Abstract
Angiotensin-converting enzyme 2 (ACE2) has a specific interaction with the coronavirus spike protein, enabling its entry into human cells. This membrane enzyme converts angiotensin II into angiotensin 1–7, which has an essential role in protecting the heart and improving lung function. Many therapeutic properties have been attributed to the human recombinant ACE2 (hrACE2), especially in combating complications related to diabetes mellitus and hypertension, as well as, preventing the coronavirus from entering the target tissues. In the current study, we designed an appropriate gene construct for the hybrid protein containing the ACE2 catalytic subunit and the B subunit of cholera toxin (CTB-ACE2). This structural feature will probably help the recombinant hybrid protein enter the mucosal tissues, including the lung tissue. Optimization of this hybrid protein expression was investigated in BL21 bacterial host cells. Also, the hybrid protein was identified with an appropriate antibody using the ELISA method. A large amount of the hybrid protein (molecular weight of ~ 100 kDa) was expressed as the inclusion body when the induction was performed in the presence of 0.25 mM IPTG and 1% sucrose for 10 h. Finally, the protein structural features were assessed using several biophysical methods. The fluorescence emission intensity and oligomeric size distribution of the CTB-ACE2 suggested a temperature-dependent alteration. The β-sheet and α-helix were also dominant in the hybrid protein structure, and this protein also displays acceptable chemical stability. In overall, according to our results, the efficient expression and successful purification of the CTB-ACE2 protein may pave the path for its therapeutic applications against diseases such as covid-19, diabetes mellitus and hypertension.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
In a renin-angiotensin (RAS) system, the first protein released by the liver into the bloodstream is angiotensinogen. This zymogenic protein is then converted to angiotensin I (Ang I) by renin which subsequently produces angiotensin II (Ang II) by the action of the angiotensin-converting enzyme (ACE) [1]. Dysfunction of the RAS system, resulting in the elevated level of Ang II, contributes to increased oxidative stress, inflammation and diabetes complications [2] (Scheme 1). The membrane enzyme ACE2, as part of the RAS system, has the highest expression levels in the lung, heart, gastrointestinal system, endothelial membrane, and kidneys [3]. The ACE2 is the homologue of the zinc-metalloprotease ACE (i.e. ∼60% similarity and ∼40% identity) and a regulator of the RAS system [4, 5]. The ACE2 has mono-carboxypeptidase activity, cleaving a single C-terminus residue from several peptides possessing important physiological functions [5]. This enzyme is encoded by the ACE2 gene (Xp22), resulting in a glycoprotein of 805 amino acid residues with an overall molecular mass of 120 kDa that contains a single HEXXH zinc-binding consensus sequence [4,5,6]. ACE2 is a type-I transmembrane protein containing a short C-terminus cytoplasmic tail, a hydrophobic transmembrane region, and an N-glycosylated N-terminal ectodomain containing the active site [5, 7]. This protein is responsible for preventing the harmful effects of angiotensin II, such as hypertension, vasoconstriction, oxidative stress, inflammation and endothelial dysfunction [1, 3, 8]. In a recently reported study, the ACE2 gene fused to a non-toxic B subunit of the cholera toxin gene and the resulting construct transferred to the probiotic bacterium Lactobacillus paracasei. Then, the bacteria produced a hybrid protein consisting of the ACE2 enzyme and the cholera toxin B subunit (CTB), which may also be capable of binding to the coronavirus. Furthermore, the non-toxic CTB enables the delivery of the ACE2 enzyme to the mucosal tissues, which is very important in fighting the virus in the lungs and gastrointestinal system that have mucous tissue [2]. The recombinant ACE2 enzyme can be potentially applied either in converting Ang II to a blood pressure-lowering peptide (Ang 1–7) or as an effective molecular tool in binding and inhibiting the coronavirus.
In the current study, the construct consists of the catalytic extracellular domain of the human ACE2 gene was designed for expression in the bacterial system as it was fused with the CTB gene. After successful expression, the hybrid protein was purified by the chromatographic method. Various spectroscopic methods were also employed to investigate the protein biophysical characteristics.
2 Materials and Methods
2.1 Materials
1-Anilino-8-naphthalene sulfonic acid (ANS), kanamycin, thioflavin T (ThT), and isopropyl-1-thio-β-D-galactopyranoside (IPTG), were purchased from Sigma. Triton X-100, sucrose, glycerol, ethylenediaminetetraacetic acid (EDTA), β-mercaptoethanol (βME), urea, and other chemicals were purchased from the Merck Company.
2.2 Methods
2.2.1 Construction of CTB-ACE2 Fusion
The hybrid protein consisting of the ACE2 catalytic subunit and CTB was prepared by designing a gene construct composed of human ACE2 extracellular domain (residues 18–740), which ligated to the cholera toxin B subunit (22–124 amino acids, with H57A mutation) (Fig. 1). As shown in Fig. 1, the CTB is separated by a hinge (Gly-Pro-Gly-Pro) and furin cleavage (Arg-Arg-Lys-Arg) sites to eliminate the steric hindrance, aiding the release of ACE2 after internalization via a ligand-receptor complex formation on the surface of the epithelial cells. The fusion gene is surrounded by two restriction enzymes, BamHI and XhoI. To produce this construct, after chemically synthesizing the fusion gene sequence with the desired restriction sites, it was cloned into a pET-28a( +) expression vector containing a His-tag.
