Abstract
Researchers developing software to predict the binding constants of small molecules for proteins have, in recent years, turned to host–guest systems as simple, computationally tractable model systems to test and improve these computational methods. However, taking full advantage of this strategy requires aqueous host–guest systems that probe a greater diversity of chemical interactions. Here, we advance the development of an experimental platform to generate such systems by building on the cyclodextrin (CD) class of hosts. The secondary face derivative mono-3-carboxypropionamido-β-cyclodextrin (CP-β-CD) was synthesized in a one-pot strategy with 87% yield, and proved to have much greater aqueous solubility than native β-CD. The complexation of anionic CP-β-CD with the cationic drug rimantadine hydrochloride was explored using one- and two-dimensional nuclear magnetic resonance; NOESY analysis showed secondary face binding of the ammonium moiety of the guest, based on cross-correlations between the amic acid functionality and the side-chain of rimantadine. Isothermal titration calorimetry was furthermore used to determine the standard Gibbs energy and enthalpy for this binding reaction, and the results were compared with those of rimantadine with native β-CD.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
A key step in many drug discovery programs is the discovery of a small, organic molecule that binds a disease-related protein target. When the three-dimensional structure of the protein is known, such as by X-ray crystallography, structure-based computational methods are often used to guide this step. A range of such methods has been developed [1], from fast docking calculations [2,3,4,5,6,7,8,9,10,11] to slower and more detailed Gibbs energy calculations based on molecular dynamics simulations [12,13,14,15,16,17,18,19,20]. As computers have become faster in recent years, the Gibbs energy approaches have become more practical, and they have yielded encouraging results in real-world applications [21,22,23,24,25]. However, errors persist, and it is essential to track down and correct their causes.
Today, researchers are increasingly turning to host–guest chemistry as a source of simple, computationally tractable model systems to troubleshoot and improve binding free energy (Gibbs energy) calculations [26,27,28,29,30,31,32,33,34,35]. In contrast to protein–ligand systems, host–guest systems are often small and simple enough that conformational sampling and setup issues can often be excluded as major sources of error, leaving the potential function, or force field, as the chief remaining source of deviations from experiment. The fact that Gibbs energy methods still can yield errors, relative to experiment, of several kJ·mol−1 for simple host–guest systems, for which sampling and setup issues can be virtually eliminated, points to a need for a new, more accurate force field. Accordingly, our group has initiated a program of using host–guest binding to test force fields and guide their improvement [34, 36, 37]. We anticipate that force fields optimized based on host–guest binding data will be better suited for computing protein–ligand binding affinities than existing force fields, for which the only experimental data used to adjust parameters are liquid solution properties, hydration Gibbs energies, and conformational distribution data [38,39,40,41,42,43,44,45,46].
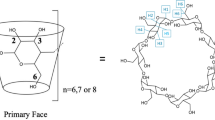
To accomplish this, we need a broad set of data that will systematically probe the variety of chemical interactions that come into play in drug–protein binding. Therefore, we aim to develop new host–guest systems, and are currently focusing on the cyclodextrins (CDs), cyclic oligosaccharide host molecules that have the shape of truncated cones, with a hydrophobic cavity and a hydrophilic outer surface (Fig. 1). These compounds are readily available from commercial suppliers, with CDs made of 6 (α-CD), 7 (β-CD) or 8 (γ-CD) glucose monomers affording containers of different size. The solubility of CDs in water, and their low toxicity, has already enabled their uses in pharmaceutical and consumer products [47,48,49]. These aqueous systems are particularly relevant as models for biomolecular modelling.
It is possible to alter the chemical and physical properties of cyclodextrins through modification of the hydroxyl groups around the smaller primary and larger secondary rims, or faces, of the hydrophobic cavity. Such modifications offer the possibility of retaining binding of guest hydrophobic moieties to the hydrophobic cavity, while introducing new interactions with substituents at the rims, which are relatively well hydrated. Currently, the most commonly used cyclodextrin derivatives are randomly methylated β-cyclodextrin and 2-hydroxypropylated-β-cyclodextrin. The aqueous solubility of these analogues is improved compared to native β-cyclodextrin, and the additional hydrophobic groups extend the binding cavity, aiding in encapsulation of highly lipophilic drugs [50]. However, random substitution is not desirable for hosts intended to improve simulation force fields, and it may also be suboptimal for applications where binding selectivity is desired.
