Abstract
Circulating exosomes are promising biomarker source in various diseases. Exosomal constituents can stably exist in the circulating plasma and serum thus making them ideal biomarkers for a number of clinical applications. Exosomes can also mediate the occurrence of many types of diseases, including distal cancerous metastasis and tumour enlargement, through encapsulated proteins or RNAs, which regulate interactions among tissues. While performing these actions, exosomes show tissue specificity. However, the mechanism for such selection is not clear. For non-small cell lung cancer (NSCLC), molecular diagnostic markers and mechanisms of exosome-mediated tumour metastasis are not well understood. Therefore, in this study, we characterized LLC exosomal proteins and mRNAs by analysing their molecular profiles, laying a foundation for exploring diagnostic markers of lung cancer. Furthermore, the interactions between exosomal membrane proteins and their target proteins were analysed and revealed a possible tissue propensity of LLC cell-derived exosomes. These findings provide a theoretical basis for studying exosome-mediated tissue targeting and distal lung cancer metastasis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Exosomes are cell derived membrane vesicles (30 to 120 nm) that are formed through inward intracellular multivesicular endosome (MVE) membrane bulging and subsequent pinching off of small membranous vesicles within the MVE [1]. Exosomes have been widely reported to mediate local and systematic cell communications through transferring informations such as nucleotide fragments, mRNAs, miRNAs, and proteins [2, 3]. They have also been used as biomarkers for several clinical diagnostic and prognostic applications [4] based on their internal components, including miRNAs, mRNA, and proteins. For example, MiRNA-1246, MiRNA-34a, MiR-29c have been reported to be potential biomarkers in oesophageal squamous cell carcinoma (ESCC) [5], Alzheimer’s disease [6], prostate cancer [7] and fibrosis [8], respectively. Despite miRNAs, exosomal proteins and mRNAs can also be used as biomarkers. CNN2 protein, CD2AP mRNAs and hnRNPH1 mRNA have been reported to be novel biomarkers for hepatic fibrosis [9], kidney diseases [10], and hepatocellular carcinoma [11], respectively.
Exosomes are reported to be involved in many important physiological processes and play important roles on the pathogenesis of cancer including tumour growth, angiogenesis, immune escape and metastasis. Exosomes derived from prostatic cancer has been identified to have effects of reduced apoptosis, increased cancer cell proliferation [12]. Exosomes derived from Mesenchymal stem cells (MSCs) have blocking effects on tumour angiogenesis and reduced activity of VEGF and NF-κB [13]. In terms of exosomes’ immune escape function, our lab early reported that tumour cell-derived miR-214 could promote tumour growth as well as host immune suppression by activating CD4+CD25highFoxp3+ regulatory T cells (Treg) [14]. And Theresa et al. have given a good summary about many important roles played by exosomes along many aspects of tumour growth process [15].
On the other hand, exosomes have been suggested to have certain tendencies to particular tissues, and the preference of exosomes towards certain tissues plays important role in cancer metastases. For example, Helen et al. have identified an interaction between CD21 on B cells and the EBV glycoprotein gp30 on lymphoblastoid cell line (LCL1)-derived exosomes [16]. Integrins differ by their expression patterns are also suggested to be involved in organ-specific metastasis [17]. In addition, pancreatic cancer exosomes can promote cancerous metastasis to liver via building up the pre-metastatic niche [18,19,20]. All these examples of distant metastasis related to tumor derived exosomes (TDEs) suggests that the interaction between exosomal surface proteins and the target tissues play an important role during this process [21, 22]. Therefore, research investigating the natural targeting properties of exosomes is important for studying interactions among tissues.
In this study, we characterized the protein and mRNA profiles of NSCLC LLC cell-derived exosomes. The relevant functions and preliminary natural targets were analysed by applying Gene Ontology (GO) annotation and Kyoto Encyclopedia for Genes and Genomes (KEGG) pathway analysis. The protein–protein interaction (PPI) analysis was carried out by applying STRING® and the resultant PPI networks were further interpreted by Cytoscape software with nodes defined based on the degree value and the edge was defined based on the Edge Betweenness. This work provides a theoretical basis for studying the molecular profiles of exosomes in NSCLC and reveal potential biomarkers for diagnosis and treatment of NSCLC.
2 Materials and Methods
2.1 Cells and Cell Culture
LLC cells lines (ATCC) were used to isolate exosomes. Cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) (ATCC 30-2002) containing 10% exosome-depleted fetal bovine serum (FBS) (ATCC 30-2020) and antibiotics (Gibco, CA, USA). Cell incubation performed at 37 °C in 5% CO2 and final exosomes were depleted using a 0.22-μm filter and then ultra-centrifuged at 110,000g for 2 h.
