Abstract
The generation of synthetic patient data that reflect the statistical properties of real data plays a fundamental role in today’s world because of its potential to (i) be enable proprietary data access for statistical and research purposes and (ii) increase available data (e.g., in low-density regions—i.e., for patients with under-represented characteristics). Generative methods employ a family of solutions for generating synthetic data. The objective of this research is to benchmark numerous state-of-the-art deep-learning generative methods across different scenarios and clinical datasets comprising patient covariates and several pharmacokinetic/pharmacodynamic endpoints. We did this by implementing various probabilistic models aimed at generating synthetic data, such as the Multi-layer Perceptron Conditioning Generative Adversarial Neural Network (MLP cGAN), Time-series Generative Adversarial Networks (TimeGAN), and a more traditional approach like Probabilistic Autoregressive (PAR). We evaluated their performance by calculating discriminative and predictive scores. Furthermore, we conducted comparisons between the distributions of real and synthetic data using Kolmogorov-Smirnov and Chi-square statistical tests, focusing respectively on covariate and output variables of the models. Lastly, we employed pharmacometrics-related metric to enhance interpretation of our results specific to our investigated scenarios. Results indicate that multi-layer perceptron–based conditional generative adversarial networks (MLP cGAN) exhibit the best overall performance for most of the considered metrics. This work highlights the opportunities to employ synthetic data generation in the field of clinical pharmacology for augmentation and sharing of proprietary data across institutions.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The value of data sharing and integration across various biopharmaceutical companies is increasingly recognized as a means to address common challenges and increase the efficiency of drug development. However, such data sharing initiatives are still challenging to set up as access to proprietary data on compounds under development cannot be provided for research purposes, given their competitive value and risks associated with data privacy. Synthetic data generation holds a promise to mitigate these issues by enabling data providers to share a synthetic version of the original data while preserving the main statistical properties.
The core idea of generative methods is to develop a model that, from a given dataset, can capture the underlying joint statistical distribution and can be used to sample new datasets that can approximate the statistics of the original one. Popular applications of generative methods include (i) data privacy: a synthetic data is less sensitive than the original as the risk of re-identification by linkage is significantly reduced by the synthetization process and (ii) data augmentation: the involved techniques may up-sample areas where the density of a given variable or set of variables is low (data imputation) and mitigate bias for under-represented groups. Gaussian-mixture models (GMMs) [1] are perhaps among the most-studied generative techniques. GMMs approximate the distribution of a collection of data points (i.e., a dataset) with a mixture of Gaussian distributions; the parameters of the approximated distribution are obtained using the expectation-maximisation algorithm [2]. Additionally, non-linear mixed-effect models [3] are the primary tool for pharmacometrics (PMX) analysis. They account for both fixed and random effects to describe the dynamics of drug effects over time at the individual subject level, which may be affected by numerous covariates (e.g., gender, race or weight) and random unexplained effects. Once a non-linear mixed-effect model is fitted to the given data, it may be used to simulate the response of patients to new doses and dosing regimens. However, the generative capabilities of such models are limited to some information (e.g., PK/PD response); moreover, complex models with assumptions are required to first characterize and then reproduce through simulations datasets including several response variables and covariates.
Although the literature on generative models spans several decades, the most-recent progress focuses on neural network–based generative models, also referred to as deep generative models. Variational autoencoders [4], Generative Adversarial Neural Networks (GANs) [5], flow-based generative models [6] and diffusion models [7] are among the prevalent deep generative models. As opposed to GMMs, deep generative models do not require the definition of an explicit distribution; their parameters are implicitly learned during the learning process. Previous studies have employed generative models in the healthcare domain. Deep generative models have been used to generate electronic health records [8], magnetic-resonance images [9], electrocardiograms [10], X-ray images [11], etc.
In the domain of model-informed drug discovery and development, GAN-based synthetic data generators can be used to enable designing optimized drug candidates by performing de novo drug design and generation of drug-like molecules [12]. Moreover, in clinical oncology, GANs can enhance the way of dealing with multimodal datasets e.g., multi-omics data. This Deep Learning (DL) approach can help to explore cancer pathogenesis, determine clinical actionability in treatment and improve patients’ survival rates [13]. Moreover, advanced Machine learning (ML) methods like deep generative models based on GAN can be utilized for generating biomarkers by learning data distributions in an unsupervised manner [14]. However, to the best of our knowledge, the application of these approaches to synthesize Pharmacokinetics (PK)/Pharmacodynamics (PD) data has been limited.