Several considerations come into play in choosing where to introduce new substituents on the CDs. The primary face, or rim, is both less hindered and more nucleophilic, making it a more facile target for synthetic modification. Accordingly, a large amount of literature related to binding of cyclodextrin analogues is focused around primary face modifications [51, 52]. However, modification of the 2 or 3 positions on the wider-rimmed secondary face seems preferable for our purpose, as this should allow bulky guests that protrude from the larger secondary face to interact with substituents added to the rim of the host. By selectively modifying the secondary face with one substituent, it should be possible to generate additional host–guest interactions without excessive steric hindrance that might result from complete substitution of the face. We also require that the resulting derivatives be water soluble and not aggregate in solution, so that accurate host–guest binding measurements can be carried out.
Accordingly, the aim of this work is to prove principle by demonstrating facile synthesis of a water-soluble CD derivative, singly substituted at the secondary face that binds a guest, with a clean isotherm. This sets the stage for generation of diverse CD derivatives that we anticipate using to generate a matrix of new experimental binding data to guide the development of enhanced force fields (Fig. 2). Herein, we present the synthesis and characterization of the novel host molecule mono-3-carboxypropionamido-β-cyclodextrin (CP-β-CD), and the characterization of its association with the guest rimantadine hydrochloride, by NMR and ITC.
2 Experimental
2.1 Materials
3-Amino-β-cyclodextrin was purchased from TCI Chemicals Ltd. Dimethylformate (99.9%) and acetone (99.5%) were used as purchased from Fisher Scientific, through the Chemistry stockroom at University of California, San Diego, without further purification or drying. Rimantadine hydrochloride (99%), deuterium oxide (99.9%), monosodium phosphate (98%) and disodium phosphate (98%) were purchased from Sigma-Aldrich Company (St. Louis, MO).
2.2 Synthesis of Mono-3-carboxypropionamido-β-cyclodextrin
Succinic anhydride (0.11 mmol) was added to a solution of 3-amino-β-cyclodextrin (0.1 mmol) in dry DMF (3 mL). The reaction mixture was stirred for 12 h at room temperature, after which it was evaporated to minimum volume, under reduced pressure at 333 K, cooled and precipitated with acetone (5 mL). The resulting precipitate was collected by filtration, and washed with excess acetone, yielding a white solid, 86%. 1H NMR (600 MHz, D2O) δ 8.03–7.33 (m, 4H), 5.07–4.70 (m, 7H), 4.11–3.26 (m, 42H). 13C DEPTQ NMR (150 MHz, D2O) δ 139.1, 101.0, 81.1, 73.3, 72.7, 72.5, 72.2, 72.1, 72.0, 71.7, 68.9, 60.4, 37.0. ESI–MS calculated for C42H75NO37, 1233.40 g·mol−1, found 1232.51 g·mol−1 [M−H]−.
2.3 Nuclear Magnetic Resonance (NMR)
NMR spectra were collected at 298 K on a 600 MHz Bruker Avance III spectrometer fitted with a 5 mm triple resonance cryoprobe with z-axis gradients. All NMR studies were run in 90% phosphate buffer (0.1 mol·L−1, pH 6.8), with 10% deuterium oxide. 1H-NMR was collected with F1 presaturation of the water peak, with 16 scans. 2D NOESY spectra were run with water suppression using excitation sculpting with gradients and the TPPI acquisition mode.