2.2 Exosome Purification, Characterization and Analyses
LLC Cells were grown with FBS-free medium for 48 h and the supernatants were then collected for subsequent centrifugation steps. The centrifugation procedures were 300 g × 5 min, 3000 g × 30 min and 10,000 g × 60 min. Then, the enriched medium was treated by applying Exosome Isolation Kit (Invitrogen) to isolate exosomes from the prepared cell culture based on the manufacture’s protocol [23, 24]. Exosomes were collected as pellets and subsequently re-suspended in phosphate-buffered saline (PBS). A 0.22-μm polyvinylidene fluoride (PVDF) membrane was used for clearing the purified exosome cell debris. At this stage, exosomes were ready for subsequently analysis including sequencing, western blot analysis and other biological studies.
2.3 RNA Isolation and Western Blot Analysis
LLC cell-derived exosomal RNA isolation procedures were carried out by using TRIzol LS Reagent (Invitrogen). Exosomes and proteins from LLC cells were quantified using antibodies against TSG101 MOUSE mAb (Santa Cruz sc-136111), CD63 Rabbit mAb (Santa Cruz sc-15363), CD9 Rabbit mAb (Santa Cruz sc-9148), Alix mouse mAb (Cell Signaling 2171s), GAPDH Mouse mAb (Santa Cruz sc-32233), KRAS Rabbit PAb (Proteintech 12063-1-AP), CDK1 Rabbit PAb (Proteintech 19532-1-AP), CDK3 Rabbit PAb (Proteintech 55103-1-AP).
2.4 LC–MS/MS and Data Analysis
The detailed methods of treatment and labelling of LLC exosomes, protein preparations and LC–MS/MS can be seen in our previous articles [25]. In terms of raw data analysis, Proteome Discoverer (Version 1.4.0.288, Thermo Fisher Scientific, Bremen, and Germany) with SEQUEST were applied as the search engine. And the UniprotKB human database (downloaded on July 20, 2018) was applied for MS/MS spectra analysis. Modification related settings were cysteine carbamidomethylation for fixed modification. N-terminal acetylation and methionine oxidation were set as variable modifications. The peptide mass was set at 10 ppm, and the fragment mass tolerances was 20 mDa. A maximum of 2 missed cleavage sites was allowed. Filtration of a 1% false discovery rate were set for Peptide identifications.
2.5 Microarrays
Human genome expression analysis was performed by applying the Affymetrix GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix, CA, USA) and the profiling analysis applied Affymetrix Expression Console software.
2.6 Functional Enrichment Analysis
Proteins identified in exosomes from either LLC or MIN6 cells were subjected to Gene Ontology (GO) and KEGG pathway analysis using Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resources 6.7 (http://david.abcc.ncificrf.gov/) [26, 27]. The protein–protein interaction (PPI) network analysis was performed by applying the STRING® programme (10.5 version) [28]. The resultant PPI images were further imported into Cytoscape_v3.6.1 for better resolution based on adjustment of the node interaction degree and scores [29].
3 Results
3.1 Extraction and Characterization of Exosomes Derived from LLC
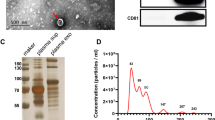
First, the LLC exosomes were purified by applying differential centrifugation methodologies and an Exosome Isolation Kit (Fig. 1a). Phenotypic features, including size and purity, were investigated using the TEM methodology (Fig. 1b). Western blot analysis proved that the exosome-specific proteins TSG101, CD63, CD9, and Alix were enriched in isolated exosomes but not found in cell lysates (Fig. 1c). Similarly, a high expression level of GAPDH was detected in cell lysates but not exosomes. Based on above results, it suggested that the exosome samples were well prepared and had a relatively high purity.
