Abstract
Quantitative systems pharmacology (QSP) is a quantitative and mechanistic platform describing the phenotypic interaction between drugs, biological networks, and disease conditions to predict optimal therapeutic response. In this meta-analysis study, we review the utility of the QSP platform in drug development and therapeutic strategies based on recent publications (2019–2021). We gathered recent original QSP models and described the diversity of their applications based on therapeutic areas, methodologies, software platforms, and functionalities. The collection and investigation of these publications can assist in providing a repository of recent QSP studies to facilitate the discovery and further reusability of QSP models. Our review shows that the largest number of QSP efforts in recent years is in Immuno-Oncology. We also addressed the benefits of integrative approaches in this field by presenting the applications of Machine Learning methods for drug discovery and QSP models. Based on this meta-analysis, we discuss the advantages and limitations of QSP models and propose fields where the QSP approach constitutes a valuable interface for more investigations to tackle complex diseases and improve drug development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
QSP and its growing role in drug development
Human cell biology is governed by complex networks of interactions between molecular structures, signaling pathways, and epigenetic remodeling in which the multiscale system governs the cell’s functionality. However, these networks and their inter-layer connections can become corrupted due to perturbations, leading to various diseases [1]. Drug development is indispensable to modern medicine; however, bringing drugs to the market is often compromised for several reasons, including lack of understanding of drug behavior at the whole system level and adverse side effects [2]. To understand the mechanism of disease networks, identify novel drug targets, and develop effective therapies requires studying individual components such as genes, RNA, or proteins as dynamic systems across scales [3]. Understanding these biological processes has been revolutionized with the development of high-throughput technologies and the accumulation of biomedical data; however, these data types demand integrative and dynamics-driven approaches to comprehend dataset repositories and accelerate novel discoveries. Another complexity to consider is drug-target and drug-drug interactions and their consequences at the system level.
Systems biology aims to address these complexities by understanding biological processes at the molecular and cellular system levels [3]. Quantitative systems pharmacology (QSP) stems from system biology and integrates pharmacological aspects with systems modeling to identify and design safer and more effective drug therapies. QSP was defined in 2011 in a National Institutes of Health white paper based on workshops and discussions with experts from academia, government, and industry [4,5,6,7,8].
One of the challenges for drug development is the increasing cost of drug developments and approvals—it costs from $1.2 to $4 billion and requires upwards of 10 years to develop and introduce a new drug [9,10,11]. QSP addresses some of these challenges by providing integrative approaches to determine mechanisms of action of the new and existing drugs, maximize therapeutic benefit, minimize toxicity and implement a procedure to improve individual patients' health [7, 12]. QSP uses mechanistic mathematical models to characterize dynamic interplays between a drug and physiopathology to explore the system at multiple scales of biological organization (molecular, cellular, organ-level networks). Incorporating mechanistic multi-scale systems aspects to classical pharmacometrics through QSP can enable novel drug target predictions, detailed studies of mechanisms of action and safety, biomarker identification, optimization of doses or regimens, compound selection, decision making, and responses considering various treatment variables [12, 13]. QSP, while relatively new, complements other modeling approaches widely adopted for preclinical and clinical studies, including the quantification of drug behavior in the body [14]. These tools include:
Pharmacokinetics (PK) focuses on studying the time-course of drugs’ absorption, distribution, metabolism, and excretion (ADME; e.g., dose-concentration relationships).
Pharmacodynamics (PD) examines the biological effects of drugs and their mechanisms of action (e.g., concentration-effect relationships) on humans, animals, microorganisms, or combinations of organisms (e.g., infection) [15].
Pharmacokinetic/pharmacodynamic (PK/PD) modeling connects PK and PD to facilitate the prediction of the time course of drug effects that result from a specific dosing regimen [14, 16,17,18]. Systems pharmacology is already widely used in the pharmaceutical industry, focusing on PK/PD modeling, predicting dose-exposure responses, and evaluating market potential [19]. This modeling assists in gaining mechanistic insights and facilitates early dose selection. In addition, population PK/PD modeling can help understand the critical PK characteristics and population-level covariates [20].
Physiologically-based pharmacokinetic (PBPK) modeling presents the pharmacokinetic behavior of a compound in the body and predicts the ADME of natural or synthetic substances in humans and other species.
