Abstract
Human cystatin C (HCC) binds and inhibits all types of cysteine proteases from the papain family, including cathepsins (a group of enzymes that participate in a variety of physiological processes), which are some of its natural targets. The affinities of diverse proteases for HCC, expressed as equilibrium binding constants (Kb), range from 106 to 1014 M−1. Isothermal titration calorimetry (ITC) is one of the most useful techniques to characterize the thermodynamics of molecular associations, making it possible to dissect the binding free energy into its enthalpic and entropic components. This information, together with the structural changes that occur during the different associations, could enable better understanding of the molecular basis of affinity. Notwithstanding the high sensitivity of modern calorimeters, ITC requires protein concentrations in at least the 10–100 μM range to obtain reliable data, and it is known that HCC forms oligomers in this concentration range. We present herein a comparative study of the structural, thermal stability, and oligomerization properties of HCC and its stabilized variant (sHCC) L47C/G69C (which possesses an additional disulfide bridge) as well as their binding thermodynamics to the protease chymopapain, analyzed by ITC. The results show that, because sHCC remains monomeric, it is a better reporter than wild-type HCC to characterize the thermodynamics of binding to cysteine proteases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
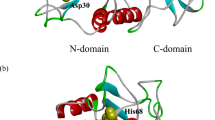
Cystatin C is an extracellular protein consisting of 120 amino acid residues that is ubiquitous in human body fluids, initially identified as an endogenous cysteine protease inhibitor [1]. In recent decades, it has been found that human cystatin C (HCC) participates in a variety of processes, such as bone resorption [2], antigen presentation [3], modulation of inflammatory responses, and fibroblast proliferation [4]. Likewise, it is associated with brain development processes such as neuropeptide activation and degradation, neurite proliferation, and neuronal survival [5, 6]. HCC is prone to form dimers and higher oligomers via a domain swapping mechanism [7]; this process is more favored in the naturally occurring L68Q HCC mutant, the major constituent of amyloid deposits in patients with hereditary amyloid angiopathy [8]. In addition, HCC has been found to colocalize with amyloid β-protein in amyloid deposits in the brain of patients with Alzheimer’s disease [9]. Domain swapping in the formation of dimers and other oligomers of cystatin C, including amyloid aggregates, totally disrupts the binding epitope, rendering all these species inactive as inhibitors of proteases of the papain family. Nilsson et al. [10] produced two variants of HCC with one additional disulfide bridge that prevents the domain swapping process; one of these monomeric variants is the double mutant L47C/G69C, hereinafter called stabilized HCC (sHCC), whose three-dimensional (3D) structure (3GAX, PDB) [11] is compared with the dimeric one (1G96, PDB) [7] obtained from wild-type HCC single crystals grown from protein solutions at concentration of 10 mg mL−1 [12] in Fig. 1. When sHCC is incubated in the presence of the reducing agent dithiothreitol (DTT) under conditions that produce oligomers from HCC, dimers and higher oligomers are also formed [13, 14]. sHCC shows the canonical structure of the cystatin family: a large α1 helix that runs across a five-stranded antiparallel β-sheet, with connectivity (N)–β1–α1–β2–L1–β3–(AS)–β4–L2–β5–(C), where AS is a large “appending structure” outside the core of the molecule, located on the opposite side of the binding epitope to papain family proteases, the wedge-like structure constituted by the loops L1 and L2 and the N-terminal region. Residues at this end that have been identified as main contributors to the affinity and selectivity for different proteases are L9, V10, and the evolutionarily conserved G11 [15, 16].
Domain-swapped dimer of human cystatin C (HCC) in green, superposed on the stabilized monomer sHCC in blue. Loops and β-strands are labeled, as well as the additional disulfide of sHCC shown in yellow. The three elements that constitute the inhibitor’s binding site to cysteine proteases, viz. two loops and the N-terminus, are highlighted in red on the sHCC backbone. Residues at this end, identified as main contributors to the affinity, are highlighted in dark blue (Color figure online)
The affinity of HCC for proteases from the papain family varies, with equilibrium binding constants (Kb) between 106 and 1014 M−1 [16,17,18,19]. This difference in affinity represents a change in the free energy of binding (ΔGb) of 46 kJ mol−1 at 300 K. One question that arises is which contributions to the binding energetics in this group of HCC–protease complexes explain this difference, the answer to which can be explored by determining the enthalpic and entropic components of ΔGb for the binding of HCC to different proteases of known structures using isothermal titration calorimetry (ITC) [20]. It is worth mentioning that it is not possible to measure the binding thermodynamics of high-affinity complexes (Kb > 108 M−1) through direct titration, even with the most sensitive microcalorimeters. However, strongly binding complexes can be characterized by displacement ITC [21]; in this mode of ITC, the receptor is first titrated with a weak ligand, then this bound ligand is displaced by titration with the high-affinity ligand. Analysis of both titrations gives the thermodynamic parameters for both binding processes [21, 22]. Thus, the characterization of the binding of HCC to a protease whose affinity could be determined by direct titration (Kb < 108 M−1) is a necessary first step. The Kb value reported for HCC and active chymopapain (CP) is 3.2 × 1010 M−1 [23]. Based on previous studies on the binding of chicken cystatin with active CP [24] and with CP blocked with iodoacetic acid [25], we decided to use CP in its carboxymethylated form to ensure that its Kb with HCC was < 108 M−1. ITC experiments require protein concentrations much higher than those previously used in fluorimetric measurements. HCC binds cysteine proteinases as a monomer; however, at concentrations higher than 0.20 mg mL−1, it forms dimers and higher oligomers. In this work, the binding thermodynamics of wild-type HCC and its stabilized monomer sHCC is studied and compared, and the use of the latter variant is proposed as a suitable substitute to characterize the binding thermodynamics of HCC to cysteine proteases.