Schematic illustration of the designated vector and fusion gene sequence. pET-28a(+) vector comprising the fusion gene is represented in this diagram. The positions of BamHI and XhoI restriction sites are also indicated. The translation frame on the vector displays the sequence of CTB-ACE2 hybrid protein. The non-toxic CTB and human ACE2 are shown in blue and red, respectively. Moreover, the hinge and cleavage site are indicated as violet. The histidine amino acid residue at position 57 of the CTB gene sequence was substituted with alanine (in green). Moreover, the amino acid sequence of the CTB-ACE2 hybrid protein is presented
The gene cloning was then confirmed by the restriction digestion using BamHI and XhoI. The resulting plasmid was subsequently transformed into strain BL21 (DE3) of Escherichia coli.
2.2.2 Expression, and Solubilization of the Recombinant CTB-ACE2
The pET-28a(+)-CTB-ACE2 vector was expressed in E. coli cells strain BL21 (DE3). The bacterial cells containing recombinant plasmid were incubated in the LB medium (10 mL) at 37 °C for overnight and then in the 1 L of the LB medium. When the strain density reaches the absorbance range of 0.6–0.8 at 600 nm, the induction of CTB-ACE2 expression was done by IPTG (0.25 mM) in the presence of 1% sucrose for 10 h at 37 °C. Then, the cells were collected by centrifugation (8000 rpm, 45 min). The hybrid protein indicated a tendency for the formation of the inclusion bodies in E. coli host cells. The bacterial pellet (2 gr) was suspended in 25 mL lysis buffer (50 mM Tris–HCl, 1% Triton X-100, pH 8.5) and then sonicated on the ice bath. This step was repeated up to 5 times. The cell debris was centrifuged (8000 rpm, for 10 min), and the collected pellet was washed by an appropriate washing buffer (50 mM Tris–HCl, pH 8.5 containing 1% Triton X-100, 2M urea, and 10 mM βME) and then sonicated. The debris was solubilized in 0.1 gr/mL of 50 mM Tris–HCl (pH 8.5), containing 10 mM βME, and 8 M urea. After centrifugation at 8000 rpm for 15 min, the supernatant was subjected to a 0.22 µm filter membrane. The protein sample in each step was also examined by SDS-PAGE analysis (gel 12%) [2, 9].
2.2.3 Purification and Refolding of the Recombinant CTB-ACE2
In order to purify the recombinant protein, the supernatant collected in the previous step was loaded onto a DEAE cellulose column (2.5 × 13 cm2). A 25 mM Tris–HCl buffer (pH 8.0), containing 5 mM βME and 4 M urea was used to equilibrate the anion exchange column. During the protein elution, those fractions that reached the highest degree of purity were collected and subjected two or three times to the same column.
The purified protein was refolded by stepwise dialysis against refolding buffer to remove the denaturation agent. The refolding buffer in the first step of dialysis was 50 mM Tris–HCl (pH 8.5), containing sucrose (5%), glycerol (10%), 2 M urea, and 10 mM βME. The same refolding was used with 0.5 M urea in the second step. Finally, the protein sample was extensively dialyzed against a large volume of 50 mM Tris–HCl buffer (pH 8.5) and then against ddH2O, respectively. The protein solution was finally lyophilized and then stored at -20 °C [2, 10].
2.2.4 Fluorescence and UV-CD Experiments
Following the procedures outlined in our prior research, we performed fluorescence assessments in many dimensions using two-dimensional (2D) excitation-emission, contour maps, three-dimensional (3D), synchronous measurements, and ANS analyses [11, 12].
The spectroscopic fluorescence assessments were performed using a temperature-controlled Varian Cary Eclipse (Australia) fluorescence spectrophotometer. The protein concentration was fixed at 0.15 mg/mL in 100 mM phosphate buffer (pH 7.8; buffer A) at two different temperatures (27 and 37 °C). In order to evaluate the fluorescence emission (2D) and synchronous measurements of Tyr and Trp residues, the protein sample was excited at 276 and 295 nm, respectively. Afterwards, the emission spectra were taken between 300 and 500 nm, with the slit widths held constant at 5/10 nm [13]. Furthermore, the wavelengths correlated with Tyr and Trp emissions were recorded during the synchronous fluorescence experiment at Δλ = 15 nm and Δλ = 60 nm, respectively (slit bandwidths fixed at 5/10 nm) [14]. Additionally, the protein surface hydrophobicity was investigated using the ANS fluorescence technique. After 30 min of ANS (100 µM) incubation with the protein sample, the resulting emission spectra were recorded (400–600 nm) when the protein/ANS solution was excited at 365 nm [15].
Further fluorescence experiments were also performed with the aim of deepening the structural studies of the protein. In order to achieve these goals, the excitation wavelength was also varied from 200 to 350 nm at a 5 nm interval, allowing for the creation of a contour map and 3D fluorescence studies of the protein samples. The emission spectra were collected in 2 nm intervals from 300 to 500 nm. The excitation and emission slit bandwidths were held constant at 5 and 10 nm [16, 17]. Sigma Plot 14.0 was also used to create the graphs and conduct the statistical analysis.
Moreover, far and near UV-CD measurements were performed as the protein concentrations were fixed at 0.2 and 1.5 mg/mL, respectively. The CD measurements were taken in buffer A at 25 °C using a JASCO 810 spectropolarimeter with the far and near cells of 0.1 and 1 cm path length. The DICHROWEB server's CONTIN algorithm was used to analyze the secondary structure content, and all data was represented as molar ellipticity [13, 18].