2.4 Isothermal Titration Calorimetry (ITC)
ITC experiments were performed using a MicroCal model VP-ITC (MicroCal, Northampton, CT, serial number 01-08-930). The instrument was positioned on a foam block to reduce vibrations that could lead to experimental errors [53]. The cell temperature was kept constant at 300 K, reference power was set to 126 µJ·s−1, and initial delay was set to 60 s. Solutions were prepared in phosphate buffer, (0.1 mol·L−1, pH 6.8), using 10 mL volumetric flasks and a Sartorius CPA225D Micro Balance. Rimantadine hydrochloride was placed in the syringe at 75.06 mmol·L−1 while CP-β-CD was in the cell with a concentration of 5.05 mmol·L−1. There were 73 injections; 34 injections of 2.5 µL, followed by 39 injections of 5 µL, with 120 s between injections. The change in injection volume is to ensure that the full binding isotherm is observed with a suitable number of injections, but without generating heat release in the initial injections that would have required increasing the reference power. Data from the first two 2.5 μL injections were discarded, to avoid possible artifacts from diffusional mixing prior to the start of the experiment. For β-cyclodextrin and rimantadine hydrochloride the solutions were prepared in the same way, except that β-cyclodextrin was in the syringe at 13.00 mmol·L−1 and rimantadine hydrochloride in the sample cell at 1.01 mmol·L−1. This was done to avoid enthalpy of dilution issues associated with injecting the cationic rimantadine into the neutral β-cyclodextrin, as described previously [54]. In the case of CP-β-CD, however, both reactants are charged, which introduces a large enthalpy of dilution for the CD as well. Therefore, for convenience, the less soluble CD derivative was chosen as the cell reactant, for which a high concentration is not required, and the enthalpy of dilution was computed and subtracted.
3 Results and Discussion
3.1 Synthesis
The synthesis of mono-3-carboxypropionamido-β-cyclodextrin (CP-β-CD) is shown in Scheme 1. This amic acid derivative was successfully made with a one-pot strategy via ring opening addition of succinic anhydride to the primary amine of mono-3-amino-β-cyclodextrin, in 87% yields, as documented by mass spectrometry (m/z = 1232.51 [M−H]−) and 1D NMR (13C and 1H). Conveniently, CP-β-CD showed improved aqueous solubility (> 100 mg·mL−1) relative to unmodified β-CD (18 mg·mL−1).
3.2 NMR Characterization of Binding with Rimantadine
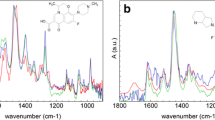
In the absence of guest molecules (Fig. 3a), the peaks associated with the cyclodextrin moiety are complex and poorly defined, due to the lack of symmetry [55]. This may be contrasted with the much simpler peaks for unmodified β-CD (Fig. S2). The broad multiplet seen at 3.6–3.8 ppm corresponds to the internal cavity protons, CH3 and CH5, as well as the CH6 protons; CH2 and CH4 are external to the cavity, and, at 3.3–3.5 ppm. The poorly defined nature of these spectra makes it hard to accurately track a specific peak during a titration. Nonetheless, NMR can be used to confirm complexation and gain in structural insights.
The hydrophobic adamantane group of rimantadine (Fig. 4) can bind tightly inside native β-CD [53], and its hydrophilic alkyl amine functionality, which should be ionized at pH 6.8, presumably projects into solution from either the primary or the secondary face. A titration NMR study, conducted in pH 6.8 phosphate buffer with 10% D2O, confirms complexation of CP-β-CD with rimantadine (Fig. 3a). No exchangeable protons are visible due to rapid exchange with the D2O, so no potential hydrogen bonding interactions were determined from NMR. The major multiplet seen for the internal surface of cyclodextrin cavity (3.6–3.8 ppm), containing the CH3, CH5 and CH6 protons, becomes shielded upon binding (Fig. 3a). More noticeably in the expanded spectra of Fig. 3b, the broad multiplet at 4.05 ppm, which is associated with CH3′ on the modified glucose unit, also becomes shielded upon binding. In contrast, no significant chemical shift is seen for CH2 and CH4; this is unsurprising as they are exterior cavity protons and should see no effect upon binding.