In this study, we explore the application of deep generative models to clinical-pharmacology datasets, including PK/PD data and patients’ covariates.
In the following sections, we first present some theoretical aspects of generative models. We then describe a design of data simulations, followed by different ML/DL simulation methods, a simulation method for (pseudo) real datasets and metrics for evaluating the generated synthetic data.
Methods
Data design
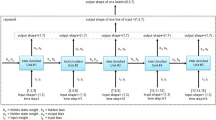
A dataset is defined as a collection of records for a set of subjects that result from a clinical study or trial. For every subject, the dataset contains a sequence of observation and dosing events recorded over the study period. For every event, there exists a record in the dataset that indicates the event time, the amount of drug (if any) administered and one or more observed PK/PD measurements. Moreover, every record includes potential covariates that may influence PK/PD measurement, such as patient’s demographics. Covariates may be time-independent (i.e., static, or constant), such as gender or race, or time-dependent, such as body weight. All records are chronologically ordered by the actual event time for an individual. For simplicity, we assume that the dataset does not contain missing values. This is presented in Fig. 1.
For every subject, the pharmacokinetics/pharmacodynamics (PK/PD) dataset contains a number of dosing and observation records. The first and second rows depict instances of dosing and observation records, respectively. The time information reflects the relative time elapsed since the first record registered for each patient. Notably, we observe intra-arm variability. Intra-arm variability refers to the (small) time discrepancies across subjects within the same treatment arm. In the absence of intra-arm variability, all the subjects within a same treatment arm follow the same exact time schedule. However, in practice the occurrence of events deviates from the designed schedule. While we acknowledge that intra-arm variability is a relevant characteristic of the data, the design of solutions to efficiently handle it is beyond the scope of this work, and we will consider it in future studies. Therefore, in the following figures, we assume all the subjects within a same treatment arm follow an identical time schedule
For this work, we group the above-mentioned information into two categories:
-
Clinical design information. It refers to the sampling time and dosing information (drug dosing regimen). Within the same study, clinical design may define one (i.e., single-arm studies) or more treatment arms (i.e., multi-arm studies). Drug combinations are not considered in the discussed examples but can similarly be captured.
-
Clinical observations. They include static and time-varying covariates as well as PK/PD measurements.
We denote each row of the dataset as \(\:{\varvec{x}}_{id}^{\left(i\right)}\), where the subscript id is the patient identifier and the superscript i indicates the i-th record in the sequence of the observation and dosing events for the patient id. We refer to \(\:\varvec{x}\) as an event record; it comprises \(\:\varvec{a}\) and \(\:\varvec{o}\), referring to the corresponding clinical design information and clinical observations, respectively. Moreover, \(\:{\varvec{x}}_{id}^{\left(id\right)}=[{\varvec{a}}_{id}^{\left(i\right)};\:{\varvec{o}}_{id}^{\left(i\right)}]\), where “;” is the concatenation operator and the identifier is not a part of the event record. The clinical observations \(\:\varvec{o}\) can be bifurcated into those that change over time \(\:\varvec{d}\) (i.e., PK/PD measurements and time-varying covariates) and those that remain unchanged \(\:\varvec{s}\) (i.e., static covariates).
Generative models
The objective of generative methods is to model the joint probability \(\:\prod\:_{id}P\left({\{\varvec{x}}_{id}^{\left(i\right)}\right)\) and is equivalent to
where N indicates the length of the sequence of events records available for the subject id.
The main differences across methods are based on (i) the factorisation applied to the above joint probability and (ii) the architecture modelling the constituents of the factorised probability distribution. In the following section, we discuss suitable generative approaches for longitudinal data with heterogeneous covariate types.
Probabilistic autoregressive model
The Probabilistic Autoregressive (PAR) [15] model defines a probability distribution over sequences of data explicitly based on conditionals (likelihood estimation), which are approximated and parameterized by neural networks. Indeed, it processes the sequence of data using a Recurrent Neural Network (RNNs), a type of DL architectures to learn patterns across the input data over time. Moreover, it models each covariate as a gaussian distribution, and its training based on the maximum likelihood estimation. PAR breaks down the learning of joint probability (Eq. (1)) into two distinct components. First, a generative component \(\:{G}_{Copula}\)—a Gaussian copula model [16]—is learned for modelling the static covariates. This is followed by the learning of a second generative model \(\:{G}_{RNN}\)—a recurrent neural network (RNN) [17]—which is conditioned on the static covariates for modelling all other information.
