Abstract
The interaction of the biodegradable ligand, l-glutamic acid N,N-diacetic acid tetrasodium salt (GLDA) with molybdenum(VI) was studied by determining stability constants at pH 6.00, T = 298.15 K, and ionic strength 0.0992 < I/mol·dm−3 < 2.5689 of sodium chloride. The ionic strength dependence of the stability constants was fitted to both extended Debye–Hückel and specific ion interaction models. Job’s method confirmed the formation of one species, MoO3GLDA4−. The values of the stability constants are in agreement with the other data in the literature for the complex formation of aminopolycarboxylic acids with molybdenum(VI). Experimental data were obtained by using UV spectrophotometric method. The formation constant in pure water is 18.96 ± 0.08 on the molal concentration scale.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Molybdenum is the most abundant transition metal in the oceans. The highly soluble molybdate ion, while clearly available, must be transported into cells by methods that differ dramatically from those used to acquire the largely cationic (or organic-bound) di- or trivalent first-row transition metal species present in the marine environment [1].
Much research has been carried out into molybdenum(VI) equilibria and complexes in the last 60 years [2,3,4,5,6,7,8,9,10,11,12,13,14,15,16]. Minubayeva et al. [2] studied molybdic acid ionization under hydrothermal conditions to 300 °C. Hamada et al. [3] investigated molybdenum(VI)–citric acid complexes in aqueous solutions. Schwarzenbach et al. [4] and Bartecki et al. [5] considered that molybdenum concentrations above 10−5 mol·dm−3 lead to the formation of polyanion species. However, Cruywagen et al. [6] have shown that at total concentration of molybdenum 7.5 × 10−5 mol·dm−3, the concentration of polyanions is negligible in comparison to the mononuclear species and it has also been shown that, at molybdate concentrations below 1 × 10−4 mol·dm−3, mononuclear species are present in the solutions [7]. Cruywagen et al. also studied several molybdenum(VI) complex formation reactions with aspartate [8], nitrilotriacetate [9], citrate [10], malate [11], mandelate [12], alizarin red S [13] and also molybdenum(VI) equilibria at high perchloric acid concentration [14]. The following scheme has been accepted for the protonation of simple tetrahedral molybdate ions in the literature [4, 15, 16]
and, for the structural changes as a result of protonation,
Investigations of the hydrolysis of the molybdenyl ion are shown below [16]
Initially it was thought that an increase in coordination number occurs during the first protonation step and, therefore, the formula of \({\text{HMoO}}_{4}^{ - }\) should be more correctly written as \({\text{MoO(OH}})_{5}^{ - }\) [4], but Cruywagen et al. [6] suggested that there is a considerable negative volume change for the second protonation, which is due to an increase in coordination number and therefore the second protonation constant should be regarded as abnormally large and the first as normal. The formulation \({\text{MoO(OH)}}_{5}^{ - }\) was also concluded to be doubtful [17]. Several “correct” formulae for molybdic acid were proposed such as Mo(OH)6, MoO2(OH)2(H2O)2 and MoO3(H2O)3. The “real” formulae of \({\text{HMoO}}_{4}^{ - }\) and H2MoO4 were proposed [18] as MoO3(OH)− and MoO2(OH)2(H2O)2, respectively, on the basis of comparisons of pK values for Mo(VI) and W(VI) with those for analogous complexes of other elements. The formula Mo(OH)6 may be used for convenience to indicate 6 coordination, but electrostatic calculations predict an increase in stability in going from Mo(OH)6 to MoO2(OH)2(H2O)2 and to MoO3(H2O)3 with a regular octahedral structure with no changes in bond length. Experiments have not been able to discriminate between the MoO3(H2O)3 and MoO2(OH)2(H2O)2 forms of molybdic acid. Using first principles molecular dynamics based pKa calculation techniques, it was identified that MoO2(OH)2(H2O)2 is the true solution structure and its OH ligands are the acidic site [19]. The following protonation scheme has also been suggested in the literature [20]
The measured equilibrium constants were reported in the literature [21] and are indicative of stronger hydrolysis of Mo(VI) in comparison to W(VI) [20]. Brown et al. [16] showed that at total concentration of molybdenum 6.0 × 10−3 mol·dm−3 and pH 6 the predominant species is \({\text{MoO}}_{4}^{2 - }\) and hydrolysis does not occur. Therefore, the species \({\text{MoO}}_{4}^{2 - }\) has been taken into account in the current work for the complexation with GLDA at pH 6.00.