2 Materials and Methods
2.1 Protein Expression and Purification
The gene encoding mature HCC [26], including the initiation codon, was synthesized and codon-optimized by GenScript. This synthetic gene was cloned between the NdeI and BamHI sites of the pET-24a vector. Plasmid construction was transformed into competent cell Top 10F′ (Invitrogen) for DNA manipulation and SHuffle® T7 Express lysY (New England Biolabs) for protein expression according to the manufacturer’s protocol. Variant L47C/G69C HCC (sHCC), which possesses a new disulfide that prevents the domain swapping process [10], was obtained by sequentially mutating residues Leu47 and Gly69 to cysteine. Mutagenesis was performed using the QuikChange II site-directed mutagenesis kit (Agilent). The mutagenic oligonucleotides were as follows: 5′-CCATTCTCGTGCGTGTCAGGTGGTTCG-3′(L47C forward) and 5′-CGAACCACCTGACACGCACGAGAATGG-3′ (L47C reverse); 5′-GGACGTCGAACTGTGTCGTACCACGTGC-3′ (G69C forward) and 5′-GCACGTGGTACGACACAGTTCGACGTCC-3′ (G69C reverse). Codon substitutions were verified by sequencing the complete genes. HCC and sHCC were expressed from a single colony, which was incubated at 30 °C in lysogeny broth (LB) medium supplemented with 25 μg mL−1 kanamycin until the OD600 reached ~ 0.6. Protein expression was induced with a final concentration of 0.4 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) for 4 h. Cells were recovered by centrifugation at 3800 × g and resuspended in buffer [40 mM Tris, 50 mM NaCl, and 1 mM ethylenediaminetetraacetic acid (EDTA), pH 8.0]. Cells were then lysed by ultrasonication (Sonifier 450; Branson Ultrasonics) employing 1.0 mM phenylmethylsulfonyl fluoride (PMSF) as protease inhibitor. The soluble fraction was obtained by centrifugation at 12,000 × g for 20 min at 4 °C. From these fractions, HCC and sHCC were purified by three chromatographic steps by fast protein liquid chromatograph (FPLC) using an ÄKTAprime plus. In the first stage, both samples were loaded onto a size-exclusion Superdex 200 column equilibrated with 50 mM glycine, 30 mM NaCl, pH 9.3; in the second stage, the samples were subjected to anion exchange chromatography using a HiPrep DEAE FF 16/60 column (40 mM Tris, pH 8.8), collecting the fractions that were not trapped (loaded) in the matrix. Finally, the proteins were obtained by size-exclusion chromatography performed on a Sephacryl 200 column (50 mM glycine, 30 mM NaCl; pH 9.3). The proteins were concentrated by ultrafiltration between each stage, while for the second stage, they were dialyzed against 40 mM Tris, pH 8.8. Proteins were stored at –70 °C.
Chymopapain (CP) was purified from a commercial extract (Sigma C-9007). One hundred milligrams of extract was dissolved in 50 mL phosphate-buffered saline (PBS) buffer (50 mM NaH2PO4/Na2HPO4; pH 7.0) in the presence of 20 mM cysteine; this mixture was stirred for 10 min at room temperature. To block the active-site cysteine of the protease, iodoacetic acid at final concentration of 30 mM was added to the solution; this reaction was left for 1 h with agitation in the dark, then the excess acid was removed by dialysis against PBS buffer. Finally, S-carboxymethyl-chymopapain (cmCP) was obtained in pure form by cation exchange chromatography using a Mono S 5/50 GL column (GE) with a NaCl gradient from 0 to 1.0 M.
