity of the crystalline regions to the action of the bacteria used and the occurrence of biodegradation processes mainly in the crystalline phase. Most of the cited works have shown the presence of cracks on biodegraded samples.
Conclusions
The study shows the impact of biodegradation time on the properties of NIPUs using the aerobic Gordonia alkanivorans strain. NIPU samples were prepared from phenolsulfonic acid, benzoic acid, PTMG 1000 or G 1000) and different amounts of formaldehyde. The tensile strength (TS) of NIPUs based on PTMG 1000 and 50% excess of formaldehyde decreased by approximately 20%, 60% and 95% after 10, 21 and 90 days of biodegradation compared to the undegraded sample. The decrease in TS was less pronounced for NIPUs prepared from G1000.
The 21 days of biodegradation caused significant changes in pH values, emulsifying activity and protein concentration. However, the increasing degree of cross-linking due to an excess of formaldehyde led to an increase in resistance to bacteria. Based on DSC, FTIR and elemental analyses, it has been shown that urea, urethane and the sulfonic groups that form the crystalline regions were mainly subject to biodegradation. SEM micrographs showed the presence of voids and micro-cracks in the samples as well as the disappearance of ordered crystalline regions of hard segments.
Both NIPU based on G 1000 or PTMG 1000 were characterized by a two-phase structure before the biodegradation process. The biodegradation took place mainly in the crystalline regions that are not resistant to microorganisms and were sensitive to hydrolysis and to lesser extent in the amorphous phase of G 1000 based NIPU. Nevertheless, NIPU synthesized from G 1000 and the stoichiometric amount of formaldehyde showed the highest biodegradability with optimal mechanical properties (best tensile strength and satisfactory strain at break).
References
Howard GT (2002) Biodegradation of polyurethane, a review. Int Biodeter Biodegr 49(4):245–252
Su SK, Gu JH, Lee HT, Wu CL, Su YR, Suen MC (2018) Biodegradable polyurethanes: Novel effects of the fluorine-containing chain extender on the thermal, physical and water vapor permeation properties. J Polym Res 25(10):227
Loredo-Treviño A, Gutiérrez-Sánchez G, Rodríguez-Herrera R, Aguilar CN (2012) Microbial enzymes involved in polyurethane biodegradation, a review. J Polym Environ 20(1):258–265
Kalita H, Mandal M, Karak N (2012) Biodegradable solvent-induced shape-memory hyperbranched polyurethane. J Polym Res 19(10):9982
Darby RT, Kaplan AM (1968) Fungal susceptibility of polyurethanes. Appl Environ Microbiol 16(6):900–905
Nakajima-Kambe T, Shigeno-Akutsu Y, Nomura N, Onuma F, Nakahara T (1999) Microbial degradation of polyurethane, polyester polyurethanes and polyether polyurethanes. Appl Microbiol Biotechnol 51(2):134–140
Kay MJ, McCabe RW, Morton LHG (1993) Chemical and physical changes occurring in polyester polyurethane during biodegradation. Int Biodeter Biodegr 31(3):209–225
Russell JR, Huang J, Anand P, Kucera K, Sandoval AG, Dantzler KW, Hickman D, Jee J, Kimovec FM, Koppstein D, Marks DH, Mittermiller PA, Núñez SJ, Santiago M, Townes MA, Vishnevetsky M, Williams NE, Vargas MPN, Boulanger LA, Bascom-Slack C, Strobel SA (2011) Biodegradation of polyester polyurethane by endophytic fungi. Appl Environ Microbiol 77(17):6076–6084
Shah Z, Krumholz L, Aktas DF, Hasan F, Khattak M, Shah AA (2013) Degradation of polyester polyurethane by a newly isolated soil bacterium, Bacillus subtilis strain MZA-75. Biodegradation 24(6):865–877
Barratt SR, Ennos AR, Greenhalgh M, Robson GD, Handley PS (2003) Fungi are the predominant micro-organisms responsible for degradation of soil-buried polyester polyurethane over a range of soil water holding capacities. J Appl Microbiol 95(1):78–85
Akutsu Y, Nakajima-Kambe T, Nomura N, Nakahara T (1998) Purification and properties of a polyester polyurethane-degrading enzyme from Comamonas acidovorans TB-35. Appl Environ Microbiol 64:62–67
Cosgrove L, McGeechan PL, Robson GD, Handley PS (2007) Fungal communities associated with degradation of polyester polyurethane in soil. Appl Environ Microbiol 73(18):5817–5824
Nakkabi A, Sadiki M, Fahim M, Ittobane N, Ibnsouda Koraichi S, Barkai H, Elabed S (2015) Biodegradation of Poly (ester urethane)s by Bacillus subtilise. Int J Environ Res 9(1): 157–162
Aldila F, Susilowati A, Setyaningsih R (2019) Polyurethane Degrading Bacteria Isolated from Decayed Teak Wood (Tectona grandis Linn. f.). Jurnal Biodjati 4(2): 225–235
Cangemi JM, Santos AMD, Neto S, Chierice GO (2008) Biodegradation of polyurethane derived from castor oil. Polímeros 18(3):201–206
De Luca BF, Santillo C, Verdolotti L, Campaner P, Minigher A, Boggioni L, Losio S, Coccia F, Iannace S, Lama GC (2020) Greener nanocomposite polyurethane foam based on sustainable polyol and natural fillers: Investigation of chemico-physical and mechanical properties. Materials 13(1):211
Oprea S (2010) Dependence of fungal biodegradation of PEG/castor oil-based polyurethane elastomers on the hard-segment structure. Polym Degrad Stabil 95(12):2396–2404