3.2 Proteomic Analysis of LLC Cell-Derived Exosomes
LC–MS/MS analysis was performed to examine the proteomic profiles of exosomes secreted by LLC cells. Two replicate samples of LLC cell-derived exosomes were studied and 1035 proteins were identified in common (S1 Table). Among these proteins, 120 proteins with an abundance value ≥ 2 were listed in Table 1. And 17 of 120 proteins with abundance value > 2 were presented in Fig. 2a. We analysed the commonly shared exosome-derived proteins using GO analysis of DAVID Bioinformatics Resources 6.7 software. Proteins were categorized by protein class, cellular component, molecular function, and biological process. The relative annotations represent specific functional enrichment of proteins (with data presentation of − log10 P value). And a high protein categorization value (%) refers to a greater functional enrichment. The protein classes of LLC cell-derived exosome were mostly associated with nucleic acid binding (21%) and hydrolase (15%) (Fig. 2b). Of the cellular component classifications, cell part (43%), organelle (27%), and macromolecular complex (22%) accounted for the majority of the contents (Fig. 2c). In terms of molecular function, the identified proteins showed high percentages of catalytic activity (40%) and binding (38%), which suggested that the LLC cell-derived exosomes possessed these two essential molecular functions (Fig. 2d). The biological processes of LLC cell-derived exosomal proteins were mainly concerned with cellular process (32%), metabolic process (25%), cellular component organization and biogenesis (10%) (Fig. 2e). On the other hand, a KEGG pathway identified through the DAVID Bioinformatics Resource. As shown in Fig. 2e, there were significant numbers of enriched proteins associated with the ribosome (119.4) and proteasome (59.34), and weaker associations with aminoacyl-tRNA biosynthesis (21.69), spliceosome (19.25), ECM-receptor interaction (18.06), and focal adhesion (15.40). Taken together, the LLC cell-derived exosomes may suggest to play functional roles involving synthesis, processing and degradation of DNA, RNA and proteins. In addition, they were involved in the maintenance of cell or tissue structure, function and carrying signals for cellular ECM. The identified pathway information was consistent with the biological process analysed above.
Proteomic analysis of LLC cell-derived exosomes. A heat map of 17 proteins with expression value > 2 derived from LLC exosome (a). A total of 1035 proteins were co-expressed among three samples. The proteins in LLC cell-derived exosomes were analysed by GO b protein classes, c cellular component, d molecular function, e biological process annotation, and f KEGG pathway g PPI network of proteins derived from LLC exosomes. Only nodes with interactions were shown and the network edges were calculated based on confidence and the rest settings were default including the minimum medium confidence of 0.4, and seven active interaction sources. Each node represents a protein and nodes with red colour are proteins with high degrees. Lines represent protein–protein associations. For sold line with thick patterns are associations with highest edge confidence. h Western blot analysis of CDK1, CDK3, KRAS and Alix protein levels in LLC, CT26 and Hepa1-6 cell-derived exosomes
The PPI network of LLC cell-derived exosomal proteins revealed 158 nodes (proteins with detected number ≥ 1 were selected) and 970 edges (Fig. 2g) (S2 Table). About 50% of proteins showed significant high degree values. Actin (ACTB) is crucial for several biological functions including cell motility, synaptic vesicle endocytosis, postsynaptic actin cytoskeleton organization, regulation of protein localization to plasma membrane. Heat shock protein HSP 90-beta (HSP90AB1) is generally involved in protein folding. And it is involved in the process of cell maturation, structural maintenance and regulation of target proteins. Cyclin-dependent kinase 1 (CDK1) is a protein kinase family and its function is tightly connected with the regulation of cell cycle control. In order to validate the potentiality of exosomal proteins to be biomarkers, we selected a group of proteins with high expression values, including CDK1, CDK3, PPP1CA, CDC42 and KRAS, Alix here were reference genes. And western blot analysis were performed (Fig. 2h). The levels of exosomal proteins of LLC cell line were compared with that of CT26 and Hepa1-6. Results show that CDK1 and CDK3 are more abundant in LLC exosomes and KRAS was abundant in exosomes of all these three types of cancer cells, while PPP1CA and CDC42 were not detected.