An identical two-step process is performed during inference. For a synthetic subject with identifier \(\:\stackrel{\sim}{id}\), PAR generates synthetic static covariates from uniformly distributed noise \(\:\varvec{u}:\:{\varvec{s}}_{\stackrel{\sim}{\:id}}\:\leftarrow\:{G}_{Copula}\left(\varvec{u}\right)\). The synthetic static covariates condition the second generative model to generate all other information: \(\:\{{{\varvec{d}}_{\stackrel{\sim}{id}}^{\left(i\right)}\}}_{i=1}^{N},\:\{{{\varvec{a}}_{\stackrel{\sim}{id}}^{\left(i\right)}\}}_{i=1}^{N}\:\leftarrow\:{G}_{RNN}\left({\varvec{s}}_{\stackrel{\sim}{\:id}}\right)\).
The main advantage of this model is its ability to deal with heterogeneous covariates effectively. By dividing the learning process into two components, the model effectively tackles static and dynamic covariates separately—this division is particularly advantageous when dealing with PK/PD datasets. The limitation of the method is that depending on data complexity, the two-step process may become computationally intensive and time-consuming.
TimeGan
TimeGan [18] is a complex framework involving 4 Neural Networks to synthesize sequential data. These architectures play distinct roles in the process of data modelling: the expected generator and the discriminator, but also, by a recovery and embedder model. TimeGan is a GAN [5] for time-series/longitudinal data. It extends the min–max loss of the standard GAN with additional unsupervised objectives. It provides consistent and significant improvements over state-of-the-art benchmarks in generating realistic time-series/longitudinal data.
Unlike PAR, the generative process of TimeGan is end-to-end. During inference, TimeGAN generates synthetic sequences from Gaussian noise.
\(\:\varvec{z}:\:\{{{\varvec{d}}_{\stackrel{\sim}{id}}^{\left(i\right)}\}}_{i=1}^{N},\:\{{{\varvec{a}}_{\stackrel{\sim}{id}}^{\left(i\right)}\}}_{i=1}^{N},\:{\varvec{s}}_{\stackrel{\sim}{\:id}}\:\leftarrow\:{G}_{TimeGAN}\left(\varvec{z}\right)\). Although its benefits have been demonstrated in several standard benchmark datasets, its complexity, which is discussed hereafter, may pose a challenge for use cases with some records/sequences and instances/subjects.
MLP-based conditional GAN
A Multi-Layer Perceptron (MLP)-based Conditional GAN) is a NN architecture that combines the power of MLPs and GANs for conditional data generation. It consists of a generator network that takes as input noise vectors concatenated with conditional information and learns to generate synthetic data conditioned on specific attributes or labels. The discriminator network is trained to distinguish between real and generated samples, incorporating conditional information to ensure that the generated samples align with the specified conditions.
To reduce the complexity of TimeGan, we explored an approach based on conditional GAN (cGAN) [19]. This approach is referred to as MLP cGAN. The generative modelling of MLP cGAN is conditioned on the clinical design (i.e., the treatment arm). This modelling choice reflects the reality that a subject is allocated to a treatment arm before the onset of a trial or clinical study. Therefore, the learning problem is reduced to modelling the conditional probability \(\:\prod\:_{id}P\left({\{\varvec{d}}_{id}^{\left(i\right)}{\}}_{i=1}^{N},\:{\varvec{s}}_{id}\right|{\varvec{a}}_{id})\). This step not only reduces the learning complexity of the problem but also provides a mechanism to select the treatment arm from which data can be synthesized. Therefore, a more accurate representation of treatment-related changes in the PK/PD parameters can be obtained.
For further simplification, the generative process of an event record is not conditioned on all previous event records. Instead, the totality of clinical observations observed over the duration of the study is generated in one step \(\:\varvec{z}:{{\varvec{d}}_{\stackrel{\sim}{id}}^{\left(i\right)}\}}_{i=1}^{N},\:\:{\varvec{s}}_{\stackrel{\sim}{id}}\:\leftarrow\:{G}_{MLP\:cGAN}\left({\varvec{a}}_{id},z\right)\). Padding is required for modelling the datasets in situations where the sequence length (N) varies among subjects.