Physiologically based pharmacokinetic/pharmacodynamic (PBPK/PD) models connect drug information with prior knowledge of the physiology and biology at the organism level to provide a mechanistic representation of the drug in biological systems [21]. PBPK models consider different organs and tissues, and assist in obtaining quantitative characterizations of concentration–time profiles in the individual compartments [22]. This modeling approach can be utilized to understand tissue-specific PK and PD and estimate drug interaction risks [20].
According to a survey across 50 pharmaceutical companies, the industry has a vague definition of QSP [13]. Although this survey showed that most pharmaceutical companies used the QSP term to describe their modeling approaches, a significant number of companies used other terms for mentioning their modeling activities. Therefore, to accelerate improvements in drug discoveries, one suggestion can be to use fixed terminologies that assist in studying and investigating QSP models. The aforementioned study also showed that the most common applications in this field are related to generating and testing hypotheses, optimizing doses or regimens, predicting clinical efficacy, and identifying biomarkers suggesting that future opportunities in the industry can be related to the usage of QSP modeling for evaluating safety and decision-making [13].
QSP modeling approaches
Pharmaceutical companies and academia utilize various approaches for drug target discovery [13, 23]. Several modeling approaches for QSP have been developed, including statistical (Bayesian), Boolean, temporal (ordinary differential equations), spatio‐temporal (partial differential equations), agent-based, integrative, empirical curve fitting, and machine learning that enable integrating molecular pathways with clinical results and pharmacology [24]. Incorporating quantitative temporal and spatial information in QSP models can provide more accurate predictions of drug discovery targets, PK/PD relationships, and clinical results [24]. Many published QSP models are constructed as multi-compartment nonlinear systems of ordinary differential equations (ODE) [25].
Recently, a diversity of software platforms have been employed to assist in developing QSP models [23, 26], such as Simbiology and toolboxes in MATLAB [27]; The R-based packages nlmixr [23], mrgsolve [28], RxODE [29], nlme [30], and Cell Collective platform [31,32,33]; Based on our review across 51 models, we note that the MATLAB environment and tools are more popular among QSP modelers [34,35,36,37,38,39,40,41,42,43,44,45,46,47].
Studies of QSP methodologies show that developing, testing, and documenting QSP models require standardization to improve the reproducibility and reusability of these models, which affect the potential impact of this approach in academia and industry [48]. Because QSP is a multidisciplinary field, the development of such models demands teamwork and collaboration of different individuals such as modeling engineers, biologists and clinicians, data programmers, statisticians, software engineers, and PK/PD scientists [49].
While standard workflows utilizing QSP continue to evolve [26], the general QSP workflow can be summarized in three main steps [49]:
Model scope The first step is to define the therapeutic field and objectives of the model by providing the physiological pathway map, which presents the incorporation of the biological and pharmacological processes associated with the model’s scope [49].
Model development Since any modeling task requires some form of data, this step starts with converting raw data to a suitable format. In this step, a modeler collects prior models, clinical and non-clinical data and develops mathematical descriptions of the processes and compartments involved in the interplay between drugs and the pathophysiology. Model development encompasses steps that can be categorized into standardizing and exploring data, parameter estimation, and simulation/analysis [26, 49, 50].
Model Qualification The modeling engineer calibrates the QSP model to relevant data from target patient populations. This step is related to collecting appropriate clinical data in patient populations that will qualify the model and calibrating the model at relevant scales of physiology and time [49].
Methods
Literature search
We surveyed recent articles for QSP models available in PubMed and published between 2019 and 2021 to identify relevant studies, providing a repository for categorizing and evaluating different research studies in this field. PubMed search term “(“Quantitative Systems Pharmacology” OR “QSP”) AND Model*” (in April 2021) resulted in 148 publications.
For QSP models, we excluded reviews and methodology-focused publications from the PubMed search results during the manual literature mining process. We selected original QSP studies that constructed QSP models utilizing clinical and experimental data, resulting in a total of 50 publications. After reviewing these models, we associated Medical Subject Headings (MeSH) terms and IDs [51] to each model. We categorized them based on their therapeutic fields (Table 1) while including various properties of the studied disease and model. We also explored the application of machine learning (ML) methods in QSP modeling and addressed modeling approaches that benefited from ML applications.