2.2.5 Fourier Transform Infrared Spectroscopy (FTIR), and Raman Assessments
To evaluate the secondary structures of the recombinant protein, the FTIR spectra of CTB-ACE2 were recorded using a Bruker ATR-FTIR spectrophotometer (Tensor II, Germany). The FTIR spectra (256 accumulated scans) were taken at 25 °C in the region of 2000–400 cm−1 with a resolution of 4 cm−1. In addition to the FTIR assessments, with the aim of obtaining a more accurate picture of the secondary and tertiary structures of this recombinant protein, Raman investigations were carried out. Raman spectroscopy was performed in the 1800–600 cm−1 range using a Raman spectrometer Lab Ram HR (Horiba, Japan) coupled with a confocal microscope and a 532 nm red laser excitation (600 g/mm grating) in combination with 50 X magnification [10]. In the FTIR and Raman analyses, the amide I region (1700–1600 cm-1) was used to determine the secondary structure contents, with curve-fitting conducted using the GRAMS/AI (9.2) software. Finally, the deconvolution analysis was performed using the Gaussian function [19, 20]. All the FTIR and Raman assessments were done on protein powder.
2.2.6 Chemical Stability Measurement of CTB-ACE2
Using an equilibrium unfolding of the protein samples, the effect of varying urea concentrations (from 0 to 8 M) on the chemical stability of CTB-ACE2 at a concentration of 0.15 mg/mL in buffer A were determined [13]. The chemical denaturation parameters were calculated from sigmoid chemical denaturation curves by fitting a three-state model to the equilibrium unfolding profile upon the assumption of the following formula:
where FN, FU, FI, and are the fluorescence emissions of Trp for native, unfolded, and intermediate protein, respectively, \(\Delta G_{1}^{0}\) is the standard free energy change between the intermediate and native states, and \(\Delta G_{2}^{0}\) is the standard free energy change between the intermediate and unfolded states.
2.2.7 Measuring Size Distribution of the CTB-ACE2 by Dynamic Light Scattering (DLS)
Using a HORIBA instrument (173° scattering angle and a laser beam of 532 nm), the oligomeric size (in diameter) of the recombinant protein was measured at 25, 37, and 47 °C after 10 min of equilibration (SZ-100, Japan) [21] . The protein sample (1 mg/mL) was dissolved in buffer A and filtered through a 0.22 µm filter membrane. The data were evaluated with an SZ-100 analyzer software running on Windows.
2.2.8 ELISA Assay
To detect the CTB-ACE2 recombinant protein, the ELISA assay was performed using an appropriate anti-His-tag antibody (Abcam, USA). The positive and negative controls used in this experiment were recombinant human insulin A-chain and human αB-crystallin. Accordingly, 25, 50, 75 and 100 µg of the purified protein was loaded into microplate wells (in three replicates) and incubated at 4 °C for 16 h. Then, PBS-T solution (0.144% NaHPO4, 0.02% KCl, 0.8% NaCl, 0.024% KH2PO4, and 50% Tween 20, PH 7.4) was used to wash the wells twice. The washing step was further repeated with 300 μL PBS solution (0.144% NaHPO4, 0.02% KCl, 0.024% KH2PO4, and 0.8% NaCl, pH 7.4). Then, bovine serum albumin (2%) in PBS-T was added to each well and incubated at 37 °C for 1 h. After that, PBS-T and PBS were used to wash the wells according to the above mentioned washing steps. The plate was then incubated at 37 °C for 1 h after adding 100 µL of 2 mg/mL of the His-tag antibody to each well. Next, we rinsed the wells with PBS-T and PBS consecutively, then we added TMB solution (100 µL; 50 µL TMB A + 50 µL TMB B) to each well and left them at room temperature in the dark condition for 1 h. Finally, 30 µL of 5M H2SO4 was added to each well to terminate the enzyme activity. The absorbance was then measured at 450 nm using a spectrophotometer; BSA (solution 2%) served as a blank.
2.2.9 Gel Electrophoresis Assessment
The supernatants and pellets of the inclusion bodies and the purified hybrid protein were evaluated under reducing condition by SDS-PAGE (12% gel) [22]. A total of 10 µg of the protein was added into each well of the gel before running the electrophoresis experiment.
2.2.10 Hydrophobicity Prediction of the Recombinant Protein
The local hydrophobicity values of the amino acid residues in the CTB-ACE2 were predicted using the Kyte-Doolittle hydrophobicity scale and the ProtScale server on the ExPASY webserver (http://web.expasy.org/protscale) [23].
2.2.11 Protein Assay
The concentration of the recombinant CTB-ACE2 protein in buffer A was evaluated by considering an extinction coefficient of 1.78 for 1 mg/mL of the protein solution at 280 nm [20].
2.2.12 The Statistical Analyses
The GraphPad Prism 8 was used for the statistical analysis, and a one-way ANOVA followed by a Bonferroni test was performed on the data. The P ≤ 0.05 was chosen to indicate the statistical significance between the groups [10, 20].