Experimental setting
We leverage publicly available PMX models in Simulx [20], an advanced PMX model simulation software part of the Lixoft suite. (Appendix A). Simulx was used to set up several experiments (Appendix C), with different level of complexity. These datasets are categorized based on the underlying PMX model used to generate the data, the number of dependent variables (DV), groups, covariates and the dose administration and sampling times. Specifically, for dose administration we considered both single administration and multiple administrations. After selecting an existing PMX model in Simulx, we proceeded as follows: (i) we manually determined the clinical design of the data that we aimed to generate, including number of treatment arms, dosing regimens, event frequency and sampling time points for each treatment arm. (ii) We defined probability distributions from which we sampled values for the covariates (e.g., sex, race, weight and/or age at baseline). (iii) We simulated the PK/PD responses, given the pharmacometric model, clinical design and generated covariates. Using this workflow, 12 datasets with different levels of complexity (measured in terms of number of treatment arms and number of PK/PD measurements) were generated (Table 1). We simulated 100 patients per each group.
The performance of the considered generative methods in synthesising the above-mentioned datasets was then assessed using several metrics:
-
Predictive and discriminative scores. These are the most used metrics to evaluate the quality of the synthetically generated time-series/longitudinal data. These metrics are proxy measures of usefulness (predictive score) and fidelity (discriminative score). For computing the predictive score, a post hoc prediction model was trained in the synthesized data to predict the next event record in the sequence. Subsequently, the trained model was tested for the original data. The mean absolute error of the trained model was returned, with a lower value indicating a better result. For computing the discriminative score, a post hoc classification model was learned to distinguish between the real and synthesized data. The classification error was then returned. Here, lower scores indicate better performance.
-
Univariate statistical tests. We performed univariate statistical hypothesis tests to compare two samples of synthetic data and real data. These tests quantified the likelihood that the two samples are drawn from the same probability distribution. We applied the Kolmogorov-Smirnov test for PK/PD responses and time-varying covariates. For static covariates, we performed the chi-squared test.
-
PMX-related metrics. We evaluated the synthetically generated time-series/longitudinal data with metrics relevant to the traditional PMX model assessment. Given that the “real” data were simulated with PMX models, their ground-truth PK/PD parameters are known to us. The PK/PD parameters can be estimated on the synthetically generated data to compare the estimated individual \(\:\stackrel{\sim}{\theta\:}\) and population parameters \(\:\stackrel{\sim}{\varvec{\varnothing\:}}\) to the real ones, θ and \(\:\varnothing\:\), respectively. For population parameters, we report the mean absolute percentage error (MAPE) [21], which is computed as follows: \(\:\frac{1}{M}\sum\:_{j=1}^{M}\frac{|{\varnothing\:}_{j}-{\stackrel{\sim}{\varnothing\:}}_{j}|}{\left|{\varnothing\:}_{j}\right|}\), where M corresponds to the number of population parameters of the PK/PD model. For individual parameters, we calculated the Kullback–Leibler (KL) divergence as follows: \(\:\sum\:_{\theta\:}P\left(\theta\:\right)\text{l}\text{o}\text{g}\left(\frac{P\left(\theta\:\right)}{P\left(\stackrel{\sim}{\theta\:}\right)}\right)\).
For PAR [14], we used the default hyperparameter values for the RNN-based generative model. For TimeGAN, the generative model consists of a three-layer stacked RNN with a hidden dimension of 24 for all three layers [18]. The architecture of the MLP cGAN is based on a five-layer MLP, comprising an intermediate and final MLPs with a leaky ReLU [22] and a hyperbolic tangent, respectively, as an activation function. Additionally, a grid search strategy was developed to find the optimal hyperparameters of the model, specifically, the learning rate in the 0.0001-0.1 range and betas coefficients in the 0.5–0.9 range. These hyperparameters are utilized within the optimizer function during the training session.
Results
In this section, we report the results for each metric category.
Predictive and discriminative scores
In terms of predictive and discriminative scores across various datasets, notably, TimeGAN and MLP cGAN had different strengths. The results are depicted in Fig. 2.