Further information regarding the three-dimensional structure of the complex was obtained by 2D NOESY experiments, where intermolecular cross-correlations identify atoms of the guest near atoms of the host. To maximize spectral resolution, 2D NMR spectra are normally obtained with pure deuterated solvent [53]. Here, however, for consistency with the 1D NMR data, we used 90% phosphate buffer, with 10% D2O. The water resonance was suppressed using excitation-sculpting, as opposed to presaturation which was used for 1D data collection [56], as this was found to give cleaner spectra. The data were collected with a 1:1 solution of CP-β-CD and rimantadine. Cross peaks can be seen between the adamantane functionality and the entirety of the protons in the CD cavity (Fig. 5), confirming binding.
As discussed above, absolute characterization of the multiplet formed for the cyclodextrin cavity is complex, so we did not attempt absolute assignment of these peaks. However, the CH8/9 protons of the amic acid functionality generate a well-resolved multiplet at 2.35 ppm, and this has clear cross correlation with the chiral rimantadine carbon, CH1, at 8.00 ppm (Fig. 5). This implies that the ammonium group of rimantadine projects out the secondary face of this modified CD host, as diagrammed in Fig. 6. This makes intuitive sense, because this orientation places the cationic ammonium near the anionic amic acid of the host.
Visualization of rimantadine (gold) binding in the secondary orientation of CP-β-CD based on 2D-NOESY NMR binding data. Intermolecular interaction between rimantadine (CH1) and CP-β-CD CH8/9 is highlighted by a black dashed line. A glucose unit has been removed from CP-β-CD for visualization purposes (Color figure online)
It has previously been reported that rimantadine preferentially binds in the secondary orientation of native β-CD [53], i.e., with its ammonium group projecting from the secondary face. As noted above, we find that this binding orientation is retained in CP-β-CD, with its anionic group at the secondary face. However, when β-CD is substituted with an amine at the three position, which is on the secondary face, rimantadine is reported to preferentially bind in the opposite, primary orientation [53], presumably due to electrostatic repulsion of the rimantadine ammonium group with the amino substituent. Thus, the binding orientation of guests inside CDs can be guided through selective modification of the host.
3.3 Thermodynamics of Binding with Rimantadine
We used isothermal titration calorimetry (ITC) to determine the binding Gibbs energy and enthalpy of CP-β-CD and rimantadine, at 300 K, in 0.1 mol·L−1 phosphate buffer at pH 6.8 to match the NMR studies above. The experimental setup provided a C value [57] of 6 and yielded a smooth, sigmoidal binding isotherm (Fig. 7), which was fitted by standard methods to provide binding Gibbs energy, enthalpy and fitted stoichiometry (N) of, respectively, − (17.87 ± 0.04) kJ·mol−1, − (18.0 ± 0.4) kJ·mol−1, and 0.9. For comparison, the binding Gibbs energy and enthalpy of native β-CD and rimantadine, measured here under the same experimental conditions, are − (26.61 ± 0.08) and − (28.9 ± 0.4) kJ·mol−1 (Fig. 7b). Thus, perhaps surprisingly, outfitting the host with a negatively charged acid that could establish favorable electrostatic interactions with the cationic guest reduces the affinity, rather than increasing it. However, the overall pattern of a large, favorable binding enthalpy and a small (< 1 kJ·mol−1) binding entropy, is retained. It is also worth noting that outfitting the secondary face with an ammonium group, which should repel the ammonium of rimantadine, leads to an even greater drop in affinity relative to native β-CD, with ∆G = − 13.8 kJ·mol−1, as well as to a flip in the binding orientation [53], as mentioned above.