3.3 Conjoint Analysis of the mRNA and Protein Profiles of LLC-Derived Exosomes
In addition to the proteomic analysis of LLC-cell derived exosomes, the transcriptomics were analysed. Two replicas of the exosome samples were analysed based on the whole-genome expression arrays, and 2531 co-expression signals were identified (Fig. 3a) (S3 Table). Molecular functional studies of these mRNA transcripts revealed that among seven kinds of molecular functions, catalytic activity (45%) accounted for the largest part, followed by binding (37%), and transport activity (10%) (Fig. 3b). The intersection analysis among LLC exosomal proteins and mRNA transcripts showed 557 genes in common (Fig. 3c). Additionally, the KEGG pathway analysis showed that these genes were mostly related to oxidative phosphorylation (59.64), Huntington’s disease (46.48), Parkinson’s disease (32.10), and Alzheimer’s disease (31.59), which was similar to the molecular function analysis (Fig. 3d). Taking this into consideration, the data suggested that both RNA and proteins identified in the exosomes were translated or not had no significant correlations. For PPI of LLC exosomal mRNAs, top 100 of 2531 mRNAs were selected for PPI analysis and 92 nodes and 2849 edges identified (Fig. 3e) (S4 Table). Majorities of mRNAs with high degree values were translated to ribosomal proteins, such as Rps3a1, RPL41, RPL29, RPL17A. These proteins were either structural constituents or functional participants of ribosomes. There were UBA52 and UBB which were ubiquitin related proteins and involved in post translational modification and ubiquitin dependent translation. On the other hand, Eukaryotic translation elongation factor (EEF1A1) as its name indicates, it played major role in mRNA binding, receptor tyrosine kinase binding, translation initiation factor activity. Nascent polypeptide-associated complex subunit alpha (NACA) has function of preventing inappropriate targeting of non-secretory polypeptides to the ER.
Conjoint analysis of the mRNA and protein profiles of LLC cell-derived exosomes. a A heat map of 2531 mRNAs derived from LLC exosomes. b Molecular functional classification of 2531 mRNAs derived LLC exosomes. c Venn diagram of LLC exosomal mRNAs and proteins. 557 genes were shared between proteins (pink) and mRNA (blue) s. d KEGG pathway analysis of LLC exosomal mRNAs. e PPI network of mRNAs derived from LLC exosomes. Only nodes with interactions were shown and the network edges were calculated based on confidence and the rest settings were default including the minimum medium confidence of 0.4, and seven active interaction sources. Each node represents a protein and nodes with red colour are proteins with high degrees. Lines represent protein–protein associations. For sold line with thick patterns are associations with highest edge confidence
3.4 Target Tissue Analysis of LLC Exosomes
Exosomes mediated tissue targeting transport possibly by interacting with relevant receptors through surface membrane proteins. Therefore, it was necessary to identify proteins that were able to mediate exosome transport, which was very important for studying the functions and mechanisms of the exosomes. A total of 1035 proteins derived from LLC exosomes were further analysed for KEGG pathway. The signaling pathways involved in cell interaction were selected for protein interaction studies, including ECM-receptor interaction (Fig. 4a), gap junction (Fig. 4b), focal adhesion (Fig. 4c), adherens junction (Fig. 4d) and tight junction (Fig. 4e), and the details were listed in supplemental Table S5. The exosome proteins are labelled with stars in the figure, proteins present on the membrane interacting with the proteins labelled with a star on the opposite site were considered to be its target protein.
Functional analysis and target tissue prediction of LLC exosomal proteins. Analysis of the specific interactions of specific membrane binding proteins derived from LLC cell-derived exosomes, respectively. The functional annotation charts obtained from DAVID Bioinformatics Resources 6.7, in terms of ECM-receptor interaction (a), gap junction (b), focal adhesion (c), and adherens junction (d), tight junction (e), target tissue analysis (f). Proteins marked with stars are the proteins derived from LLC exosomes
To exert further efforts on the distribution of these target proteins, we applied the UP-TISSUE tool from the DAVID Bioinformatics Resource to investigate the tissue distribution of our protein targets of interest. The target proteins of LLC cell-derived exosomes were mainly associated with brain, mammary gland, bone marrow, placenta, lung, heart and liver (Fig. 4f) (S6 Table).
4 Discussion
Exosomes play vital roles in the pathogenesis of diseases, especially in the occurrence, angiogenesis and metastasis of cancer. They are naturally secreted exocrine bodies with characteristics of low immunogenicity and high efficiency. Compared with traditional drug delivery methods, including virus carriers and nano-particles, exosomes tend to have advantages of easy release, avoiding immune clearance, and high working efficiency. Therefore, exosomes have been broadly studied as drug delivery cargo in vivo.
The molecular composition of exosomes are rich in different molecules including mRNAs, microRNAs, and proteins. Many studies have found out that they are functionally important in applied as diagnostic markers [30], mediating cell–cell interaction and natural targeting. For example in HCC, exosomal IncRNA-RP11-513I15.6, miR-1262, and RAB11A have been suggested as potential diagnostic factors [31]. Exosomes derived from fibroblast are suggested to promote breast cancer cell metastasis through activating Wnt-planar cell polarity (PCP) signaling [32]. Secreted exosomal miR-150 has ability to promote angiogenesis in both vitro and vivo [33]. What’s more, exosome integrins α6β1 and αvβ5 were suggested to tightly associate with the cancerous metastasis of bone-tropic tumour to lung and liver, respectively [17]. Therefore, studies of the molecular composition of exosomes are very important to further clarify the function and use of exosomes as tools to treat diseases.