Predictive and discriminative scores rely on post hoc ML models built upon RNNs. Therefore, these scores may be positively biased towards techniques that are also built upon RNNs (i.e., PAR and TimeGan). This bias is well illustrated in the predictive score: where MLP cGAN rarely outperforms the PAR model and TimeGAN across most datasets. Surprisingly, the MLP cGAN is much more competitive in terms of discriminative score, and it outperforms the other two techniques in almost all datasets. The good discriminative ability of this model may be attributed to its effective generative process in clinical design.
A comparative assessment of these models underscores the importance of choosing the most appropriate generative model to align with the specific requirements of a given PK/PD study. These scores also show that the benchmarked techniques have different robustness to the dataset characteristics. For instance, the predictive and discriminative scores of MLP cGAN often deteriorate in datasets with multiple arms and PK/PD responses (e.g., PK/PD effect, TMDD).
Univariate statistical tests
The results of univariate statistical tests (Fig. 3) reflect the quality of the synthetic data generated by the three models. A higher p-value in the tests indicates a higher likelihood of both synthetic and real data samples being derived from the same distribution. The quality of the synthetically generated covariates appears to be relatively consistent across all three models. However, notable discrepancies are noted in an examination of the quality of the generated PK/PD responses. The MLP cGAN model is remarkable in that it delivers the highest quality of synthetic PK/PD responses with almost no exceptions. To obtain these results, we classified the PK/PD responses based on their individual sampling times and calculated the Kolmogorov-Smirnov test for each category of responses. The test results estimated for each sampling time were averaged.
Performance of the selected methods measured in terms of univariate statistical tests. The test scores range from 0 to 1, with higher values indicating better performance. The Kolmogorov-Smirnov test was used for PK/PD measurements and time-varying covariates. For static covariates the chi-squared test was applied. For datasets with more than one PK/PD measurement and/or one covariate, we report the average of the corresponding statistical test. For each dataset, statistical tests are performed for each treatment arm independently and averaged out
The synthetically generated covariates for the four datasets without covariates were not assessed (Table 1).
PMX-related metrics
To assess whether the synthetic data provide similar modelling results in a PMX task, KL divergence and MAPE were derived for all scenarios (Fig. 4). The KL divergence is a measure of the statistical distance between the derived probability distributions of individual PK/PD parameters and is obtained by comparing the real data with the synthetically generated data. MAPE facilitates a comparison between the population parameters drawn from both real and synthetic data. While MAPE provides a combined view of the PK/PD parameters represented in different units, it is notably sensitive to small values. As opposed to univariate statistical tests, these two metrics provide a more holistic evaluation of data quality.
With a few exceptions, the MLP cGAN model delivers consistently better performance compared to the other models across most datasets and metrics. This model also exhibits notable time efficiency, as further detailed in Appendix B. In contrast, TimeGAN, which is an established model used in generative methods for time-series and longitudinal data, frequently delivers poorer performance than the other techniques in our specific case study. An in-depth analysis of TimeGAN revealed that it involves a higher number of learnable parameters compared to the other methods, increasing its susceptibility to overfitting. In addition, its historical performance evaluations were based on longitudinal data with an abundance of sampling points. We posit that these factors contribute to the underperformance of TimeGAN in scenarios with short sequence lengths, as in our current use case.
Discussion
Different applications of generative models are emerging within the healthcare sector [23], with recent works proposing applications of GANs for constructions of virtual populations [24, 25] and more advanced methodologies employing ChatGPT Large Language Models (LLMs) to analyse PK data [26]. The use of generative models for simulating virtual patients’ covariates and PK/PD profiles have not been fully explored yet. In this context, we investigated different architectures to generate PK/PD synthetic data.
Our study provides a comprehensive evaluation of three leading deep generative techniques, namely, PAR, TimeGAN and MLP cGAN, across 12 clinical datasets comprising patient covariates and PK/PD endpoints. By using an array of metrics that emphasise different statistical attributes of the data, we can explain various aspects of performance of each model. Furthermore, the integration of metrics grounded in PMX analytic tools enhanced the robustness of our evaluation. The MLP cGAN model demonstrated all-round performance, excelling in most metrics and datasets. This result strongly highlights the power of this DL-based approach to generate synthetic data that accurately reflect the original sources while enabling data sharing in a privacy-preserving manner. Furthermore, such approaches can be adopted for data augmentation—that is, to increase the sample size—for under-represented data or to enhance the robustness of modelling analyses.