Wiseman plots and enthalpograms. a CP-β-CD and rimantadine hydrochloride; floating N, ∆S = – 0.4 J·K−1·mol−1, ∆H = – 18.0 kJ·mol−1, K = 1280 mol·L−1, and N = 0.9. The step in peaks seen near 70 min in the Wiseman plot is due to an increase in injection volume from 2.5 to 5 µL at that point in the experiment. The x-axis shows the molar ratio of rimantadine (the syringe reactant) to CP-β-CD (the cell reactant). b β-CD and rimantadine hydrochloride; floating N, ∆S = – 8.8 J·K−1·mol−1, ∆H = – 18.9 kJ·mol−1, K = 43,285 mol·L−1, and N = 0.9. Here, the x-axis shows the molar ratio of CD (the syringe reactant) to rimantadine (the cell reactant)
4 Conclusion
We have reported a successful, one-pot synthesis of a new, highly water-soluble CD derivative, mono-3-carboxypropionamido-β-cyclodextrin (CP-β-CD). ITC binding studies show that this compound binds the guest rimantadine hydrochloride in a 1:1 stoichiometry with a ∆G of − 18.0 kJ·mol−1, and analysis of the bound complex in solution, via 2D-NOESY NMR, reveals that the amic acid moiety lies near the chiral carbon of rimantadine hydrochloride, as anticipated. These results support the feasibility of further efforts aimed at building out the matrix of host–guest interactions diagrammed in Fig. 2. This promises to be an efficient means of generating a database of binding thermodynamics that will probe a range of chemical interactions, and thus inform the development of more accurate force fields for use in computer-aided drug design.
References
Gilson, M.K., Zhou, H.-X.: Calculation of protein–ligand binding affinities. Ann. Rev. Biophys. Biomol. Struct. 36, 21–42 (2007)
Kuntz, I.D., Blaney, J.M., Oatley, S.J., Langridge, R., Ferrin, T.E.: A geometric approach to macromolecule–ligand interactions. J. Mol. Biol. 161, 269–288 (1982)
Goodsell, D.S., Olson, A.J.: Automated docking of substrates to proteins by simulated annealing. Protein Struct. Funct. Gen. 8, 195–202 (1990)
Abagyan, R., Totrov, M., Kuznetsov, D.: ICM—a new method for protein modeling and design. Applications to docking and structure prediction from the distorted native conformation. J. Comput. Chem. 15, 488–506 (1994)
Apostolakis, J., Pluckthun, A., Caflisch, A.: Docking small ligands in flexible binding sites. J. Comput. Chem. 19, 21–37 (1998)
Charifson, P.S., Corkery, J.J., Murcko, M.A., Walters, W.P.: Consensus scoring: a method for obtaining improved hit rates from docking databases of three-gimensional structures into proteins. J. Med. Chem. 42, 5100–5109 (1999)
Clark, K.P.: Ajay: flexible ligand docking without parameter adjustment across four ligand–receptor complexes. J. Comput. Chem. 16, 1210–1226 (1995)
David, L., Luo, R., Gilson, M.K.: Ligand–receptor docking with the Mining Minima optimizer. J. Comput. Aided Mol. Des. 15, 157–171 (2001)
Trott, O., Olson, A.J.: AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010). https://doi.org/10.1002/jcc.21334
Jain, A.N.: Surflex: fully automatic flexible molecular docking using a molecular similarity-based search engine. J. Med. Chem. 46, 499–511 (2003). https://doi.org/10.1021/jm020406h
Halgren, T.A., Murphy, R.B., Friesner, R.A., Beard, H.S., Frye, L.L., Pollard, W.T., Banks, J.L.: Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 47, 1750–1759 (2004). https://doi.org/10.1021/jm030644s