The protein concentration was determined at 280 nm with a NanoDrop 2000 (Thermo Scientific) using extinction coefficients (ε) of 11,100 M−1 cm−1 for both HCC and sHCC [17] and 51,000 M−1 cm−1 for cmCP [27]. The homogeneity of the proteins was verified by sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis (PAGE) and matrix-assisted laser desorption–ionization (MALDI) time-of-flight (TOF) mass spectrometry.
2.2 Mass Spectrometry
Molecular weight measurements were performed using a MALDI-TOF mass spectrometer (AutoFlex Speed MALDI-TOF/TOF) equipped with a nitrogen ultraviolet (UV) laser (337 nm). Mass spectra were obtained in positive ion and linear mode with an accelerating voltage of 20 kV. α-Cyano-4-hydroxycinnamic acid (CHCA) was prepared as a saturated solution in 0.1% trifluoroacetic acid (TFA) in water/acetonitrile (1/1, v/v) and employed as a matrix for the analysis.
2.2.1 Molecular Weight of Recombinant HCC and sHCC
Both 1 μL CHCA matrix solution and 1 μL pure protein sample (HCC or sHCC, 1 mg mL−1) were applied onto a stainless-steel plate, mixed, and air-dried.
2.2.2 Confirmation of Formation of New Disulfide Bridge
Two aliquots of sHCC were dried by vacuum centrifugation and dissolved in 10 µL 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 5.0 M guanidinium chloride, and 1.0 mM EDTA pH 9.0 buffer. Samples were denatured at 85 °C for 15 min. One aliquot was used to determine the presence of free cysteines, for which the alkylating agent iodoacetamide was added to final concentration of 10 mM and incubated for 1 h at room temperature in the dark. The other aliquot was used to obtain the mass of the reduced and alkylated protein; in this case, the sample was first treated with 10 mM DTT for 15 min at 80 °C, then sulfhydryl groups were alkylated with iodoacetamide at final concentration of 25 mM and incubated for 1 h at room temperature in the dark. This last sample was loaded onto Zip-Tip C18 tips that had been previously activated and equilibrated and eluted in a mixture of 70% acetonitrile and 0.1% trifluoracetic acid. HCCA was also used as the matrix.
2.3 Circular Dichroism
To evaluate the structural integrity of the recombinant proteins, their circular dichroism (CD) spectra in the far-UV region were obtained. Previously, HCC and sHCC samples were dialyzed against 10 mM phosphate buffer (NaH2PO4/Na2HPO4), pH 7.0, and their concentrations were adjusted to 0.10 mg mL−1. CD spectra were obtained at 25 °C in a Jasco J715 spectropolarimeter using a 0.10-cm cell. The spectrum corresponding to the buffer was used as a blank.
2.4 Dynamic Light Scattering
Dynamic light scattering (DLS) analysis of HCC and sHCC solutions was performed at 25, 35, 50, and 65 °C using a Zetasizer NANO (Malvern, UK) equipped with temperature control. Proteins were dialyzed against 20 mM phosphate buffer with NaCl (56 mM, pH 7.0) and filtered through a membrane with pore size of 0.1 µm. The samples were analyzed at concentration of 0.40 mg mL−1. Prior to DLS measurements, the samples were equilibrated for 3 min at each desired temperature. At least three replicates of each sample were performed with multiple (≥ 10) data acquisitions.
2.5 Size-Exclusion Chromatography
Both HCC and sHCC solutions were dialyzed in membranes with a molecular cutoff of 6–8 kDa against 20 mM phosphate buffer with 100 mM NaCl, pH 7.0 and filtered through a pore size of 0.22 µm. Solutions with two protein concentrations, viz. 0.20 and 0.40 mg mL−1, were prepared for both inhibitors. Aliquots of both proteins at 0.40 mg mL−1 were incubated at 65 °C for 45 min, then cooled to room temperature. The samples were loaded into a Superdex 75 HR 10/30 column previously equilibrated with 20 mM phosphate buffer with 100 mM NaCl, pH 7.0, with a flow rate of 0.75 ml min−1 using an ÄKTA FPLC system.
2.6 Differential Scanning Calorimetry
Protein denaturation is a process that can be thermally activated and can be followed by differential scanning calorimetry (DSC) by analysis of the first heating. The thermal denaturation of HCC and sHCC was followed by DSC by heating at a rate of 90 K h−1 in a VP‐DSC capillary (GE Healthcare) automated with a 0.1-mL cell volume. Sweeps were obtained from 15 to 110 °C. For these experiments, the samples were dialyzed in a cellulose membrane with a molecular cutoff of 6–8 kDa against 20 mM phosphate buffer, with 60 mM NaCl, pH 7.0, then filtered through a membrane with pore size of 0.22 µM. Both samples were taken at concentration of 0.40 mg mL−1. Protein solutions were degassed with vacuum agitation, as was the end-of-dialysis buffer, which was used as a reference. The data obtained were analyzed using software developed by MicroCal implemented in Origin 7 (OriginLab, Northampton, MA). The “non-2-state model,” which is a two-state model that differentiates the calorimetric enthalpy (ΔHcal) from the van’t Hoff enthalpy (ΔHvH), was used for the analysis of the data.