GLDA is one of a new generation of chelating agents that have been designed to be a biodegradable, cost-effective product with a high solubility [22]. GLDA or Dissolvine® GL (market name) has received great attention in recent years due to its various applications in different fields as a complexing agent to remove many metal cations. GLDA has been also used successfully in the oil industry, printing ink, industrial cleaning agents, textiles, polymer production, metal plating and electronics, gas sweetening, and pulp and paper treatment due to its versatility in chelation technology [23]. Formulators are able to make very highly concentrated products due to the high solubility of GLDA, which reduces packaging and transport costs [22].
Studies of the thermodynamic properties and applications of electrolytes and salt solutions, which occur in nature, the chemical industry and biology, are very important. Several papers have reported acid–base properties of aminopolycarboxylates in aqueous solution [24,25,26,27,28,29], in particular the study of protonation constants in different media and temperatures, while for their complexation with molybdenum(VI) and tungsten(VI) few data are found and some of them are at a single ionic strength [30,31,32,33,34,35,36,37,38,39,40,41,42,43]. The current research work has been done in order to improve the knowledge of behavior of GLDA, and for this reason we studied the complexation with molybdenum(VI) at pH 6.00, T = 298.15 K, and different ionic strengths 0.0992 < I/mol·dm−3 < 2.5689 of sodium chloride.
2 Experimental Section
2.1 Reagents
Double-distilled water with specific conductance equal to (1.3 ± 0.1) µS·cm−1 was used for the preparation of the solutions. Analytical reagent grade chemicals: 99.5% (w–w) sodium molybdate, 99.5% (w–w) anhydrous sodium carbonate, 99.5% (w–w) sodium chloride, hydrochloric acid titrazole (1 mol·dm−3), sodium hydroxide titrazole (1 mol·dm−3), and potassium hydrogen phthalate, minimum 99.9% (w–w), were purchased from E. Merck and Dissolvine GL-PD-S, 78–82% (w–w) (Scheme I) from Akzonobel and were used without further purification. The NaOH solutions were prepared from the titrazole solutions and their concentrations were determined by several titrations with potassium hydrogen phtalate. The HCl solution was standardized with sodium carbonate solution. Sodium chloride, potassium hydrogen phthalate and sodium carbonate were dried in an oven at T = 383.15 K for two h.
2.2 Measurements
All measurements were performed at T = 298.15 K and different ionic strengths (0.0992 < I/mol·dm−3 < 2.5689) of sodium chloride. A Metrohm pH-meter, model 827, was used for pH measurements. The hydrogen ion concentration was measured with a Metrohm combination electrode, model 6.0228.010. A 0.01 mol·dm−3 hydrochloric acid solution containing 0.0892 mol·dm−3 sodium chloride (for adjusting the ionic strength to 0.0992 mol·dm−3) was employed as a standard solution of hydrogen ion concentration; the same procedure was performed for the other ionic strengths. The calibration was done for the whole pH (pH = – log10[H+]) range used. It was assumed that the hydrogen ion activity coefficient is constant, and therefore the hydrogen ion concentration was calculated from Eq. 1:
The change in liquid junction potential was calculated from Eq. 2:
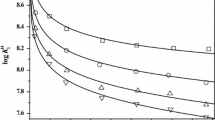
where a and b were determined by measurement of the hydrogen ion concentration for two different solutions of HCl with sufficient NaCl to adjust the ionic media. This was repeated for the other ionic strengths so that the total concentration should be equal to the required ionic strength. Spectrophotometric measurements were performed with a Perkin Elmer Lambda 25 UV–Vis spectrophotometer between 245 and 280 nm in thermostatted 10 mm quartz cells at T = 298.15 K. The measurement cell was of the flow type. A Masterflux pump was used for the circulation of the solution under study between the potentiometric and spectrophotometric cells and simultaneous measurement of the absorbance and pH of the solution. The total concentration (metal + ligand) of 0.004 mol·dm−3 for the UV measurements in the UV range (260–270) nm at constant pH 6.00 and different ionic strengths (0.0992 < I/mol·dm−3 < 2.5689) of sodium chloride gave the best results for the calculations according to Job’s method [44]. The Job procedure is a well-known method in coordination solution chemistry for the determination of the ligand to metal ratio and the details have been reported in the literature [33]. Since the hydrolysis of molybdenum does not occur at pH 6.00, UV measurements for the complex formation reaction have been done at that pH; on the other hand we can be sure to have had only the \({\text{MoO}}_{4}^{2 - }\) species in the solution. Corrected absorbance data for different ionic strengths are gathered in Tables 6, 7, 8, 9 and 10 in the Appendix and are plotted in Fig. 1 for I = 1.0 mol·kg−1.