Abel, R., Wang, L., Mobley, D.L., Friesner, R.A.: A critical review of validation, blind testing, and real-world use of Alchemical protein–ligand binding free energy calculations. Curr. Top. Med. Chem. 17, 2577–2585 (2017)
Gallicchio, E., Lapelosa, M., Levy, R.M.: Binding energy distribution analysis method (BEDAM) for estimation of protein–ligand binding affinities. J. Chem. Theory Comput. 6, 2961–2977 (2010). https://doi.org/10.1021/ct1002913
Gilson, M.K., Given, J.A., Bush, B.L., McCammon, J.A.: The statistical-thermodynamic basis for computation of binding affinities: a critical review. Biophys. J. 72, 1047–1069 (1997)
Hansen, N., van Gunsteren, W.F.: Practical aspects of free-energy calculations: a review. J. Chem. Theory Comput. 10, 2632–2647 (2014). https://doi.org/10.1021/ct500161f
Jorgensen, W.L., Buckner, J.K., Boudon, S., Tirado-Rives, J.: Efficient computation of absolute free energies of binding by computer simulations. Application to the methane dimer in water. J. Chem. Phys. 89, 3742–3746 (1988). https://doi.org/10.1063/1.454895
Lau, F.T.K., Karplus, M.: Molecular recognition in proteins: simulation analysis of substrate binding by a tyrosyl-tRNA synthetase mutant. J. Mol. Biol. 236, 1049–1066 (1994). https://doi.org/10.1016/0022-2836(94)90011-6
Simonson, T., Archontis, G., Karplus, M.: Free energy simulations come of age: protein–ligand recognition. Acc. Chem. Res. 35, 430–437 (2002). https://doi.org/10.1021/ar010030m
Tembe, B.L., McCammon, J.A.: Ligand–receptor interactions. Comput. Chem. 8, 281–283 (1984)
Woo, H.-J., Roux, B.: Calculation of absolute protein–ligand binding free energy from computer simulations. Proc. Natl. Acad. Sci. USA 102, 6825–6830 (2005). https://doi.org/10.1073/pnas.0409005102
Homeyer, N., Stoll, F., Hillisch, A., Gohlke, H.: Binding free energy calculations for lead optimization: assessment of their accuracy in an industrial drug design context. J. Chem. Theory Comput. 10, 3331–3344 (2014). https://doi.org/10.1021/ct5000296
Kuhn, B., Tichý, M., Wang, L., Robinson, S., Martin, R.E., Kuglstatter, A., Benz, J., Giroud, M., Schirmeister, T., Abel, R., Diederich, F., Hert, J.: Prospective evaluation of free energy calculations for the prioritization of cathepsin L inhibitors. J. Med. Chem. 60, 2485–2497 (2017). https://doi.org/10.1021/acs.jmedchem.6b01881
Williams-Noonan, B.J., Yuriev, E., Chalmers, D.K.: Free energy methods in drug design: prospects of “Alchemical Perturbation” in medicinal chemistry. J. Med. Chem. (2017). https://doi.org/10.1021/acs.jmedchem.7b00681
Cournia, Z., Allen, B., Sherman, W.: Relative binding free energy calculations in drug discovery: recent advances and practical considerations. J. Chem. Inf. Model. 57, 2911–2937 (2017). https://doi.org/10.1021/acs.jcim.7b00564
Sherborne, B., Shanmugasundaram, V., Cheng, A.C., Christ, C.D., DesJarlais, R.L., Duca, J.S., Lewis, R.A., Loughney, D.A., Manas, E.S., McGaughey, G.B., Peishoff, C.E., van Vlijmen, H.: Collaborating to improve the use of free-energy and other quantitative methods in drug discovery. J. Comput. Aided Mol. Des. 30, 1139–1141 (2016). https://doi.org/10.1007/s10822-016-9996-y
Lybrand, T.P., McCammon, J.A., Wipff, G.: Theoretical calculation of relative binding affinity in host–guest systems. Proc. Natl. Acad. Sci. USA 83, 833–835 (1986)
Pitera, J., Kollman, P.: Designing an optimum guest for a host using multimolecule free energy calculations: predicting the best ligand for Rebek’s “tennis ball”. J. Am. Chem. Soc. 120, 7557–7567 (1998). https://doi.org/10.1021/ja973028s