3 Results
3.1 Expression, Purification and ELISA Detection of the CTB-ACE2 Recombinant Protein
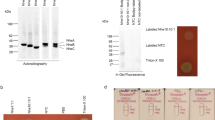
The recombinant CTB-ACE2 protein was expressed using the procedure explained in the experimental section. The expression process was optimized at different times and in the presence of some additives (Figs. S1 and S2), which led to the selection of the best time of 10 h and 1% glucose. As mentioned earlier, this hybrid protein has the propensity to appear in the inclusion bodies; for this reason, most of it was observed as the pellet (Fig. S3). Therefore, the pellet was considered for the further purification step. The solubilization of the recombinant protein (Fig. S4) and its purity (Fig. 2) was examined by SDS-PAGE analyses. About 30.81% of the CTB-ACE2 protein is found in the solubilized inclusion bodies. In the end, using these methods, about 97.2 mg of the CTB-ACE2 was collected from one liter of the LB medium. Approximately 100 kDa was the molecular weight assigned to the purified hybrid protein (Fig. 2) that corresponds to the total molecular weight of the non-toxic CTB (11.6 kDa) and the extracellular domain of ACE2 (85.3 kDa).
We proved the identity of the hybrid protein (CTB-ACE2) by the ELISA method, using an appropriate antibody that recognizes the histidine tag. The CTB-ACE2 contains C-terminal His-tag, while the A-chain of human insulin used as a positive control has an N-terminal His-tag in its primary structure. In this study, we also used recombinant human αB-crystallin as a negative control. The absorbance of CTB-ACE2 and positive control (A-chain of human insulin) was increased in a concentration-dependent manner (Fig. 3).
Detection and confirmation of CTB-ACE2 by ELISA assay. Recognition of the recombinant protein was performed using anti His-tag antibody. ELISA was carried out using 100, 75, 50, 25, and 5 µg concentrations of the recombinant protein into wells for the assay. Correspondingly, the recombinant purified αB-crystallin protein and A-chain of insulin containing His-tagged peptide was used as negative and positive controls. The samples were investigated in three replicates and deviations included. The absorbance of each well was read at 450 nm
Both His-tagged proteins represented high specificity with anti-His-tag antibodies. In addition, the protein without His-tagged displayed a very low absorbance as compared to the His-tagged proteins.
3.2 Structural Analyses of the Recombinant CTB-ACE2 Protein by Fluorescence and CD Spectroscopy
The tertiary structural alteration of the recombinant protein was assessed by contour map (topological view of the fluorescence spectrum) and 3D fluorescence spectroscopy at 27 °C and 37 °C (Fig. 4).
The spectra of the recombinant CTB-ACE2 indicated two significant peaks associated with Trp and/or Tyr side-chain emission (λex/λem = 230/345 nm and 280/347 nm at 27 °C). In this evaluation, any emission from a Tyr residue is dominated by the Trp fluorescence, and the emission from Phe can be disregarded [11, 13]. The “a” and “b” peaks in the spectra represent the Rayleigh and the second-order scattering, respectively (Fig. 4) [12, 16, 17].
The polarity of the microenvironment affects the fluorescence emission from Trp [16, 17]. The protein Trp fluorescence intensity was lower at 37 °C compared to that obtained at 27 °C. The changes in the protein structure account for the decreased Trp fluorescence intensity [16]. Consistent with the findings of the 2D Trp, Tyr, and synchronous fluorescence evaluations, the protein fluorescence emission intensities were shown to be lower at 37 °C compared to 27 °C (Fig. 5). Meanwhile, the fluorescence peak for Trp of the recombinant protein at both temperatures in the contour map, 3D spectra (27 °C, λex/λem = 230/345 nm and 280/347 nm, 37 °C, λex/λem = 230/344 nm and 280/347 nm) and 2D (27 °C, λmax = 347 nm, 37 °C, λmax = 344 nm) remain unchanged.
The fluorescence spectra of Tyr, Trp, synchronous, and ANS fluorescence measurements for CTB-ACE2 protein at 27 °C and 37 °C. The excitation wavelengths of Tyr (Tyr panel) and Trp (Trp panel) were 276 nm and 295 nm, respectively. The wavelength difference in the synchronous spectra was set at 15 nm for the tyrosine and 60 nm for the tryptophan (Syn panel). The surface hydrophobicity assessment was done by ANS fluorescent probe, λex = 365 (ANS panel). The protein sample was prepared at a concentration of 0.15 mg/mL in buffer A
The surface hydrophobicity of the protein was further evaluated by an ANS fluorescence spectroscopy at 27 °C and 37 °C [24]. The ANS fluorescence intensity of the CTB-ACE2 protein was decreased without a detectable shift in the emission maximum, implying a decrease in the solvent exposed hydrophobic surface of the protein. Based on the hydrophobicity calculation (last panel) in Fig. 5, by the ProtScale online tool on the Expasy web server, the proportion of the hydrophilic region of the CTB-ACE2 protein is significantly high (around 72%), classifying it as a hydrophilic protein.
The far-UV CD was performed to assess the protein secondary structure. Moreover, the structure contents of the recombinant protein were predicted by PROTEUS2 web server [25]. The secondary structures of CTB-ACE2 protein were estimated to be 5% β-sheet, 49% α-helix and 46% random coil. In addition to far-UV CD, the near-UV CD was also used to evaluate the tertiary structures of the CTB-ACE2 protein (Fig. 6).
Far and near UV-CD spectroscopic analyses of the protein secondary and tertiary structures. A and B correspond to the far and near UV-CD spectra of the CTB-ACE2 protein. The protein concentration was fixed at 0.2 and 1.5 mg/mL in buffer A, with the path lengths of 0.1 and 1 cm for the far and near UV-CD, respectively
As shown in Fig. 6, the far-UV CD spectrum exhibits two minimum wavelengths, at 208 nm and 222 nm, which are indicative of the distinctive wavelengths of the α-helix structure. Table 1 displays the quantitative findings from the far-UV CD study. According to this study, the α-helix structure has the highest percentage in this protein. The β-sheet, turn, and unordered structures showed contents around 27.5%, 21.5% and 16.8%, respectively.