Corrected absorbance data, Ac, for MoO3GLDA4−, versus the mole fraction of Mo(VI), xmetal, at T = 298.15 K, an ionic strength of 1.0 mol·kg−1 NaCl, and different wavelengths: circles, 260 nm; squares, 265 nm; triangles, 270 nm. All of the lines have been obtained on the basis of the best fit to corrected absorbance data
3 Results and Discussion
3.1 Complexation of Molybdenum(VI) with GLDA at Different Ionic Strengths of Sodium Chloride: Data Refinement
Table 1 summarizes the values of the GLDA protonation constants at T = 298.15 K at different ionic strengths of sodium chloride from our published paper [24]. The following equilibria represent the protonation constant reactions
The values of protonation constants from Ref. [24] have been used for the calculation of stability constants in the current work. Three titrations were performed at each ionic strength, and approximately 110 points have been used for calculations at each ionic strength. The binding of \({\text{MoO}}_{4}^{2 - }\) with GLDA as a 1:1 complex occurs on the basis of the following reaction
where x = 1, y = 0 and z = 1. Therefore the stability constants for the formation of the MoO3GLDA4− species, have been calculated at different ionic strengths of sodium chloride by combination of the following equations:
The values of logarithm of stability constants (log10 β101) for the complexation of molybdenum(VI) with GLDA (which have been calculated by the Microsoft Excel 2010 program according to Eqs. 5–9) at different ionic strengths are shown in Table 2. The corrected absorbance of the complex, the observed absorbance and the molar absorptivity of \({\text{MoO}}_{4}^{2 - }\) are designated by Ac, Aobs, and є0, respectively. Calculations at xmetal = 1.0 give the values of є0. The molar absorptivity values of the complex, є1, were calculated at low mole fraction of the metal, where all the metal ions are in the form of a complex. [MoO3GLDA4−] was calculated at xmetal = 0.5, where the maximum absorbance is observed (Fig. 1). The value of [GLDA] has been used for the calculation of [GLDA4−] according to the determined protonation constants. Insertion of the values of [MoO3GLDA4−], \(\left[ {{\text{MoO}}_{4}^{2 - } } \right]\), [H+] and [GLDA4−] in Eq. 5 made it possible to calculate the values of the stability constants. Conversion of log10β101 (mol·dm−3) to log10β101 (mol·kg−1) has been done according to Eq. 10 [45]:
where Σν is the sum of stoichiometric coefficients of the reaction species.
3.2 Ionic Strength Dependence of Dissociation and Stability Constants on the Basis of EDH and SIT Models
The origin of the negative square root dependence on ionic strength of activity coefficients for electrolyte solutions was first solved in the work of Debye and Hückel [46] whose equation is valid at small salt concentrations. In the Debye–Hückel model the ions and counter ions will form distributions that are different from a random mixture. The ionic strength dependence of the dissociation and stability constants can be obtained on the basis of an EDH model as:
where log10β101 is the formation constant at ionic strength I and log10 \(\beta_{101}^{\text{T}}\) is \({\text{the}}\) formation constant at infinite dilution; C and D are empirical parameters; Δz2 represents the Σ(charges)2 reactants − Σ(charges)2 products, \(\Delta z^{ 2} = \, \left( {{-}\, 4x\, + z\,{-}\,y} \right)^{ 2} \,{-}\,\left( { 6x\, + \,y\, + \, 9z} \right) ({\text{where}}\,x = \, 1,y = \,0{\text{ and}}\,z = \, 1\,\,{\text{in}}\,{\text{this}}\,{\text{work}})\). The Microsoft Excel 2010 program has been used for the calculation of the C and D empirical coefficients which were obtained by minimizing the error function, (U), using the Gauss–Newton nonlinear least-squares method
where ai is an experimental quantity and bi is a calculated one. The results on the basis of the EDH for the stability constants are shown in Fig. 2 and Table 2. The specific ion interaction (SIT) model also uses a Debye–Hückel term for the description of long range electrostatic forces and a virial series expansion in the molalities of the electrolytes to model short range interactions, with specific interaction terms for each type of pair or triple interaction [47]. The SIT describes the activity coefficient γj of an ion j of charge zj in an aqueous solution of ionic strength I [47, 48] as:
DH is a particular form of the Debye–Hückel term used in the SIT model, while ε(j, k, I) is an aqueous species interaction coefficient which describes the specific short range interactions between aqueous species j and k; the molality concentration scale is used.