Kaminski, G.A., Jorgensen, W.L.: Host–guest chemistry of rotaxanes and catenanes: application of a polarizable all-atom force field to cyclobis(paraquat-p-phenylene) complexes with disubstituted benzenes and biphenyls. J. Chem. Soc. Perkin Trans. 2, 2365–2375 (1999). https://doi.org/10.1039/A905160K
Chen, W., Chang, C.E., Gilson, M.K.: Calculation of cyclodextrin binding affinities: energy, entropy, and implications for drug design. Biophys. J. 87, 3035–3049 (2004). https://doi.org/10.1529/biophysj.104.049494
Chang, C.-E., Gilson, M.K.: Free energy, entropy, and induced fit in host-guest recognition: calculations with the second-generation mining minima algorithm. J. Am. Chem. Soc. 126, 13156–13164 (2004)
Rogers, K.E., Ortiz-Sánchez, J.M., Baron, R., Fajer, M., de Oliveira, C.A.F., McCammon, J.A.: On the role of dewetting transitions in host–guest binding free energy calculations. J. Chem. Theory Comput. 9, 46–53 (2013). https://doi.org/10.1021/ct300515n
Wickstrom, L., He, P., Gallicchio, E., Levy, R.M.: Large scale affinity calculations of cyclodextrin host–guest complexes: understanding the role of reorganization in the molecular recognition process. J. Chem. Theory Comput. 9, 3136–3150 (2013). https://doi.org/10.1021/ct400003r
Bell, D.R., Qi, R., Jing, Z., Xiang, J.Y., Mejias, C., Schnieders, M.J., Ponder, J.W., Ren, P.: Calculating binding free energies of host–guest systems using AMOEBA polarizable force field. Phys. Chem. Chem. Phys. 18, 30261–30269 (2016). https://doi.org/10.1039/c6cp02509a
Henriksen, N.M., Gilson, M.K.: Evaluating force field performance in thermodynamic calculations of cyclodextrin host-guest binding: water models, partial charges, and host force field parameters. J. Chem. Theory Comput. 13, 4253–4269 (2017). https://doi.org/10.1021/acs.jctc.7b00359
Mobley, D.L., Gilson, M.K.: Predicting binding free energies: frontiers and benchmarks. Ann. Rev. Biophys. 46, 531–558 (2017)
Gao, K., Yin, J., Henriksen, N.M., Fenley, A.T., Gilson, M.K.: Binding enthalpy calculations for a neutral host–guest pair yield widely divergent salt effects across water models. J. Chem. Theory Comput. 11, 4555–4564 (2015)
Yin, J., Fenley, A.T., Henriksen, N.M., Gilson, M.K.: Toward improved force-field accuracy through sensitivity analysis of host–guest binding thermodynamics. J. Phys. Chem. B 119, 10145–10155 (2015). https://doi.org/10.1021/acs.jpcb.5b04262
Bradshaw, R.T., Essex, J.W.: Evaluating parametrization protocols for hydration free energy calculations with the AMOEBA polarizable force field. J. Chem. Theory Comput. 12, 3871–3883 (2016). https://doi.org/10.1021/acs.jctc.6b00276
Buck, M., Bouguet-Bonnet, S., Pastor, R.W., MacKerell, A.D.: Importance of the CMAP correction to the CHARMM22 protein force field: dynamics of hen lysozyme. Biophys. J. 90, L36–L38 (2006). https://doi.org/10.1529/biophysj.105.078154
Jorgensen, W.L., Tirado-Rives, J.: The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J. Am. Chem. Soc. 110, 1657–1666 (1988). https://doi.org/10.1021/ja00214a001
Kang, M., Smith, P.E.: A Kirkwood-Buff derived force field for amides. J. Comput. Chem. 27, 1477–1485 (2006). https://doi.org/10.1002/jcc.20441
Oostenbrink, C., Villa, A., Mark, A.E., van Gunsteren, W.F.: A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. 25, 1656–1676 (2004). https://doi.org/10.1002/jcc.20090