Lung cancer is a kind of highly malignant cancer and endangers human health due to its increased mortality and morbidity. In order to explore the role of lung cancer derived exosomes during the process of cancerous pathogenesis and metastasis, it is necessary to study the function of lung cancer-secreted endocrine bodies. Among genes identified from LLC cell derived proteins and mRNAs, only few proteins have been reported including FN1 [22] as a diagnostic marker for gastric cancer and FASN [34] as a candidate biomarker for prostate cancer. Since exosome are potential diagnostic markers for lung cancer, its surface proteins have attracted much attentions. Several lung cancer derived exosomal proteins have been reported as diagnostic markers for lung cancer, such as LRG1 [35], EGFR [36], CD9, CD63, CD81 [37], GRB2, SRC [38], CD151, CD171, tetraspanin8 [39], TCF21 [40], NY-ESO-1, PLAP, EpCam, Alix [41] and Tim-3 and Galectin-9 (Exo-T/G) [42]. In our study, majorities of identified proteins are not being reported yet, and we strongly believe that these proteins have great potentials to be diagnostic markers for lung cancer. ACTB, HSPA8, HSP90AB1, KRAS, CDK1, CFL1, RAB11A, YWHAE, CDC42, CLDN1, ACTR3, TUBB4B, EIF4A1, PPP1CA, TUBB1, and ENO1 are proteins which is highly correlated with lung cancer. Among these proteins, our results find that CDK1 and CKD3 are enriched specifically in exosomes derived from lung cancer cell line, suggesting that they may serve as potential markers for lung cancer. Intriguingly, KRAS was somehow globally abundant in exosomes derived from cancer cells, indicating that it may be a common indicator of cancer.
Our results identified that LLC exosomal proteins and mRNAs mainly possess catalytic and binding activity. And the protein KEGG pathways were dominantly ribosome and proteasome-related. This may suggest that LLC cell-derived exosomes could interact with the targeted cells and thus mediate the regulatory effects caused by LLC cells. Therefore, analysis of these results offers an innovative research direction for studying the regulatory effects of cancer tissues to their target tissues via exosomes.
Additionally, exosomes tend to have preferences for their target tissues. They play important role in the aspects of target tissues selection, regulatory effects of lung cancer on target tissues and metastasis. Thus, we further examined the natural targeting characteristics of LLC cell-derived exosomes. Previous studies have reported that lung cancer is easily metastasized to the brain [43], liver [44], lymph node [45], and bone [46]. In this study, the target proteins of the proteins derived from LLC exosomes are dominantly associated with brain, mammary gland, bone marrow, placenta, heart and liver, which are consistent with earlier reports. Based on our results, recognition of LLC exosomal CD47 and ECM protein THBS1/THBS2 may lead to exosome targeting to brain and therefore promote brain metastasis. Exosomal surface DAG1 binds to LAMA4 from ECM may facilitate exosome targeting to mammary gland, further leading to lung cancer mammary gland metastasis. In addition, our finding identified that recognition of exosomal surface ITGA5 and ECM COL5A2 could cause lung cancer bone marrow metastasis. In terms of the metastasis to liver, interactions between COL1A1/COL1A2 and SDC1 could cause the metastasis transfer to liver.
These studies provide a theoretical basis for investigating exosomal mediated distal metastasis. Moreover, certain exosomes containing highly expressed proteins and mRNAs could serve as potential prognostic and diagnostic biomarkers for lung cancer. Of course, these predicted results need to be confirmed by a large number of experiments. And these results provided new research directions for study of the natural targeting of exosomes and the metastasis of lung cancer mediated by exosomes.
Moreover, these data provide a basis for the selection of LLC cell exosomes as drug delivery system. As important drug delivery system, exosomes have been extensively studied for its roles in the pathogenesis of diseases, especially in the development, angiogenesis and metastasis of cancers. The molecules delivered by exosomes, such as proteins, mRNAs and miRNAs, are involved in these processes. And these disease-promoting molecules will have side effects on patients when exosomes were used as drug delivery vehicles. A comprehensive analysis of the components of LLC exosomes will provide a better understanding for biology of LLC cell-derived exosomes. And the same time, by studying the natural targeting of exosomes, drugs can be delivered to the target tissues more effectively.