2.7 Isothermal Titration Calorimetry
To evaluate the activity of the HCC and sHCC inhibitors, their binding to cmCP at 35 °C was studied using a MicroCal iTC200 calorimeter. Samples of HCC, sHCC, and cmCP were simultaneously dialyzed against 10 mM phosphate buffer (NaH2PO4/Na2HPO4), pH 7.0, with 78 mM NaCl to give an ionic strength of 0.10 M. The solutions were filtered through a membrane with pore size of 0.22 µm and degassed under vacuum with mild agitation before each experiment. For the HCC/cmCP system (cell/syringe), the concentrations of the samples were 0.020 mM and 0.135 mM, respectively, while for the cmCP/sHCC system, the concentrations were 0.020 mM and 0.140 mM, respectively. The calorimeter cell was filled with 200 µL HCC or cmCP, while the syringe was filled with 40 µL cmCP or sHCC as appropriate. The titration consisted of 21 injections of variable volume with constant agitation at 300 rpm. The data obtained were analyzed using Origin software with the model of one binding site. Prior to analysis, the data were corrected by subtracting the heats of dilution of the injectant into the buffer solution in the cell using the same injection program.
2.8 Molecular Dynamics Simulation
The 3D structure of monomeric HCC was constructed from the sHCC PDB file (3GAX) [11], changing Cys47 and 69 to Leu and Gly, respectively, followed by an energy-minimization protocol. Prior to molecular dynamics (MD) simulations, the side-chain ionization states in both HCC and sHCC at pH 7.0 were established using the pKa values as estimated using the PROPKA subroutine in the PDB2PQR server [28]. MD simulations were performed using GROMACS 5.0.7 software [29] with the optimized potential for liquid simulations (OPLS) force field [30]. Both proteins were placed at the center of a cubic box with its nearest edge 12 Å away from the protein. Approximately 20,500 extended simple point-charge (SPC/E) water molecules were required to fill the box, and one chloride ion was used to neutralize the net charge of each protein. The total number of atoms was 63,232 and 63,229 for the HCC and sHCC systems, respectively. Energy minimization was performed using the steepest-descent algorithm for 1000 steps followed by thermal equilibration (100 ps), restricting the position of the heavy atoms of the proteins by fixing a harmonic force constant of 1000 kJ mol−1. MD simulation was performed in an NPT ensemble with time step of 2 fs at temperature of 350 K or 450 K at 1.0 bar for 200 ns, using a V-rescale thermostat and Parrinello–Rahman barostat. A linear constraint solver (LINCS) [31] algorithm was applied to limit the length of all covalent bonds. A cutoff of 1.0 nm was applied for short-range electrostatic and van der Waals interactions, while long-range electrostatic forces were treated with the particle mesh Ewald method [32]. Two replicates were performed for each system.
3 Results
3.1 Structural Analysis of HCC and sHCC
The molecular mass of the recombinant inhibitors, wild-type cystatin C (HCC) and its disulfide-stabilized double mutant L47C/G69C (sHCC), determined by MALDI-TOF mass spectrometry, was 12,537 Da and 12,575 Da, respectively (Fig. S1, Online Resource 1), consistent with the backbones of both proteins being cleaved at their N-termini, losing their first eight amino acids. This unexpected cleavage of HCC has been reported previously [33, 34], although the full-length protein has also been obtained [10, 16,17,18,19]. Both inhibitors folded with identical secondary structures as judged from their far-UV CD spectra (Fig. 2). Using this spectrum and the K2D2 program [35], a content of 34% β structure and 9% α was predicted, while that reported for the crystallographic structure of sHCC (3GAX, PDB) [11] is 47% β and 13% α.
The presence of the additional disulfide introduced into sHCC, in addition to the two present in the wild-type HCC, was verified by mass spectrometry, as shown in Fig. 3. After confirming the absence of free cysteines in the nonreduced protein, the mass increment due to the reaction of iodoacetamide with the denatured and reduced protein corresponds to the carbamidomethylation of six cysteines, indicating the presence of three disulfides in sHCC [36].