In the SIT model, the relation between the stability constant of \(( {\text{MoO}}_{3} )_{x} {\text{H}}_{y} {\text{GLDA}}_{z}^{(4z - y) - }\), βxyz, determined in an ionic medium (1:1 salt, NaCl) of the ionic strength, I, and \(\beta_{xyz}^{0}\) (at zero ionic strength) is defined by Eq. 15:
where
∆z2 = – 6, thus the following formula was obtained for the MoO3GLDA4− complex according to the SIT model
The values of log10 \(\beta_{101}^{0}\) at infinite dilution together with ionic strength dependence parameters are summarized in Table 3.
3.3 Case Study
Molybdenum is a metal widely present in foods, water, in multivitamin supplements and used in a variety of industrial operations [49]. This metal is essential for humans at low levels because it is present in the molybdenum enzyme sulfite oxidase, but exposure at high levels can be very dangerous. In a study published by Lewis et al. [50] some biomarkers of molybdenum and several other metals were associated with altered testosterone in men of reproductive age. They report some real data of concentration of molybdenum in blood, urine and serum, and showed that the mean concentration of molybdenum in the urine is around 10−6 mol·dm−3. We used this concentration to model the speciation of molybdenum in urine. The chemical composition of urine is variable and depends on several factors (age, sex, pathologies, etc.) [51]. The composition of the urine in this case study was taken from unpublished data from this laboratory. The components considered are: Na+ (94.6 mmol·dm−3), K+ (28.6 mmol·dm−3), Mg2+ (1.79 mmol·dm−3), and Ca2+ (2.63 mmol·dm−3) as cations, and Cl− (90.4 mmol·dm−3), \({\text{SO}}_{4}^{2 - }\) (9.67 mmol·dm−3), \({\text{PO}}_{4}^{3 - }\) (36.4 mmol·dm−3), citrate (1.48 mmol·dm−3) and oxalate (0.23 mmol·dm−3) as anions; in addition are urea (192 mmol·dm−3) and ammonia (16.4 mmol·dm−3). The pH of the urines vary as a function of the factors above and it can be considered to be in the range 5–7; the ionic strength of this medium, with the above composition, is I (~ 0.3 mol·dm−3). If we considered pH 6 when only the natural components are present, then Mg2+ is present as free cation (~ 40%), MgSO4 (~ 5%), MgCit (~ 20%), MgUrea (~ 4%), MgHPO4 (~ 1%), and MgOx (~ 26%) (Fig. 3), while Ca2+ is present as free cation (~ 42%), CaCit (32%), CaSO4 (~ 5%), CaOx (~ 12%), CaUrea and CaHCit (< 1%). If we consider the presence in urine of molybdenum (1 × 10−6 mol·dm−3), we have the following speciation: Mo(VI) as free ion (~ 50%), CaMo and H2MoCit (~ 20%), MgMo (~ 2.5%), HMo, HMoCit and H3MoCit (1–2%). When we consider the simultaneous presence of Mo(VI) (1 × 10−6 mol·dm−3) and GLDA (1 × 10−6 mol·dm−3) in the urine model at pH 6, then we have the 100% of the Mo(VI) complexed as MoO3GLDA.