The near-UV CD spectrum of the recombinant CTB-ACE2 protein displays the local environments of the aromatic residues, Phe (255–270 nm), Tyr (275–282 nm), and Trp absorption (290 nm), in its tertiary structure.
3.3 Protein Structural Analysis of the CTB-ACE2 Protein Using FTIR and Raman Analyses
Protein secondary structures were further evaluated using the ATR-FTIR method. FTIR analysis within the amide band I was utilized for this investigation (Fig. 7), and the secondary structure contents (%) are shown in Fig. 7 and Table 1.
FTIR structural assessment of the recombinant CTB-ACE2 protein. The FTIR spectrum was recorded at 25 °C. Deconvolution analysis with the Gaussian function in the amide I region of FTIR spectra (1700–1600 cm−1) was done using the following four peaks: 1600–1635 cm−1 (β-sheet), 1635 to 1650 cm−1 (unordered), 1650 to 1665 cm−1 (α-helix), and 1665 to 1700 cm−1 (turn). Furthermore, the percentage of the secondary structure content was shown
The FTIR spectra of this recombinant protein demonstrated two significant peaks at 1653 cm−1 and 1627 cm−1. These prominent peaks were the main feature of the α-helix and β-sheet secondary structures, respectively. Similar to the far UV-CD results, α-helix and the β-sheet secondary structures were significant in this recombinant protein. The percentage of turn content in both far UV-CD and FTIR is around 20%, and the amount of unordered structure has a minimum portion in both analyses.
The CTB-ACE2 protein was also analyzed using its Raman spectra and the band assignment in the fingerprint region (1800–600 cm−1) to determine the secondary and tertiary structures of this protein (Fig. 8).
Raman secondary and tertiary structural analyses of CTB-ACE2 protein. A The Raman spectrum of the protein sample with band assignments in the fingerprint region was shown (1800–600 cm−1). The spectrum was recorded with 532 nm red laser excitation (600 g/mm grating), power of 17 MW, and optical microscope objective 50 × . B The secondary structure deconvolution analyses in the amide I region were done by Gaussian function using GRAMS/AI software (version 9.2). Moreover, the percentage of the secondary structure content was shown
The characteristic bands corresponding to the α-helix (in 1665–1650 cm−1 region) and β-structure (in 1635–1600 cm−1 and 1700–1665 regions) were indicated in the amide I of the Raman spectra [26, 27]. In addition, we calculated the percentage of secondary structures using the amide I region of the Raman spectrum of the recombinant hybrid protein (Fig. 8B and Table 1). The Raman spectroscopy's findings for evaluating the secondary structure content are in good agreement with those obtained from the far UV-CD and FTIR analyses (Fig. 9).
The Raman spectra of protein describe the aromatic residues' surroundings by assigning specific bands to Trp, Tyr, Phe, and the Fermi doublets [19, 26, 27]. As seen in Fig. 8A, only the Tyr Fermi doublet is observed in the Raman spectrum of this protein. The Tyr Fermi doublet intensity ratio (I850/I830) of CTB-ACE2 protein is calculated to be 1.06. Since this ratio is between 0.3 (phenolic OH; strong hydrogen bond donor) and 2.5 (strong hydrogen acceptor), our finding means that the tyrosine side chain in the CTB-ACE2 protein serves as hydrogen bond donors [28, 29].
3.4 Stability Assessment of the Recombinant CTB-ACE2 Protein
By analyzing the denatured protein and detecting the fluorescence emission of Trp at varying urea concentrations, we were able to assess the chemical stability of the protein (Fig. 10). Fluorescence intensity was measured in both fully unfolded (FU) and native (FN) states and plotted as a function of urea concentrations.
Assessment of the chemical stability. The equilibrium urea denaturation profiles of proteins (0.15 mg/mL, in buffer A) were obtained by intrinsic Trp fluorescence measurement at 25 °C. The plot (Trp fluorescence ratio (IU/IN) against various concentrations of urea) was fitted according to a three-state model by Eq. (1). The chemical unfolding parameters were the average of three individual experiments
In order to determine the thermochemical parameters (transition midpoint (C1/2) and ΔG° values), a three-state (folded, unfolded, and intermediate states) fitting approach was used (Eq. (1)). Table 2 displays these quantitative findings.
3.5 Oligomeric Size Distribution Analysis of the CTB-ACE2 Protein
DLS measurements were used to examine the protein size distribution at 27, 37, and 47 °C. The CTB-ACE2 protein demonstrated an average hydrodynamic diameter of 79.8 ± 21.8 nm at 27 °C (Fig. 11).
The quaternary structural analyses of the recombinant CTB-ACE2 protein by dynamic light scattering measurement. DLS analyses of the protein (1 mg/mL in buffer A) were done at 25 °C, 37 °C, and 47 °C. The proteins were exposed to a laser light at wavelength of 532 nm and an irradiation angle of 173°. The inset graphs display the intensity of the light scattered on the particles, display the size of protein oligomers
The oligomeric size of this protein was decreased with increasing temperature (37 °C, 75.2 ± 20.6 nm, 47 °C, 76.3 ± 20.3 nm). Also, there was no significant change in the hydrodynamic diameter of the protein at 37 °C and 47 °C.