3.2 Oligomerization of HCC
To test the capacity of HCC to dimerize and to confirm the elimination of this phenomenon in sHCC [10], size-exclusion chromatography (SEC) experiments were performed in a previously calibrated Superdex 75 column (Fig. 4). At 25 °C and protein concentration of 0.20 mg mL−1, the chromatogram of HCC showed a main peak associated with the monomer and a smaller peak related to the dimer (cyan curve). Doubling the concentration of HCC to 0.40 mg mL−1 caused a rise in the proportion of the dimer peak (blue line). In addition, heating an aliquot of this last sample for 40 min at 65.0 °C and cooling to 25 °C led to further dimer formation (red curve). On the other hand, a concentrated (0.40 mg mL−1) and heated sample of sHCC was eluted as a single peak in the zone corresponding to the monomer (black line), the same peak observed for a dilute (0.20 mg mL−1) and nonheated sample (not shown). Solutions of HCC at 0.40 mg mL−1 formed higher-order oligomers upon heating at 65 °C, according to DLS experiments, while under the same conditions, sHCC remained a monomer (Fig. S2, Online Resource 1).
Size-exclusion chromatograms of HCC and sHCC in a Superdex 75 column: HCC at 0.20 mg mL−1 (cyan line and triangles), HCC at 0.40 mg mL−1 (blue line and circles), HCC at 0.40 mg mL−1 heated at 65 °C and cooled (red line and circles), and sHCC at 0.40 mg mL−1 heated at 65 °C and cooled (black line and circle). The continuous line in panel B is the linear regression of the green data points, representing the peak positions and molecular weights of γ-globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa), and vitamin B12 (1.35 kDa) (Color figure online)
3.3 Thermal Stability of HCC and sHCC
The thermal stability of both inhibitors was analyzed by DSC. Endotherms obtained at the same protein concentration and solvent conditions as used in the SEC and DLS experiments are shown in Fig. 5. The thermal denaturation of both inhibitors was irreversible; i.e., after the first heating scanning, no significant change in the heat capacity of the solution was observed upon cooling and reheating. Even though equilibrium equations are not applicable under these conditions, the calculation of the van’t Hoff enthalpy (ΔHvH) and its comparison with the calorimetric enthalpy (ΔHcal) can help to compare the molecularity of the processes [37]. Table 1 presents the values of ΔHcal and ΔHvH obtained from the analysis of two independent scans, such as those in Fig. 5, assuming an equilibrium two-state model. The last column presents the factor by which ΔHvH is greater than ΔHcal.
3.4 Molecular Dynamics Simulations of Thermal Unfolding of the Inhibitors
During the 200-ns MD simulations of both proteins in explicit solvent at 450 K, unfolding of both polypeptides was observed. However, while HCC lost the α-helix in the first 15–30 ns, sHCC took more than 100 ns (Fig. 6a). The mobility of both chains was very similar at 350 K, as indicated by their almost superimposable root-mean-square fluctuation (RMSF) profiles (Fig. S3, Online Resource 1), which are directly related to the profiles of B-factors according to the equation \(B_{i} = \frac{{8\pi^{2} }}{3}\text{RMSF}_{i}^{2}\) [38]; at 450 K, the mobility was only slightly greater in HCC, even near the location where the new disulfide bridge was introduced in sHCC. A notable exception was the N-terminus, including the α-helix, which was much more mobile in the wild-type cystatin C (Fig. 6b).
3.5 Protease–Inhibitor Binding Thermodynamics
The binding thermodynamics of the protease cmCP to both HCC and sHCC was studied by means of ITC. To obtain reliable ITC measurements with this protein–protein system, we found the minimum concentration of protein in the cell to be approximately 0.020 mM and the minimum concentration of protein in the syringe to be approximately 0.140 mM. Figure 7 presents the binding isotherms for the HCC/cmCP and cmCP/sHCC systems (cell/syringe) at 35 °C and pH 7.0.
The thermodynamic parameters for the formation of both complexes, viz. cmCP–CCH and cmCP–sCCH, are presented in Table 2. The equilibrium binding constant (Kb) of the protease was three times larger for sHCC than for HCC. The binding enthalpies for both complexes were very similar, so the moderately higher affinity of chymopapain for sHCC was mainly due to a lower entropy diminution in the binding reaction. It is worth mentioning the low and similar value of n obtained for both complexes, which we cannot explain but which could be due to a fraction of chymopapain molecules that lost their ability to bind to the inhibitors.
4 Discussion
Recombinant human cystatin C (HCC) and its variant L47C/G69C with a new disulfide bridge (sHCC) were obtained in their folded and active conformations, both able to bind the cysteine protease S-carboxymethyl-chymopapain (cmCP). As eventually happens for HCC [33, 34], both proteins were cleaved at the peptide bond between R8 and L9, losing their first eight amino acids; their CD spectra were identical (Fig. 2) and quite similar to that previously reported for HCC [39].