3.4 Conclusions and Comparison with Literature Data
According to our knowledge there is no publication in the literature about the complexation of GLDA with molybdenum(VI) at pH 6.00, T = 298.15 K, and 0.0992 < I/mol·dm−3 < 2.5689 of sodium chloride and the dependence of the complexation constant on ionic strength by using EDH and SIT models. As can be observed in Fig. 2, the value of stability constants decreases with the increasing of ionic strength, and around ionic strength of 2.0 mol·dm−3 they tend to increase. The values of log10 \(\beta_{101}^{0}\) (Table 3) on the basis of the EDH and SIT models obtained in the current study are close to each other. Only few data of formation constants with molybdate are present for some aminopolycarboxylates that are listed in Table 4, and as can be seen in Fig. 4, we tried to do a correlation with the numbers of nitrogens and carboxylates groups involved in the protonation and the value of the formation constants (ionic strength 0.1–0.2 mol·dm−3); we found a fairly good correlation Y = 17.95 + 0.2083X with a R2 = 0.9213 (Y represents the values of formation constants while X represents nN*nCOOH). If we apply the correlation for instance in our case with GLDA, we found a value of 18.78 (at ~ 0.1 mol·dm−3) that is in quite acceptable agreement with the value that we found experimentally, 18.38. We applied the correlation also to other ligands (asparagine, glutamine, glutamic acid and cysteine) in the literature, which are gathered in Table 5. The values that we found are fairly acceptable. The aim of this research was not to find out the geometry of the molybdenum complex with GLDA, but we can assume that it is similar to the molybdenum complex with ethylenediaminetetra acetic acid (EDTA), the three coordination sites of MoO3 core are occupied by one nitrogen and two acetate oxygen atoms of GLDA which leads to the formation of the MoO3GLDA4− species but we did not take into account the possible formation of the dimeric species, which is in consistent with the stability constant value (18.76 ± 0.12) for the 1:1 molybdenum complex with EDTA [40].
References
Bertini, I., Gray, H.B., Stiefel, E.I., Valentine, J.S.: Biological Inorganic Chemistry, Structure and Reactivity, 1st edn. University Science Books, Sausalito (2007)
Minubayeva, Z., Seward, T.M.: Molybdic acid ionisation under hydrothermal conditions to 300 °C. Geochim. Cosmochim. Acta 74, 4365–4374 (2010)
Hamada, Y.Z., Bayakly, N., George, D., Greer, T.: Speciation of molybdenum(VI)–citric acid complexes in aqueous solutions. Synth. React. Inorg. Met. Org. Nano. Metal. Chem. 38, 664–668 (2008)
Schwarzenbach, G., Meier, J.: Formation and investigation of unstable protonation and deprotonation products of complexes in aqueous solution. J. Inorg. Nucl. Chem. 8, 302–312 (1958)
Bartecki, A., Dembicka, D.: Spectroscopy of molybdenum(VI) compounds. J. Inorg. Nucl. Chem. 29, 2907–2916 (1967)
Cruywagen, J.J., Heyns, J.B.B.: Equilibria and UV spectra of mono- and polynuclear molybdenum(VI) species. Inorg. Chem. 26, 2569–2572 (1987)
Cruywagen, J.J.: Protonation, oligomerization, and condensation reactions of vanadate(V), molybdate(VI), and tungstate(VI). Adv. Inorg. Chem. 49, 127–182 (1999)
Cruywagen, J.J., Heyns, J.B.B., Rohwer, E.A.: Molybdenum(VI) complex formation. Part 6. Reactions with aspartate in 1.0 mol dm−3 sodium chloride medium at 25 °C. J. Chem. Soc. Dalton Trans. 11, 1713–1717 (1993)
Cruywagen, J.J., Heyns, J.B.B., Rohwer, E.A.: Molybdenum(VI) complex formation. Part 7. Equilibria and thermodynamic quantities for the reactions with nitrilotriacetate. J. Chem. Soc. Dalton Trans. 1, 45–49 (1994)
Cruywagen, J.J., Rohwer, E.A., Wessels, G.F.S.: Molybdenum(VI) complex formation—8. Equilibria and thermodynamic quantities for the reactions with citrate. Polyhedron 14, 3481–3493 (1995)
Cruywagen, J.J., Rohwer, E.A., Van de Water, R.F.: Molybdenum(VI) complex formation. Equilibria and thermodynamic quantities for the reactions with malate. Polyhedron 16, 243–251 (1997)