In conclusion, this study examined the expression profiles of proteins and mRNAs of exosomes derived from LLC cells. And these results provide a theoretical basis for investigating the function of lung cancer cells secreted exosomes on the processes of lung cancer. Moreover, certain exosomes containing highly expressed proteins and mRNAs could serve as potential prognostic and diagnostic biomarkers for lung cancer. And the predicted results need to be confirmed by further experiments.
References
Gutierrez-Vazquez C, Villarroya-Beltri C, Mittelbrunn M, Sánchez-Madrid F (2013) Transfer of extracellular vesicles during immune cell-cell interactions. Immunol Rev 251(1):125–142
Street JM, Barran PE, Mackay CL, Weidt S, Balmforth C, Walsh TS, Chalmers RT, Webb DJ, Dear JW (2012) Identification and proteomic profiling of exosomes in human cerebrospinal fluid. J Transl Med 10:5
Fruhbeis C, Frohlich D, Kuo WP, Krämer-Albers EM (2013) Extracellular vesicles as mediators of neuron-glia communication. Front Cell Neurosci 7:182
Sharma R, Huang X, Brekken RA, Schroit AJ (2017) Detection of phosphatidylserine-positive exosomes for the diagnosis of early-stage malignancies. Br J Cancer 117(4):545–552
Takeshita N, Hoshino I, Mori M, Akutsu Y, Hanari N, Yoneyama Y, Ikeda N, Isozaki Y, Maruyama T, Akanuma N, Komatsu A, Jitsukawa M, Matsubara H (2013) Serum microRNA expression profile: miR-1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. Br J Cancer 108(3):644–652
Liu CG, Song J, Zhang YQ, Wang PC (2014) MicroRNA-193b is a regulator of amyloid precursor protein in the blood and cerebrospinal fluid derived exosomal microRNA-193b is a biomarker of Alzheimer’s disease. Mol Med Rep 10(5):2395–2400
Corcoran C, Rani S, O’driscoll L (2014) miR-34a is an intracellular and exosomal predictive biomarker for response to docetaxel with clinical relevance to prostate cancer progression. Prostate 74(13):1320–1334
Sole C, Cortes-Hernandez J, Felip ML, Vidal M, Ordi-Ros J (2015) miR-29c in urinary exosomes as predictor of early renal fibrosis in lupus nephritis. Nephrol Dial Transplant 30(9):1488–1496
Charrier A, Chen R, Chen L, Kemper S, Hattori T, Takigawa M, Brigstock DR (2014) Exosomes mediate intercellular transfer of pro-fibrogenic connective tissue growth factor (CCN2) between hepatic stellate cells, the principal fibrotic cells in the liver. Surgery 156(3):548–555
Lv LL, Cao YH, Pan MM, Liu H, Tang RN, Ma KL, Chen PS, Liu BC (2014) CD2AP mRNA in urinary exosome as biomarker of kidney disease. Clin Chim Acta 428:26–31
Xu H, Dong XY, Chen YM, Wang X (2018) Serum exosomal hnRNPH1 mRNA as a novel marker for hepatocellular carcinoma. Clin Chem Lab Med (CCLM) 56:479–484
Hosseini-Beheshti E, Choi W, Weiswald L-B, Kharmate G, Ghaffari M, Roshan-Moniri M, Hassona MD, Chan L, Chin MY, Tai IT, Rennie PS, Fazli L, Tomlinson Guns ES (2016) Exosomes confer pro-survival signals to alter the phenotype of prostate cells in their surrounding environment. Oncotarget 7(12):14639–14658
Alcayaga-Miranda F, González PL, Lopez-Verrilli A, Varas-Godoy M, Aguila-Díaz C, Contreras L, Khoury M (2016) Prostate tumor-induced angiogenesis is blocked by exosomes derived from menstrual stem cells through the inhibition of reactive oxygen species. Oncotarget 7(28):44462–44477
Yin Y, Cai X, Chen X, Liang H, Zhang Y, Li J, Wang Z, Chen X, Zhang W, Yokoyama S, Wang C, Li L, Li L, Hou D, Dong L, Xu T, Hiroi T, Yang F, Ji H, Zhang J, Zen K, Zhang C-Y (2014) Tumor-secreted miR-214 induces regulatory T cells: a major link between immune evasion and tumor growth. Cell Res 24:1164
Whiteside TL (2016) Tumor-derived exosomes and their role in cancer progression. Adv Clin Chem 74:103–141