Monomeric wild-type HCC forms dimers [7] and higher oligomers [10] through a domain swapping mechanism. Here, we clearly observed the formation of dimers at HCC concentrations ≥ 0.20 mg mL−1 (Fig. 4). Additionally, heating HCC solutions to 65 °C increased the formation of dimers (Fig. 4) and promoted the generation of high-molar-mass aggregates (Fig. S2), as also observed previously [39]. However, sHCC maintained its monomeric state even at high concentrations and temperatures (Figs. 4 and S2) due to its additional disulfide link that precludes dissociation of domains.
The thermal unfolding of both inhibitors was irreversible. The DSC thermogram obtained for HCC was similar to that previously reported [40]. Using a concentration of 0.40 mg mL−1, we determined values for Tm and for the ratio (ΔHvH/ΔHcal) of 78.6 °C and 2.92, respectively, comparable to the values of 82.0 °C and 2.67 obtained previously with equally truncated HCC at 1.0 mg mL−1 [40]. The new disulfide bond in sHCC increased the conformational stability of the protein, shifting the Tm to 89.5 °C and decreasing the ratio ΔHvH/ΔHcal to 1.86 (Table 1). The stabilizing effect of the S–S crosslink results from a decrease in the conformational entropy of the unfolded chain, which has been estimated by the equation [41]
where n is the number of residues in the loop forming the disulfide bond, equal to 23 for the C47–C69 disulfide in sHCC. Thus, the entropy diminution caused by this bond (ΔΔS) calculated using Eq. 1 is 48 J K−1 mol−1. As a reference, assuming that both denaturations were reversible and the energetic stabilization of the new disulfide was entirely due to this entropic effect (i.e., ΔΔH = 0), a Tm value of 93.9 °C would be expected for sHCC (see Online Resource 1). On the other hand, the higher ΔHvH/ΔHcal ratio of 2.92 for HCC compared with 1.86 for sHCC reveals the higher-oligomeric nature of the species undergoing thermal denaturation in the case of HCC [37].
Molecular dynamics simulations of the thermal unfolding of both inhibitors also reflected the higher stability of sHCC, particularly of the helix region, which took approximately five times longer to unfold than in HCC (Fig. 6a).
Regarding ITC experiments, note that the propensity of HCC to dimerize limits its use at concentrations above 0.20 mg mL−1, hindering its application as a titrant; thus, it was necessary to place it in the calorimetric cell and add the protease cmCP. In contrast, for the complex with sHCC, we filled the syringe with inhibitor at 1.76 mg mL−1 and titrated it with the cmCP solution within the cell. The measured affinities of cmCP for both inhibitors were very similar, with Kb values of 4.0 × 106 and 12 × 106 M−1 for HCC and sHCC, respectively; this slightly greater affinity of the protease for sHCC compared with HCC was also observed previously with cathepsin B, with Ki values of 0.23 nM and 0.37 nM, respectively [10].
The mean value of Kb for the complexes cmCP–HCC and cmCP–sHCC is 8.0 × 106 M−1, which, compared with the Kb of the active-CP–HCC complex of 3.2 × 1010 M−1 [23], gives a decrease in affinity of 20.7 kJ mol−1 at 300 K (\(\Delta \Delta G_{\rm b} = RT \ln K_{\rm b1} /K_{\rm b2}\)). This reduction is mainly caused by the carboxymethyl group introduced at the sulfur atom of the catalytic Cys of CP, which hinders the interaction with HCC. In the case of the complexes of HCC with actinidin and with carboxymethyl-actinidin, the respective Kb values are 5.3 × 107 M−1 and 8.3 × 105 M−1 [17], giving a ΔΔGb value of 10.4 kJ mol−1, which is only half the decrease in energy obtained with the chymopapain complexes. This dissection of the binding free energy of these complexes into their enthalpic and entropic components using ITC analysis will help to improve understanding of the molecular basis of these processes.
The ΔGb value for the complex of cmCP with sHCC was only 7.8% more negative than that with HCC (Table 2), and this difference was mainly due to the entropic component ΔSb, which was –32.4 J K−1 mol−1 for HCC and –14.3 J K−1 mol−1 for sHCC. This lesser entropic decrease can be explained by restrictions imposed by the new disulfide in the free sHCC. Assuming that the final bound state in both complexes is equally restricted, those bond rotations already restrained by the new link in the free sHCC will not contribute to the diminution of the binding entropy.
In conclusion, because stabilized human cystatin C (sHCC) cannot form dimers or higher oligomers, it represents a better model than the wild-type inhibitor for studying the wide variation in affinity for different cysteine proteases. Characterization of the binding thermodynamics of sHCC to diverse cysteine proteases of the papain family could shed new light on the structural basis that determines the protein–protein affinities.