4 Discussion
It has been shown that ACE2 (EC 3.4.17.23), a homologue of an angiotensin-converting enzyme, is expressed in most human tissues [30]. Since the main product of ACE2, angiotensin 1–7, has anti-inflammatory properties, antioxidant, and blood vessel dilating effects, human recombinant ACE2 has many therapeutic potentials [31]. Several studies suggested that hrACE2 may be essential for patients with diabetes complications and lung injuries [32]. Animal studies suggested that therapy with rACE2 dramatically alleviated the acute lung injury symptoms in the wild-type and ACE2 gene-knockout mice [33]. Injecting a high amount of the recombinant ACE2 into the bloodstream may bind competitively with the SARS-CoV-2 virus. In this way, it will not only play a role in neutralizing the coronavirus but also increases the activity level of the cellular ACE2 enzyme that regulates the RAS system to prevent lung damage [34, 35].
Furthermore, the sites of interaction between ACE2 and coronavirus SARS-CoV2 virus have been discovered on an atomic scale [35]. Therefore, blocking the surface of the ACE2 receptor with anti-ACE2 antibodies or peptides is another important strategy that can be considered against SARS-CoV-2 infection. In a study whose results were recently reported, the bioavailability of ACE2 has been improved with the use of CTB as a fusion carrier by enhancing its uptake with mucosal tissues such as the lungs [2]. It has also been observed that this enzyme has significant activity in blood and animal tissues when fed to laboratory mice with the recombinant bacteria containing the ACE2 gene. These findings also indicated that ACE2, which is expressed in the bacterial host and lacks its normal post-translational modifications (glycation) has bio-activity.
In the current study, we designed the hybrid protein containing the catalytic/extracellular domain of human ACE2 as fused to the CTB gene for expression in the E. coli strain BL21, which is used mostly as bacterial host for the large-scale production of recombinant proteins (Fig. 1). This protein might be used therapeutically to fight coronavirus infection while preserving its ability to bind to the new coronavirus. After protein optimizing expression at different times and in the presence of some additives (Figs. S1 and S2), the solubilization of the hybrid protein from the inclusion body was investigated (Figs. S3 and S4). The purity of the recombinant CTB-ACE2 hybrid protein was also analyzed by SDS-PAGE (reducing conditions) (Fig. 2). Our research showed that the hybrid protein has a molecular weight of ~ 100 kDa (without glycation), which is also supported by the other studies [7, 36, 37]. This molecular weight represents the sum of the mass of human ACE2 (85 kDa for deglycated ACE2) and CTB (11.6 kDa). In the following, while using the antibody that identified the histidine tag of the hybrid protein, it was well confirmed that the designed gene construct expressed the desired protein in the bacterial host (Fig. 3). Finally, the biophysical properties of this protein were studied using several spectroscopic techniques. In the primary structure, the recombinant CTB-ACE2 protein contains 22 Trp and 35 Tyr amino acid residues. According to the results in Figs. 4 and 5, the intensity of the fluorescence emission at 37 °C shows a decrease compared to room temperature; this observation suggests that this protein's tertiary structure is notably sensitive to temperature changes. As we examined the hydropathy index by ProtScale program/Kyke-Doolittle we observed that CTB-ACE2 protein is a hydrophilic protein. Our findings are consistent with the other study that identifies the pig counterpart protein as a hydrophilic protein [9]. The ACE2 metallopeptidase domain has nine 310 helices and 20 α-helical segments constituting 62% of its regular structures. Also, this protein has six short β-structures that account for 3.5% of the structure [38]. Also, the B-subunit of the cholera toxin has two α-helices and ten β-strands [39].
Considering the sum of secondary structures of the two components of the hybrid protein, it is estimated that it will be rich in α-helix and β-sheet with a lower content of turn and random coil structures. Our hybrid protein's calculated secondary structural contents by CD, FTIR and Raman assessments were similar (Figs. 6, 7, 8, 9 and Table 1). These results displayed that the secondary structural content of CTB-ACE2 protein decreases according to the following order: α-helix > β-sheet > turn > unordered structures. In agreement with the temperature-dependent tertiary structural changes as observed in the fluorescence studies (Figs. 4 and 5), the hydrodynamic size of the hybrid protein's oligomers also showed a significant decrease with increasing temperature (Fig. 11). Probably, the thermal dependence of the tertiary structure can explain the basis of the change in the oligomeric structure of this protein with temperature (Scheme 2).
5 Conclusions
ACE2 has an essential role in converting angiotensin II into angiotensin 1–7, protecting the heart and improving lung function. Also, this protein has a high potential for specific interaction with the coronavirus spike protein, enabling its entry into cells. Recently, the therapeutic properties of hrACE2 have been reported. Our designed construct, CTB-ACE2, indicated an efficient expression in E. coli strain BL21 as optimized, considering different incubation times and some additives. The purification of this protein was successfully performed in a way that makes it possible to produce it in a high amount for potential therapeutic uses. The secondary structural analyses of the recombinant protein showed that the α-helix and β-sheet secondary structures have the highest amount. Moreover, the fluorescence emission intensity and oligomeric size distribution of CTB-ACE2 protein exhibited a temperature-dependent change. Overall, the hybrid CTB-ACE2 protein may have therapeutic applications against the complications associated with diabetes and hypertension, and it can also be considered in the treatment regimen for Covid-19.