Vallhov H, Gutzeit C, Johansson SM, Nagy N, Paul M, Li Q (2011) Exosomes containing glycoprotein 350 released by EBV-transformed b cells selectively target B cells through CD21 and block EBV infection in vitro. J Immunol 186(1):73
Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, Singh S, Williams C, Soplop N, Uryu K, Pharmer L, King T, Bojmar L, Davies AE, Ararso Y, Zhang T, Zhang H, Hernandez J, Weiss JM, Dumont-Cole VD, Kramer K, Wexler LH, Narendran A, Schwartz GK, Healey JH, Sandstrom P, Labori KJ, Kure EH, Grandgenett PM, Hollingsworth MA, de Sousa M, Kaur S, Jain M, Mallya K, Batra SK, Jarnagin WR, Brady MS, Fodstad O, Muller V, Pantel K, Minn AJ, Bissell MJ, Garcia BA, Kang Y, Rajasekhar VK, Ghajar CM, Matei I, Peinado H, Bromberg J, Lyden D (2015) Tumour exosome integrins determine organotropic metastasis. Nature 527(7578):329–335
Zhang Y, Wang XF (2015) A niche role for cancer exosomes in metastasis. Nat Cell Biol 17(6):709–711
Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK, Becker A, Hoshino A, Mark MT, Molina H, Xiang J, Zhang T, Theilen TM, Garcia-Santos G, Williams C, Ararso Y, Huang Y, Rodrigues G, Shen TL, Labori KJ, Lothe IM, Kure EH, Hernandez J, Doussot A, Ebbesen SH, Grandgenett PM, Hollingsworth MA, Jain M, Mallya K, Batra SK, Jarnagin WR, Schwartz RE, Matei I, Peinado H, Stanger BZ, Bromberg J, Lyden D (2015) Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol 17(6):816–826
Yu Z, Zhao S, Ren L, Wang L, Chen Z, Hoffman RM, Zhou J (2017) Pancreatic cancer-derived exosomes promote tumor metastasis and liver pre-metastatic niche formation. Oncotarget 8(38):63461–63483
Yue S, Mu W, Erb U, Zöller M (2015) The tetraspanins CD151 and Tspan8 are essential exosome components for the crosstalk between cancer initiating cells and their surrounding. Oncotarget 6(4):2366–2384
Arita T, Ichikawa D, Konishi H, Komatsu S, Shiozaki A, Ogino S, Fujita Y, Hiramoto H, Hamada J, Shoda K, Kosuga T, Fujiwara H, Okamoto K, Otsuji E (2016) Tumor exosome-mediated promotion of adhesion to mesothelial cells in gastric cancer cells. Oncotarget 7(35):56855–56863
Lee C, Carney RP, Hazari S, Smith ZJ, Knudson A, Robertson CS, Lam KS, Wachsmann-Hogiu S (2015) 3D plasmonic nanobowl platform for the study of exosomes in solution. Nanoscale 7(20):9290–9297
Liu Y, Li D, Liu Z, Zhou Y, Chu D, Li X, Jiang X, Hou D, Chen X, Chen Y, Yang Z, Jin L, Jiang W, Tian C, Zhou G, Zen K, Zhang J, Zhang Y, Li J, Zhang C-Y (2015) Targeted exosome-mediated delivery of opioid receptor Mu siRNA for the treatment of morphine relapse. Scientific Rep 5:17543
Li J, Chen X, Yi J, Liu Y, Li D, Wang J, Hou D, Jiang X, Zhang J, Wang J, Zen K, Yang F, Zhang CY, Zhang Y (2016) Identification and Characterization of 293T Cell-Derived Exosomes by Profiling the Protein, mRNA and MicroRNA Components. PLoS ONE 11(9):e0163043
Huang DW, Sherman BT, Lempicki RA (2008) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 44:44–57
Huang DW, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37(1):1–13
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C (2015) STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43(D1):D447–D452
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Benno S, Trey I (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504
Muller L, Muller-Haegele S, Mitsuhashi M, Gooding W, Okada H, Whiteside TL (2015) Exosomes isolated from plasma of glioma patients enrolled in a vaccination trial reflect antitumor immune activity and might predict survival. OncoImmunology 4(6):e1008347
Abd El Gwad A, Matboli M, El-tawdi A, Habib E, Shehata H, Ibrahim D, Tash F (2018) Role of exosomal competing endogenous RNA in patients with hepatocellular carcinoma. J Cell Biochem 119:8600–8610