Although the robust performance of the MLP cGAN across several metrics underscores the capability of this approach in our setting, the potential utility of the other tested models should not be discarded. The difference in performance indicates that the utility of each generative method may differ depending on the precise requirements of the task at hand. In addition, the specificity of the datasets used may influence the outcomes. According to data complexity, some datasets are represented with and without covariates; this context may increase or decrease the bias to synthetically generate data with high fidelity to the original one. Hence, future work on exploring these aspects and pursuing advances in generating synthetic data that preserve the richness of real-world information while addressing data privacy concerns can be advantageous to the present field of study.
GAN has been used in wide range of context. But some questions remained open about the possible theoretical limitations. In theory, the main assumption behind GAN is that they can generate any probability distribution from the input data. Nevertheless, a new result based on Extreme Value Theory proofs [27] suggests that GAN cannot generate heavy tailored distributions. In addition, the performance of these models in relation to the size of the dataset (e.g., number of subjects) should be assessed. Future work may also focus on comparing GAN architectures with models like GMMs, Copula and VAEs.
By further validating the feasibility of synthetic data generation in the PK/PD domain, we can envisage the efficient generation of clinical-pharmacology datasets characterized by multiple and diverse clinical endpoints and patients’ covariates, in an unsupervised and mechanism-agnostic manner. This can ultimately enable data sharing across institutions while ensuring mitigation of privacy risks.
Conclusions
We applied generative models to PK/PD data, starting with a literature review to understand the deep generative model techniques. We established a foundational experiment using the PAR models. Although we attempted to optimize TimeGAN, an advanced generative model, it underperformed on our datasets. Drawing from our experiences, we developed the MLP cGAN model, which effectively transforms time-series data into vector inputs. This model performed better than the other models examined in this study. The performance of the MLP cGAN model was especially better when benchmarked against multiple evaluation metrics. By training our models on diverse datasets, we confirmed that the MLP cGAN framework has good capability for synthesising PK/PD data.
Moving forward, exploration of other generative models or refinement of the current models to optimize their performance will be pivotal in this field. This endeavour can pave the way for the further use of synthetics in clinical research, providing meaningful insights while preserving data privacy. Ultimately, our work underlines the potential of synthetic data generation in the clinical pharmacology and drug development field towards advancing research and data sharing in the digital age of artificial intelligence.
Data availability
Not applicable because only simulated data are included in this manuscript.
References
Reynolds DA (2009) Gaussian mixture models. Encyclopedia Biometr 741:659–663
Elkan C (2010) Expectation maximization algorithm. Encyclopedia Machine Learning. https://cseweb.ucsd.edu/~elkan/250Bfall2007/em.pdf. Accessed 12 December 2023
Lavielle M (2014) Mixed effects models for the population approach: models, tasks, methods and tools. CRC, Boca Raton, Florida
Kingma DP, Welling M (2013) Auto-encoding variational bayes. Clin Orthop Relat Res arXiv Preprint arXiv:1312.6114.
Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville AC, Bengio Y (2014) Generative adversarial nets. Adv Neural Inf Process Syst 27
Rezende DJ, Mohamed S (2015) Variational inference with normalizing flows. In: International Conference on Machine Learning, PMLR, pp. 1530–1538
Dhariwal P, Nichol A (2021) Diffusion models beat gans on image synthesis. Adv Neural Inf Process Syst 34:8780–8794
Ghosheh GO, Li J, Zhu T (2022) A review of generative adversarial networks for electronic health records: applications, evaluation measures and data sources. arXiv preprint arXiv:2203.07018.
Haarburger C, Horst N, Truhn D, Broeckmann M, Schrading S, Kuhl CK, Merhof D (2019) Multiparametric magnetic resonance image synthesis using generative adversarial networks. In: Eurographics Workshop on Visual Computing for Biomedicine, VCBM, pp 11–15
Delaney AM, Brophy E, Ward TE (2019) Synthesis of realistic ECG using generative adversarial networks. arXiv preprint arXiv:1909.09150.