References
Atlas SA (2007) The renin-angiotensin aldosterone system: pathophysiological role and pharmacologic inhibition. J Manag Care Pharm JMCP 13:9–20. https://doi.org/10.18553/jmcp.2007.13.s8-b.9
Verma A, Xu K, Du T, Zhu P, Liang Z, Liao S, Zhang J, Raizada MK, Grant MB, Li Q (2019) Expression of human ACE2 in Lactobacillus and Beneficial effects in diabetic retinopathy in mice. Mol Ther Methods Clin Dev 14:161–170. https://doi.org/10.1016/j.omtm.2019.06.007
Hamming I, Timens W, Bulthuis MLC, Lely AT, Navis GJ, van Goor H (2004) Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol 203:631–637. https://doi.org/10.1002/path.1570
Lippi G, Lavie CJ, Henry BM, Sanchis-Gomar F (2020) Do genetic polymorphisms in angiotensin converting enzyme 2 (ACE2) gene play a role in coronavirus disease 2019 (COVID-19)? Clin Chem Lab Med 58(2020):1415–1422. https://doi.org/10.1515/cclm-2020-0727
Lambert DW, Yarski M, Warner FJ, Thornhill P, Parkin ET, Smith AI, Hooper NM, Turner AJ (2005) Tumor necrosis factor-alpha convertase (ADAM17) mediates regulated ectodomain shedding of the severe-acute respiratory syndrome-coronavirus (SARS-CoV) receptor, angiotensin-converting enzyme-2 (ACE2). J Biol Chem 280:30113–30119. https://doi.org/10.1074/jbc.M505111200
Aragão DS, Cunha TS, Arita DY, Andrade MCC, Fernandes AB, Watanabe IKM, Mortara RA, Casarini DE (2011) Purification and characterization of angiotensin converting enzyme 2 (ACE2) from murine model of mesangial cell in culture. Int J Biol Macromol 49:79–84. https://doi.org/10.1016/j.ijbiomac.2011.03.018
Tipnis SR, Hooper NM, Hyde R, Karran E, Christie G, Turner AJ (2000) A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J Biol Chem 275:33238–33243. https://doi.org/10.1074/jbc.M002615200
Crackower MA, Sarao R, Oudit GY, Yagil C, Kozieradzki I, Scanga SE, Oliveira-dos-Santos AJ, da Costa J, Zhang L, Pei Y, Scholey J, Ferrario CM, Manoukian AS, Chappell MC, Backx PH, Yagil Y, Penninger JM (2002) Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature 417:822–828. https://doi.org/10.1038/nature00786
Xiao H, Nie X-T, Ji X-X, Yan S, Zhu B, Zhang Y-S (2020) Establishing prokaryotic expression system of angiotensin-converting enzyme 2 (ACE2) gene in pigs. BioRxiv. https://doi.org/10.1101/2020.03.12.988634
Ghahramani M, Yousefi R, Niazi A, Kurganov B (2020) The congenital cataract-causing mutations P20R and A171T are associated with important changes in the amyloidogenic feature, structure and chaperone-like activity of human αB-crystallin. Biopolymers 111:e23350
Teale FWJ, Weber G (1957) Ultraviolet fluorescence of the aromatic amino acids. Biochem J 65:476
Zhang Y-Z, Zhou B, Liu Y-X, Zhou C-X, Ding X-L, Liu Y (2008) Fluorescence study on the interaction of bovine serum albumin with p-aminoazobenzene. J Fluoresc 18:109–118
Khoshaman K, Oryan A, Moosavi-Movahedi AA, Tamaddon AM, Kurganov BI, Yousefi R, Abolmaali SS (2017) impact of different mutations at Arg54 on structure, chaperone-like activity and oligomerization state of human αA-crystallin: The pathomechanism underlying congenital cataract-causing mutations R54L, R54P and R54C. BBA-Proteins Proteom 1865:604–618
Maltas E (2014) Binding interactions of niclosamide with serum proteins. J Food Drug Anal 22:549–555
Sharma KK, Kumar GS, Murphy AS, Kester K (1998) Identification of 1, 1′-bi (4-anilino) naphthalene-5, 5′-disulfonic acid binding sequences in α-crystallin. J Biol Chem 273:15474–15478
Bortolotti A, Wong YH, Korsholm SS, Bahring NHB, Bobone S, Tayyab S, van de Weert M, Stella L (2016) On the purported “backbone fluorescence” in protein three-dimensional fluorescence spectra. Rsc Adv 6:112870–112876
Grigoryan KR, Shilajyan HA (2017) Fluorescence 2D and 3D spectra analysis of tryptophan, tyrosine and phenylalanine. Chem Biol 51:3–7
Whitmore L, Wallace BA (2004) DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res 32:W668–W673
Sadat A, Joye IJ (2020) Peak fitting applied to Fourier transform infrared and Raman spectroscopic analysis of proteins. Appl Sci 10:5918
Khaleghinejad SH, Shahsavani MB, Ghahramani M, Yousefi R (2023) Investigating the role of double mutations R12C/P20R, and R12C/R69C on structure, chaperone-like activity, and amyloidogenic properties of human αB-crystallin. Int J Biol Macromol 242:124590
Zhou C, Qi W, Lewis EN, Carpenter JF (2015) Concomitant Raman spectroscopy and dynamic light scattering for characterization of therapeutic proteins at high concentrations. Anal Biochem 472:7–20
Schägger H, Von Jagow G (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166:368–379
Gasteiger E, Hoogland C, Gattiker A, et al (2005) Protein identification and analysis tools on the ExPASy server. Springer
Hawe A, Sutter M, Jiskoot W (2008) Extrinsic fluorescent dyes as tools for protein characterization. Pharm Res 25:1487–1499