Luga V, Zhang L, Viloria-Petit AM, Ogunjimi AA, Inanlou MR, Chiu E, Buchanan M, Hosein AN, Basik M, Wrana JL (2012) Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell 151(7):1542–1556
Li J, Zhang Y, Liu Y, Dai X, Li W, Cai X, Yin Y, Wang Q, Xue Y, Wang C, Li D, Hou D, Jiang X, Zhang J, Zen K, Chen X, Zhang CY (2013) Microvesicle-mediated transfer of microRNA-150 from monocytes to endothelial cells promotes angiogenesis. J Biol Chem 288(32):23586–23596
Duijvesz D, Burnum-Johnson KE, Gritsenko MA, Hoogland AM, Vredenbregt-van den Berg MS, Willemsen R, Luider T, Pasa-Tolic L, Jenster G (2013) Proteomic profiling of exosomes leads to the identification of novel biomarkers for prostate cancer. PLoS ONE 8(12):e82589
Li Y, Zhang Y, Qiu F, Qiu Z (2011) Proteomic identification of exosomal LRG1: a potential urinary biomarker for detecting NSCLC. Electrophoresis 32(15):1976–1983
Yamashita T, Kamada H, Kanasaki S, Maeda Y, Nagano K, Abe Y, Inoue M, Yoshioka Y, Tsutsumi Y, Katayama S, Inoue M, Tsunoda S (2013) Epidermal growth factor receptor localized to exosome membranes as a possible biomarker for lung cancer diagnosis. Pharmazie 68(12):969–973
Jakobsen KR, Paulsen BS, Baek R, Varming K, Sorensen BS, Jørgensen MM (2015) Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma. J Extracell Vesicles 4:26659. https://doi.org/10.3402/jev.v4.26659
Clark DJ, Fondrie WE, Yang A, Mao L (2016) Triple SILAC quantitative proteomic analysis reveals differential abundance of cell signaling proteins between normal and lung cancer-derived exosomes. J Proteom 133:161–169
Christian R (2016) Exosomal proteins in lung cancer: the last frontier in liquid biopsies. J Thorac Oncol 11(10):1609–1611
Wu H, Zhou JC, Zeng C, Wu D, Mu Z, Chen B, Xie Y, Ye Y, Liu J (2016) Curcumin increases exosomal TCF21 thus suppressing exosome-induced lung cancer. Oncotarget 7(52):87081–87090
Sandfeld-Paulsen B, Aggerholm-Pedersen N, Baek R, Jakobsen KR, Meldgaard P, Folkersen BH, Rasmussen TR, Varming K, Jorgensen MM, Sorensen S (2016) Exosomal proteins as prognostic biomarkers in non-small cell lung cancer. Mol Oncol 10(10):1595–1602. https://doi.org/10.1016/j.molonc.2016.10.003
Gao J, Qiu X, Li X, Fan H, Zhang F, Lv T, Song Y (2018) Expression profiles and clinical value of plasma exosomal Tim-3 and Galectin-9 in non-small cell lung cancer. Biochem Biophys Res Commun 498(3):409–415
Alexander CaR K (2010) Treatment of brain metastasis from lung cancer. Cancers (Basel) 2(4):2100–2137
Riihimaki M, Hemminki A, Fallah M, Thomsen H, Sundquist K, Sundquist J, Hemminki K (2014) Metastatic sites and survival in lung cancer. Lung Cancer 86(1):78–84
Hayama M, Chida M, Karube Y, Tamura M, Kobayashi S, Oyaizu T, Honma K (2014) One-step nucleic acid amplification for detection of lymph node metastasis in lung cancer. Ann Thorac Cardiovasc Surg 20(3):181–184
Liu M, Sun W, Liu Y, Dong X (2016) The role of lncRNA MALAT1 in bone metastasis in patients with non-small cell lung cancer. Oncol Rep 36(3):1679–1685
Acknowledgements
We thank for Pei He and Shibei Wang collecting exosomes of LLC cells and running assays for following bioinformatics investigations in this study. We thanks Yujing Zhang designing and supervising this work.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 31771666, 31741066, 31200969) and the Industrialization of Mongolian Medicine of “Prairie Talents” engineering (NEI ZU TONG ZI No. 2015-56).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, W., He, P., Wang, S. et al. Characterization of Protein Profiling and mRNA Expression of LLC Exosomes. Protein J 38, 586–597 (2019). https://doi.org/10.1007/s10930-019-09849-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-019-09849-0