Motamed S, Rogalla P, Khalvati F (2021) Data augmentation using generative adversarial networks (gans) for Gan-based detection of pneumonia and Covid-19 in chest x-ray images. Inf Med Unlocked 27:100779. https://doi.org/10.1016/j.imu.2021.100779
Abbasi M et al (2022) Designing optimized drug candidates with generative Adversarial Network. J Cheminform 14(1):40
Yang H et al (2021) Subtype-GAN: a deep learning approach for integrative cancer subtyping of multi-omics data. Bioinformatics 37(16):2231–2237
Pauley M et al (2023) T1dCteGui: a user-friendly clinical trial Enrichment Tool to optimize T1D Prevention studies by leveraging AI/ML based Synthetic Patient Population. Clinical Pharmacology & Therapeutics
Patki N, Wedge R, Veeramachaneni K (2016) The synthetic data vault. In: IEEE International Conference on Data Science and Advanced Analytics (DSAA), 2016, pp 399–410. https://doi.org/10.1109/DSAA.2016.49. https://doi.org/10.1214/12-AOS1037
Zwep LB et al (2022) Virtual patient simulation using copula modeling. Clinical Pharmacology & Therapeutics
Hochreiter S, Schmidhuber J (1997) Long short-term memory. Neural Comput 9:1735–1780. https://doi.org/10.1162/neco.1997.9.8.1735
Yoon J, Jarrett D, Van der Schaar M (2019) Time-series generative adversarial networks. Adv Neural Inf Process Syst 32
Mirza M, Osindero S (2014) Conditional generative adversarial nets. arXiv preprint arXiv:1411.1784
Simulx is an easy efficient and flexible application for clinical trial simulations. https://lixoft.com/products/simulx/. Accessed 29 November 2023
de Myttenaere A, Golden B, Le Grand B, Rossi F (2016) Mean absolute percentage error for regression models. Neurocomputing 192:38–48. https://doi.org/10.1016/j.neucom.2015.12.114
Xu B, Wang N, Chen T, Li M (2015) Empirical evaluation of rectified activations in convolutional network. arXiv Preprint arXiv:1505.00853.
Jacobs F, D’Amico S, Benvenuti C, Gaudio M, Saltalamacchia G, Miggiano C, Zambelli A (2023) Opportunities and challenges of synthetic data generation in oncology. JCO Clinical Cancer Informatics. 7:e2300045
Parikh J, Rumbell T, Butova X, Myachina T, Acero JC, Khamzin S, Gurev V (2022) Generative adversarial networks for construction of virtual populations of mechanistic models: simulations to study Omecamtiv Mecarbil action. J Pharmacokinet Pharmacodyn 49(1):51–64
D’amico S, Dall’Olio D, Sala C, Dall’Olio L, Sauta E, Zampini M, G. Della Porta M (2023) Synthetic data generation by artificial intelligence to accelerate research and precision medicine in hematology. JCO Clin Cancer Inf 7:e2300021
Shin E, Ramanathan M (2023) Evaluation of prompt engineering strategies for pharmacokinetic data analysis with the ChatGPT large language model. J Pharmacokinet Pharmacodyn, 1–8
Oriol B, Miot A (2021) On some theoretical limitations of Generative Adversarial Networks. arXiv preprint arXiv:2110.10915.
Acknowledgements
Editorial support was provided by Krunal Vasant Kavathiya of Merck Specialities Pvt. Ltd., Bengaluru, India, an affiliate of Merck KGaA.
Funding
No funding was received for this work. The internship of YJ was sponsored by Merck KGaA.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design, material preparation, data collection, analysis, review and editing. YJ, AGD, PG and NT contributed to conceptualization, methodology, formal analysis and investigation. YJ, AGD, IBL and NT contributed to original draft preparation.
Corresponding author
Ethics declarations
Conflict of interest
N.T. is an employee of Ares Trading SA (an affiliate of Merck KGaA, Darmstadt, Germany), Lausanne, Switzerland. Y.J. (the School of Computer and Communication Science of EPFL) was performing an internship at the Merck Institute of Pharmacometrics, Lausanne, Switzerland, when part of this work was performed. A.G.D and P.G were employees of Ares Trading SA (an affiliate of Merck KGaA, Darmstadt, Germany), Lausanne, Switzerland, when this work was performed. I.B.L. is an employee of Randstad and contributed as a paid contractor for Merck Quantitative Pharmacology, Ares Trading SA., Lausanne.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jiang, Y., García-Durán, A., Losada, I.B. et al. Generative models for synthetic data generation: application to pharmacokinetic/pharmacodynamic data. J Pharmacokinet Pharmacodyn (2024). https://doi.org/10.1007/s10928-024-09935-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10928-024-09935-6