Piana, S., Donchev, A.G., Robustelli, P., Shaw, D.E.: Water dispersion interactions strongly influence simulated structural properties of disordered protein States. J. Phys. Chem. B 119, 5113–5123 (2015). https://doi.org/10.1021/jp508971m
Ploetz, E.A., Smith, P.E.: A Kirkwood-Buff force field for the aromatic amino acids. Phys. Chem. Chem. Phys. PCCP 13, 18154–18167 (2011). https://doi.org/10.1039/c1cp21883b
Song, D., Wang, W., Ye, W., Ji, D., Luo, R., Chen, H.-F.: ff14IDPs force field improving the conformation sampling of intrinsically disordered proteins. Chem. Biol. Drug Des. 89, 5–15 (2017). https://doi.org/10.1111/cbdd.12832
Weerasinghe, S., Gee, M., Kang, M., Bentenitis, N., Smith, P.: Developing Force Fields from the Microscopic Structure of Solutions: The Kirkwood-Buff Approach. Modelling Solvent Environment, pp. 55–76. Wiley-VCH, Weinheim (2010). https://doi.org/10.1002/9783527629251.ch3
Davis, M.E., Brewster, M.E.: Cyclodextrin-based pharmaceutics: past, present and future. Nat. Rev. Drug Discov. 3, 1023–1035 (2004). https://doi.org/10.1038/nrd1576
Brewster, M.E., Loftsson, T.: Cyclodextrins as pharmaceutical solubilizers. Adv. Drug Deliv. Rev. 59, 645–666 (2007). https://doi.org/10.1016/j.addr.2007.05.012
Loftsson, T., Brewster, M.E.: Pharmaceutical applications of cyclodextrins: basic science and product development. J. Pharm. Pharmacol. 62, 1607–1621 (2010). https://doi.org/10.1111/j.2042-7158.2010.01030.x
Yanli, C., Caixia, W., Jianwei, M., Yongping, Y.: A facile and practical approach to randomly methylated beta-cyclodextrin. J. Chem. Technol. Biotechnol. 85, 248–251 (2010). https://doi.org/10.1002/jctb.2295
Khan, A.R., Forgo, P., Stine, K.J., D’Souza, V.T.: Methods for selective modifications of cyclodextrins. Chem. Rev. 98, 1977–1996 (1998). https://doi.org/10.1021/cr970012b
Ashton, P.R., Königer, R., Stoddart, J.F., Alker, D., Harding, V.D.: Amino acid derivatives of β-cyclodextrin. J. Org. Chem. 61, 903–908 (1996). https://doi.org/10.1021/jo951396d
Carrazana, J., Jover, A., Meijide, F., Soto, V.H., Tato, J.V.: Complexation of adamantyl compounds by β-cyclodextrin and monoaminoderivatives. J. Phys. Chem. B 109, 9719–9726 (2005). https://doi.org/10.1021/jp0505781
Kantonen, S.A., Henriksen, N.M., Gilson, M.K.: Accounting for apparent deviations between calorimetric and van’t Hoff enthalpies. Biochim. Biophys. Acta (2017). https://doi.org/10.1016/j.bbagen.2017.11.020
Dodzuik, E.: Cyclodextrins and Their Complexes. Wiley-VCH, Weinheim (2006)
Hwang, T.L., Shaka, A.J.: Water suppression that works. Excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J. Magn. Reson. A 112, 275–279 (1995). https://doi.org/10.1006/jmra.1995.1047
Wiseman, T., Williston, S., Brandts, J.F., Lin, L.N.: Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 179, 131–137 (1989). https://doi.org/10.1016/0003-2697(89)90213-3
Acknowledgements
M.K.G. has an equity interest in, and is a cofounder and scientific advisor of, VeraChem LLC. NMR spectra were collected at the UCSD Skaggs School of Pharmacy and Pharmaceutical Sciences NMR Facility.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kellett, K., Kantonen, S.A., Duggan, B.M. et al. Toward Expanded Diversity of Host–Guest Interactions via Synthesis and Characterization of Cyclodextrin Derivatives. J Solution Chem 47, 1597–1608 (2018). https://doi.org/10.1007/s10953-018-0769-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10953-018-0769-1