Change history
24 June 2019
The original publication of this article contained a number of grammatical errors. Unfortunately, an incorrect version of the file that did not include some final language editing was inadvertently published online. The original article has been corrected.
References
Turk B, Turk D, Turk V (2000) Lysosomal cysteine proteases: more than scavengers. Biochim Biophys Acta 1477:98–111
Lerner UH, Grubb A (1992) Human cystatin C, a cysteine proteinase inhibitor, inhibits bone resorption in vitro stimulated by parathyroid hormone and parathyroid hormone-related peptide of malignancy. J Bone Miner Res 7:433–440
Kopitar-Jerala N (2006) The role of cystatins in cells of the immune system. FEBS Lett 580:6295–6301
Sun Q (1989) Growth stimulation of 3T3 fibroblasts by cystatin. Exp Cell Res 180:150–160
Sastre M, Calero M, Pawlik M, Mathews PM, Kumar A, Danilov V, Schmidt SD, Nixon RA, Frangione B, Levy E (2004) Binding of cystatin C to Alzheimer’s amyloid beta inhibits in vitro amyloid fibril formation. Neurobiol Aging 25:1033–1043
Palm DE, Knuckey NW, Primiano MJ, Spangenberger AG, Johanson CE (1995) Cystatin C, a protease inhibitor, in degenerating rat hippocampal neurons following transient forebrain ischemia. Brain Res 691:1–8
Janowsky R, Kozak M, Jankowska E, Grzonka Z, Grubb A, Abrahamson M, Jaskolski M (2001) Human cystatin C, an amyloidogenic protein, dimerizes through three-dimensional domain swapping. Nat Struct Biol 8:316–320
Abrahamson M (1996) Molecular basis for amyloidosis related to hereditary brain hemorrhage. Scand J Clin Lab Invest Suppl 226:47–56
Levy E, Sastre M, Kumar A, Gallo G, Piccardo P, Ghetti B, Tagliavini F (2001) Codeposition of cystatin C with amyloid-β protein in the brain of Alzheimer disease patients. J Neuropathol Exp Neurol 60:94–104
Nilsson M, Wang X, Rodziewicz-Motowidlo S, Janowski R, Lindström V, Önnerfjord P, Westermark G, Grzonka Z, Jaskolski M, Grubb A (2004) Prevention of domain swapping inhibits dimerization and amyloid fibril formation of cystatin C. J Biol Chem 279:24236–24245
Kolodziejczyk R, Michalska K, Hernandez-Santoyo A, Wahlbom M, Grubb A, Jaskolski M (2010) Crystal structure of human cystatin C stabilized against amyloid formation. FEBS J 277:1726–1737
Kozak M, Jankowska E, Janowski R, Grzonka Z, Grubb A, Alvarez Fernandez M, Abrahamson M, Jaskolski M (1999) Expression of a selenomethionyl derivative and preliminary crystallographic studies of human cystatin C. Acta Crystallogr D 55:1939–1942
Wahlbom M, Wang X, Lindström V, Carlemalm E, Jaskolski M, Grubb A (2007) Fibrillogenic oligomers of human cystatin C are formed by propagated domain swapping. J Biol Chem 282:18218–18326
Östner G, Lindström V, Christensen PH, Kozak M, Abrahamson M, Grubb A (2013) Stabilization, characterization, and selective removal of cystatin C amyloid oligomers. J Biol Chem 288:16438–16450
Hall A, Håkansson K, Mason RW, Grubb A, Abrahamson M (1995) Structural basis for the biological specificity of cystatin C. J Biol Chem 270:5115–5121
Björk I, Pol E, Raub-Segall E, Abrahamson M, Rowan AD, Mort JS (1994) Differential changes in the association and dissociation rate constants for binding of cystatins to target proteinases occurring on N-terminal truncation of the inhibitors indicate that the interaction mechanism varies with different enzymes. Biochem J 299:219–225
Lindahl P, Abrahamson M, Björk I (1992) Interaction of recombinant human cystatin C with the cysteine proteinases papain and actinidin. Biochem J 281:49–55
Brieditis I, Raub-Segall E, Pol E, Hakansson K, Abrahamson M (1996) The importance of the second hairpin loop of cystatin C for proteinase binding. Characterization of the interaction of Trp-106 variants on the inhibitor with cysteine proteinases. Biochemistry 35:10720–10726
Turk V, Stoka V, Vasiljeva O, Renko M, Sun T, Turk B, Turk D (2012) Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim Biophys Acta 1824:68–88
Wiseman T, Williston S, Brandts JF, Lin L-N (1989) Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem 179:131–137
Sigurskjold BW (2000) Exact analysis of competition ligand binding by displacement isothermal titration calorimetry. Anal Biochem 277:260–266
Velazquez-Campoy A, Freire E (2006) Isothermal titration calorimetry to determine association constants for high-affinity ligands. Nat Protoc 1:186–191
Buttle DJ, Abrahamson M, Barret AJ (1986) The biochemistry of the action of chymopapain in relief of sciatica. Spine 11:688–694