Montgomerie S, Cruz JA, Shrivastava S, Arndt D, Berjanskii M, Wishart DS (2008) PROTEUS2: a web server for comprehensive protein structure prediction and structure-based annotation. Nucleic Acids Res 36:W202–W209. https://doi.org/10.1093/nar/gkn255
Bursell S-E, Yu N-T (1990) Fluorescence and Raman spectroscopy of the crystalline lens. Noninvasive Diagn Tech Ophthalmol 319–341
Rygula A, Majzner K, Marzec KM, Kaczor A, Pilarczyk M, Baranska M (2013) Raman spectroscopy of proteins: a review. J Raman Spectrosc 44:1061–1076
Benevides JM, Overman SA, Thomas GJ Jr (2003) Raman spectroscopy of proteins. Curr Protoc Protein Sci 33:17–18
Wen Z (2007) Raman spectroscopy of protein pharmaceuticals. J Pharm Sci 96:2861–2878
Lubbe L, Cozier GE, Oosthuizen D, Acharya KR, Sturrock ED (2020) ACE2 and ACE: structure-based insights into mechanism, regulation and receptor recognition by SARS-CoV. Clin Sci Lond Engl 1979 134:2851–2871. https://doi.org/10.1042/CS20200899
Zhang H, Baker A (2017) Recombinant human ACE2: acing out angiotensin II in ARDS therapy. Crit Care Lond Engl 21:305. https://doi.org/10.1186/s13054-017-1882-z
Oudit GY, Liu GC, Zhong J, Basu R, Chow FL, Zhou J, Loibner H, Janzek E, Schuster M, Penninger JM, Herzenberg AM, Kassiri Z, Scholey JW (2010) Human recombinant ACE2 reduces the progression of diabetic nephropathy. Diabetes 59:529–538. https://doi.org/10.2337/db09-1218
Tikellis C, Thomas MC (2012) Angiotensin-converting enzyme 2 (ACE2) is a key modulator of the renin angiotensin system in health and disease. Int J Pept 2012:256294. https://doi.org/10.1155/2012/256294
Imai Y, Kuba K, Rao S, Huan Y, Guo F, Guan B, Yang P, Sarao R, Wada T, Leong-Poi H, Crackower MA, Fukamizu A, Hui C-C, Hein L, Uhlig S, Slutsky AS, Jiang C, Penninger JM (2005) Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature 436:112–116. https://doi.org/10.1038/nature03712
Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS (2020) Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med 46:586–590. https://doi.org/10.1007/s00134-020-05985-9
Tada T, Fan C, Chen JS, Kaur R, Stapleford KA, Gristick H, Dcosta BM, Wilen CB, Nimigean CM, Landau NR (2020) An ACE2 microbody containing a single immunoglobulin fc domain is a potent inhibitor of SARS-CoV-2. Cell Rep 33:108528. https://doi.org/10.1016/j.celrep.2020.108528
D’Onofrio N, Scisciola L, Sardu C, Trotta MC, De Feo M, Maiello C, Mascolo P, De Micco F, Turriziani F, Municinò E, Monetti P, Lombardi A, Napolitano MG, Marino FZ, Ronchi A, Grimaldi V, Hermenean A, Rizzo MR, Barbieri M, Franco R, Pietro Campobasso C, Napoli C, Municinò M, Paolisso G, Balestrieri ML, Marfella R (2021) Glycated ACE2 receptor in diabetes: open door for SARS-COV-2 entry in cardiomyocyte. Cardiovasc Diabetol 20:99. https://doi.org/10.1186/s12933-021-01286-7
Towler P, Staker B, Prasad SG, Menon S, Tang J, Parsons T, Ryan D, Fisher M, Williams D, Dales NA, Patane MA, Pantoliano MW (2004) ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J Biol Chem 279:17996–18007. https://doi.org/10.1074/jbc.M311191200
Chaudhuri K, Chatterjee SN (2009) Cholera toxin (CT): structure. Cholera toxins. Springer, Berlin, pp 105–123
Acknowledgements
The financial support of the National Institute of Genetic Engineering and Biotechnology (NIGEB) is greatly appreciated. Also, the authors appreciate and acknowledge the financial support of the Shiraz University Research Council, and Iran National Science Foundation (INSF).
Funding
This work was also partially supported by INSF (grant number 99032483).
Author information
Authors and Affiliations
Contributions
MG: Software, Formal analysis, Investigation, Data Curation, Writing-Original Draft. MBS: Writing and Editing, Software, Investigation, Data Curation. SHK: Software, Investigation, Data Curation. AN: Methodology, Investigation Validation, Data Curation, Resources. AAM-Movahedi: Writing-Review and Editing, Visualization, Supervision, Funding acquisition. RY: Conceptualization, Methodology, Validation, Resources, Writing—Review and Editing, Visualization, Supervision, Project administration, Funding acquisition.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of Interest
The authors declare that they have no conflicts of interest with the contents of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ghahramani, M., Shahsavani, M.B., Khaleghinejad, S.H. et al. Efficient Expression in the Prokaryotic Host System, Purification and Structural Analyses of the Recombinant Human ACE2 Catalytic Subunit as a Hybrid Protein with the B Subunit of Cholera Toxin (CTB-ACE2). Protein J 43, 24–38 (2024). https://doi.org/10.1007/s10930-023-10164-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-023-10164-y