Yeganeh H, Hojati-Talemi P (2007) Preparation and properties of novel biodegradable polyurethane networks based on castor oil and poly (ethylene glycol). Polym Degrad Stabil 92(3):480–489
Orgilés-Calpena E, Arán-Aís F, Torró-Palau AM, Orgilés-Barceló C (2014) Biodegradable polyurethane adhesives based on polyols derived from renewable resources. Proc Inst Mech Eng L 228(2):125–136
Gogoi S, Karak N (2014) Biobased biodegradable waterborne hyperbranched polyurethane as an ecofriendly sustainable material. ACS Sustain Chem Eng 2(12):2730–2738
Sawpan MA (2018) Polyurethanes from vegetable oils and applications: A review. J Polym Res 25(8):184
Stanzione M, Russo V, Oliviero M, Verdolotti L, Sorrentino A, Di Serio M, Tesser R, Iannace M, Lavorgna S (2018) Synthesis and characterization of sustainable polyurethane foams based on polyhydroxyls with different terminal groups. Polymer 149:134–145
Hatakeyama H, Hirose S, Hatakeyama T, Nakamura K, Kobashigawa K, Morohoshi N (1995) Biodegradable polyurethanes from plant components. J Macromol Sci A 32(4):743–750
Jalilian S, Yeganeh H (2015) Preparation and properties of biodegradable polyurethane networks from carbonated soybean oil. Polym Bull 72(6):1379–1392
Farhadian A, Ahmadi A, Omrani I, Miyardan AB, Varfolomeev MA, Nabid MR (2018) Synthesis of fully bio-based and solvent free non-isocyanate poly (ester amide/urethane) networks with improved thermal stability on the basis of vegetable oils. Polym Degrad Stabil 155:111–121
Ye S, Xiang X, Wang S, Han D, Xiao M, Meng Y (2020) Nonisocyanate CO2-based poly (ester-co-urethane)s with tunable performances: A potential alternative to improve the biodegradability of PBAT. ACS Sustain Chem Eng 8(4):1923–1932
Białkowska A, Mucha K, Przybyłek M, Bakar M (2018) Effect of hard segments content on the properties, structure and biodegradation of nonisocyanate polyurethane. Polym Polym Compos 26(8–9):423–430
Białkowska A, Bakar M, Marchut-Mikołajczyk O (2019) Biodegradation of linear and branched nonisocyanate condensation polyurethanes based on 2-hydroxy-naphthalene-6-sulfonic acid and phenol sulfonic acid. Polym Degrad Stabil 159:98–106
Bakar M, Białkowska A, Szymańska J (2013) Synthesis and evaluation of mechanical and thermal properties of segmented condensation polyurethanes. Plast Rubber Compos 42(5):203–209
Bakar M, Białkowska A (2012) Preparation and properties evaluation of nonisocyanate condensation polyurethanes based on phenol sulfonic and hydroxybenzoic acids J Plast Film Sheet 28(3):257–272
Gautam R, Bassi AS, Yanful EK, Cullen E (2007) Biodegradation of automotive waste polyester polyurethane foam using Pseudomonas chlororaphis ATCC55729. Int Biodeter Biodegr 60(4):245–249
Nakajima-Kambe T, Onuma F, Akutsu Y, Nakahara T (1997) Determination of the polyester polyurethane breakdown products and distribution of the polyurethane degrading enzyme of Comamonas acidovorans strain TB-35. J Ferment Bioeng 83(5):456–460
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72(1–2):248–254
Oprea S, Potolinca VO, Gradinariu P, Joga A, Oprea V (2016) Synthesis, properties, and fungal degradation of castor-oil-based polyurethane composites with different cellulose contents. Cellulose 23(4):2515–2526
Pilch-Pitera B, Wojturska J (2016) Biodegradacja poli(estrouretanów) w symulowanych warunkach kompostowania (in Polish) Polimery 57(11–12): 852–860
Aranguren MI, González JF, Mosiewicki MA (2012) Biodegradation of a vegetable oil based polyurethane and wood flour composites. Polym Test 31(1):7–15
Tatai L, Moore TG, Adhikari R, Malherbe F, Jayasekara R, Griffiths I, Gunatillake PA (2007) Thermoplastic biodegradable polyurethanes: The effect of chain extender structure on properties and in-vitro degradation. Biomaterials 36(28):5407–5417
McCarthy SJ, Meijs GF, Mitchell N, Gunatillake PA, Heath G, Brandwood A, Schindhelm K (1997) In-vivo degradation of polyurethanes: Transmission-FTIR microscopic characterization of polyurethanes sectioned by cryomicrotomy. Biomaterials 18(21):1387–1409
Nakkabi A, Sadiki M, Fahim M, Ittobane N, Ibnsouda Koraichi S, Barkai H, El Abed S (2015) Biodegradation of Poly (ester urethane)s by Bacillus subtilis. Int J Environ Res 9(1):157–162
Dale R, Squirrell DJ (1990) A rapid method for assessing the resistance of polyurethanes to biodeterioration. Int Biodeterior Biodegrad 26:355–367
Acknowledgement
This study was carried out with the support of the CPS project “Strengthening research capacity” (reg. number: CZ.1.05/2.1.00/19.0409) and Program NPU I (LO1504) by the Ministry of Education, Youth and Sports of the Czech Republic and research grant 3269/32/P from the Ministry of Science and Higher Education in Poland.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Bakar, M., Białkowska, A., Hanulikova, B. et al. Effect of structure of nonisocyanate condensation polyurethanes based on benzoic acid on its susceptibility to biodegradation. J Polym Res 27, 379 (2020). https://doi.org/10.1007/s10965-020-02353-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10965-020-02353-5