Björk I, Ylinenjarvi K (1990) Interaction between chicken cystatin and the cysteine proteinases actinidin, chymopapain A, and ficin. Biochemistry 29:1770–1776
Reyes-Espinosa F, Arroyo-Reyna A, García-Gutiérrez P, Serratos IN, Zubillaga RA (2015) Effects of pH on the association between the inhibitor cystatin and the proteinase chymopapain. Protein Pept Lett 22:239–247
Abrahamson M, Grubb A, Olafsson I, Lundwall Å (1987) Molecular cloning and sequence analysis of cDNA coding for the precursor of the human cysteine proteinase inhibitor cystatin C. FEBS Lett 216:229–233
Buttle DJ, Barret AJ (1984) Chymopapain. Chromatographic purification and immunological characterization. Biochem J 223:81–88
Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA (2004) PDB2PQR: an automated pipeline for the setup, execution, and analysis of Poisson–Boltzmann electrostatics calculations. Nucleic Acids Res 32:W665–W667
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindhal E (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2:19–25
Jorgensen WL, Maxwell DS, Tirado-Rives J (1996) Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc 118:11225–11236
Hess B, Bekker H, Berendsen HJC, Fraaije JGEM (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18:1463–1472
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092
Carrette O, Burkhard PR, Hughes S, Hochstrasser DF, Sanchez JC (2005) Truncated cystatin C in cerebrospinal fluid: technical artefact or biological process? Proteomics 5:3060–3065
Del Boccio P, Pieragostino D, Lugaresi A, Di Ioia M, Pavone B, Travaglini D, D’Aguanno S, Bernardini S, Sacchetta P, Federici G, Di Ilio C, Gambi D, Urbani A (2007) Cleavage of cystatin C is not associated with multiple sclerosis. Ann Neurol 62:201–204
Perez-Iratxeta C, Andrade-Navarro MA (2008) K2D2: estimation of protein secondary structure from circular dichroism spectra. BMC Struct Biol 8:25
Calvete JJ (2007) Determinación del número de grupos sulfidrilo y de enlaces disulfuro mediante espectrometría de masas. Proteómica 0:21–28. https://helvia.uco.es/bitstream/handle/10396/8926/pro4.pdf?sequence=1&isAllowed=y
Marky LA, Breslauer KJ (1987) Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers 26:1601–1620
Kuzmanic A, Zagrovic B (2010) Determination of ensemble-average pairwise root mean-square deviation from experimental B-factors. Biophys J 98:861–871
Perlenfein TJ, Murphy RM (2016) Expression, purification, and characterization of human cystatin C monomers and oligomers. Protein Expr Purif 117:35–43
Žerovnik E, Cimerman N, Kos J, Turk V, Lohner K (1997) Thermal denaturation of human cystatin C and two of its variants; comparison to chicken cystatin. Biol Chem 378:1199–1203
Pace CN, Grimsley GR, Thomson JA, Barnet BJ (1988) Conformational stability and activity of ribonuclease T1 with zero, one, and two intact disulfide bonds. J Biol Chem 263:11820–11825
Acknowledgments
This work was supported by the National Council of Science and Technology (CONACyT, México) by grants no. 181049 and 237256, by scholarship no. 283296 to D.O. Tovar-Anaya, and by a postdoctoral scholarship to M.T. Vieyra-Eusebio. The authors thank the Laboratorio de Supercómputo y Visualización en Paralelo and the Laboratorio Divisional de Espectrometría de Masas at the Universidad Autónoma Metropolitana-Iztapalapa for the use of their facilities. We are grateful to Miguel Costas from the Facultad de Química, UNAM for providing the VP‐DSC capillary facility to perform the differential scanning calorimetry experiments. We also thank Georgina Garza-Ramos from the Facultad de Medicina, UNAM for her expert help and facilitating our use of the FPLC for SEC experiments.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
All the authors declare that they have no conflicts of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original version of this article was revised: The grammatical errors in the article have been corrected.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tovar-Anaya, D.O., Vera-Robles, L.I., Vieyra-Eusebio, M.T. et al. Stabilized Human Cystatin C Variant L47C/G69C Is a Better Reporter Than the Wild-Type Inhibitor for Characterizing the Thermodynamics of Binding to Cysteine Proteases. Protein J 38, 598–607 (2019). https://doi.org/10.1007/s10930-019-09839-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-019-09839-2