Cruywagen, J.J., Rohwer, E.A.: Equilibria and thermodynamic quantities for the reactions of molybdenum(VI) and tungsten(VI) with mandelate (α-hydroxybenzeneacetate). J. Chem. Soc. Dalton Trans. 21, 3433–3438 (1995)
Cruywagen, J.J., Heyns, J.B.B., Rohwer, E.A.: Spectrophotometric investigation of the complex formation of molybdate and tungstate with Alizarin Red S. S. Afr. J. Chem. 52, 15–19 (1999)
Cruywagen, J.J., Heyns, B.: Molybdenum(VI) equilibria at high perchloric acid concentration. Polyhedron 19, 907–911 (2000)
Sasaki, Y., Sillen, L.G.: On equilibria in polymolybdate solutions. Acta Chem. Scand. 18, 1014–1019 (1964)
Brown, P.L., Shying, M.E., Sylva, R.N.: The hydrolysis of metal ions. Part 10. Kinetic and equilibrium measurements of molybdenum(VI). J. Chem. Soc. Dalton Trans. 9, 2149–2157 (1987)
Cruywagen, J.J., Heyns, J.B.B.: Spectrophotometric determination of the thermodynamic parameters for the first two protonation reactions of molybdate: an advanced undergraduate laboratory experiment. J. Chem. Educ. 66, 861–863 (1989)
Tytko, K.H.: Structure, bonding and acid–base properties of protonated monometalate ions and polymetalate ions forming chains or rings of corner-sharing MO4 tetrahedra of transition metals of groups V and VI. A theoretical approach. Polyhedron 5, 497–503 (1986)
Liu, X., Cheng, J., Sprik, M., Lu, X.: Solution structures and acidity constants of molybdic acid. J. Phys. Chem. Lett. 4, 2926–2930 (2013)
Pershina, V., Kratz, J.V.: Solution chemistry of element 106: theoretical predictions of hydrolysis of group 6 cations Mo, W, and Sg. Inorg. Chem. 40, 776–780 (2001)
Crea, F., De Stefano, C., Irto, A., Milea, D., Pettignano, A., Sammartano, S.: Modeling the acid–base properties of molybdate(VI) in different ionic media, ionic strengths and temperatures, by EDH, SIT and Pitzer equations. J. Mol. Liq. 229, 15–26 (2017)
Dissolvine Chelates Product Guide. Akzo Nobel Functional Chemicals B.V., Amsterdam (2017)
Kolondyska, D.: The effect of the novel complexing agent in removal of heavy metal ions from waters and waste waters. Chem. Eng. J. 165, 835–845 (2010)
Bretti, C., Majlesi, K., De Stefano, C., Sammartano, S.: Thermodynamic study on the protonation and complexation of GLDA with Ca2+ and Mg2+ at different ionic strengths and ionic media at 298.15 K. J. J. Chem. Eng. Data 61, 1895–1903 (2016)
Bretti, C., De Stefano, C., Foti, C., Sammartano, S.: Acid–base properties, solubility, activity coefficients and Na+ ion pair formation of complexons in NaCl(aq) at different ionic strengths. J. Solution Chem. 42, 1452–1471 (2013)
Bretti, C., De Stefano, C., Foti, C., Sammartano, S.: Total and specific solubility and activity coefficients of neutral species of (CH2)2i–2Ni(CH2COOH)i+2 complexons in aqueous NaCl solutions at different ionic strengths, (0 ≤ I ≤ 5) mol L−1, and 298.15 K. J. Chem. Eng. Data 56, 437–443 (2011)
Bretti, C., Cigala, R.M., De Stefano, C., Lando, G., Sammartano, S.: Understanding the bioavailability and sequestration of different metal cations in the presence of a biodegradable chelant S, S-EDDS in biological fluids and natural waters. Chemosphere 150, 341–356 (2016)
Bretti, C., Cigala, R.M., De Stefano, C., Lando, G., Sammartano, S.: Thermodynamic solution properties of a biodegradable chelant (MGDA) and its Interaction with the major constituents of natural fluids. Fluid Phase Equilib. 434, 63–73 (2017)
Anderegg, G.: Critical survey of stability constants of EDTA complexes. Pure Appl. Chem. 54, 2693–2758 (1982)
Gharib, F., Zare, K., Majlesi, K.: Ionic strength dependence of formation constants, complexation of molybdenum(VI) with glutamic acid. J. Chem. Eng. Data 45, 833–836 (2000)
Majlesi, K., Rezaienejad, S.: Application of specific ion interaction theory and parabolic models for the molybdenum(VI) and tungsten(VI) complexes with NTA and IDA at different ionic strengths. Chin. Chem. Lett. 20, 759–762 (2009)
Majlesi, K.: Ionic strength dependence patterns for the Mo(VI) + NTA and Mo(VI) + EDTA systems. Rev. Inorg. Chem. 26, 507–520 (2006)
Majlesi, K., Gholamhosseinzadeh, M., Rezaienejad, S.: Interaction of molybdenum(VI) with methyliminodiacetic acid at different ionic strengths by using parabolic, extended Debye–Hückel and specific ion interaction models. J. Solution Chem. 39, 665–679 (2010)
Majlesi, K., Rezaienejad, S.: Study on the complexation of molybdenum(VI) with iminodiacetic acid and ethylenediamine-N,N′-diacetic acid by specific ion interaction and Debye-Hückel theories. Chin. J. Chem. 25, 1815–1820 (2007)
Majlesi, K., Rezaienejad, S.: Investigation on the complexation of molybdenum(VI) with d-(−)-quinic acid at different ionic strengths. J. Chem. Eng. Data 56, 3194–3200 (2011)
Majlesi, K.: Determination of solvatochromic regression coefficients for the molybdenum(VI) complex with ethylenediamine-N, N′-diacetic acid by using Kamlet–Abboud–Taft equation. Chin. J. Chem. 28, 1973–1977 (2010)
Majlesi, K., Rezaienejad, S.: Application of specific ion interaction theory and parabolic models for the molybdenum(VI) and tungsten(VI) complexes with NTA and IDA at different ionic strengths. Chin. Chem. Lett. 20, 759–762 (2009)
Majlesi, K., Rezaienejad, S.: Complexation of tungsten(VI) with d-(−)-quinic acid at different ionic strengths. J. Solution Chem. 42, 1770–1781 (2013)
Majlesi, K., Hajali, N.: Complexation of tungsten(VI) with methyliminodiacetic acid at different ionic strengths. J. Solution Chem. 41, 1889–1905 (2012)
Zare, K., Lagrange, P., Lagrange, J.: Determination and comparison of stability constants of vanadium(V), molybdenum(VI) and tungsten(VI) aminocarboxylate complexes. J. Chem. Soc. Dalton Trans. 9, 1372–1376 (1979)
Santos, A., Gama, S., Pessoa, J.C., Oliveira, M.C., Tóth, I., Farkas, E.: Complexation of molybdenum(VI) with bis(3-hydroxy-4-pyridinone) amino acid derivatives. Eur. J. Inorg. Chem. 12, 1728–1737 (2007)
Lund, W.: Stability constants of the molybdenum(VI) complexes of DTPA and TTHA. Anal. Chim. Acta 53, 295–302 (1971)
Kula, R.J.: Solution equilibria and structures of molybdenum(VI) chelates. (ethylenedinitrilo)tetraacetic acid. Anal. Chem. 38, 1581–1584 (1966)
Job, P.: Formation and stability of inorganic complexes in solution. Ann. Chim. Applicata 9, 113–203 (1928)
Weast, R.C.: Handbook of Chemistry and Physics. CRC Press Inc., Boca Raton, FL (1986)
Debye, P., Hückel, E.: The theory of electrolytes. I. Lowering of freezing point and related phenomena. Phys. Z. 24, 185–206 (1923)
Guggenheim, E.A., Turgeon, J.C.: Specific interaction of ions. Trans. Faraday Soc. 51, 747–761 (1955)
Bretti, C., Foti, C., Porcino, N., Sammartano, S.: SIT parameters for 1:1 electrolytes and correlation with Pitzer coefficients. J. Solution Chem. 35, 1401–1415 (2006)
Turnlund, J.R.: Molybdenum metabolism and requirements in humans. Met. Ions Biol. Syst. 39, 727–739 (2002)
Lewis, R.J., Meeker, M.S., Meeker, J.D.: Biomarkers of exposure to molybdenum and other metals in relation to testosterone among men from the United States national health and nutrition examination survey 2011–2012. Fertil. Steril. 103, 172–178 (2015)
Lentner, C.: Geigy Scientific Tables, 8th edn. CIBA-Geigy, Basilea (1983)
Tewari, R.C., Srivastava, M.N.: Potentiometric determination of stepwise stability constants of vanadium, molybdenum and tungsten chelates formed with asparagine and glutamine. Talanta 20, 133–134 (1973)
Cavaleiro, A.M.V.S.V., Gil, V.M.S., Pedrosa De Jesus, J.D., Gillard, R.D., Williams, P.A.: Molybdenum(VI) complexes of (R)-cysteine in aqueous solution. Rev. Port. Quim. 27, 305–306 (1985)
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix
Rights and permissions
About this article
Cite this article
Majlesi, K., Bretti, C., De Stefano, C. et al. Complexation of Molybdenum(VI) with GLDA at Different Ionic Strengths. J Solution Chem 47, 1965–1979 (2018). https://doi.org/10.1007/s10953-018-0825-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10953-018-0825-x