Considering the Pubmed search term results and manual literature mining to find relevant studies, for this review, we were able to gather and analyze a resource addressing recent original QSP researches and their applications in different diseases, which provides an insight into potential future directions of QSP studies.
Results
Categories of recently published original QSP models
We categorized original QSP models based on different properties after the PubMed literature search. Because the biological questions motivating a study play a critical role in selecting the methodology and other properties of the project, we classified the corresponding publications from the most to the least represented biological field that utilized QSP models in the last three years. This analysis can assist in summarizing domains that have been intensively investigated in QSP and help find areas that need to be explored by QSP approaches. Table 1 presents the literature mining results for recent QSP model original publications between 2019 and 2021, including the name, PubMed ID, title, the year of publication, a MeSH term, and a unique ID associated with each based on the underlying biological question. We specifically utilized MeSH terms and unique IDs, official words or phrases selected to represent particular biomedical concepts.
We found that 24 different diseases categorized in nine major disease areas are represented by the 51 identified QSP model (Fig. 1). Below we describe several QSP applications to different biological questions for the top three categories: Immuno-oncology, nutritional and metabolic diseases, and nervous system diseases. The models described in these categories were identified with n# referring to Table 1.
The recently published QSP models and their disease areas. The bar chart presents the number of articles published between 2019 and 2021 for developing original QSP models. Categorizing these articles based on the biological questions they focused on (presented by their MeSH terms), revealed that most models are related to neoplasms
Immuno-oncology (IO)/neoplasms QSP models
According to the World Health Organization, cancer is among the preeminent diseases worldwide, causing globally 10 million deaths in 2020. The most frequent cancer is breast cancer (2.26 million cases), but the most lethal is lung cancer, with 1.8 million deaths and nearly 2.21 million cases in 2020. In the last decade, understanding the cancer tumor microenvironment (TME) and immunosurveillance has led to promising strategies that can harness immune cells to fight cancer [52]. Many immunotherapies focus on the immune T cell population for their cytotoxic function and anti-tumoral response. Current main immunotherapies include immune checkpoint inhibitors (CPIs) and chimeric antigen receptor T cells (CAR-T) [53]. These immunotherapies used in several clinical trials show tremendous response to a wide range of solid and blood neoplasms.
Immune checkpoints regulate the immune system and are principal targets for cancer immunotherapy in different cancer types [54, 55]. FDA-approved CPIs target CTLA-4, PD-1, and PD-L1 to prevent T cell inhibition by cancer cells. Blocking these receptors increases the activation and proliferation of effector cells following stimulation and antigen recognition, and consequently, is more effective to remove cancer cells [47]. CAR-T, however, is a cellular therapy that employs genetic modifications of autologous T cells to maximize tumor antigen recognition and intracellular signaling pathways in T cell activation. Despite the enthusiasm around these strategies, they have been associated with unique side effects, such as autoimmune reactions, lethal cytokine release, immune cell dysfunction, and organ failures [56]. In addition to immunotherapies, other strategies focus more on the cancer side, such as identifying tumor antigens or neoantigens expressed solely by cancer cells or developing small molecules that target the signaling landscape for more personalized approaches with minimal side effects. Nevertheless, these therapies exhibit several challenges (e.g., efficacy, heterogeneity in response, drug resistance, etc. [57]) that need to be addressed to maximize immune response and minimize lethal side effects. We first describe immunotherapies aided with QSP models, and we later categorize studies based on cancer type.
Immunotherapies
In the case of cellular therapy, model #41 addresses the complex relationships between CAR-T cell doses and the magnitude of cytokine release syndrome (CRS), one of the side effects following CAR-T cell therapy [58]. Interestingly, this quantitative model indicates that CAR-T injection does not cause severe CRS; however, the magnitude of cytokines at the baseline operates as an auspicious accelerator of CRS after CAR-T administration. Thus, this tool may serve as a personalized model of CAR-T cell therapy to interrogate dosing and clinical toxicity [59].
Bispecific antibodies, a new generation of engineered antibodies that can simultaneously bind two different antigens—one side to a tumor antigen and the other side to immune cells—has emerged as a promising novel therapy for cancer treatment [60]. Because T cells are key effector cells in immune response, a potent procedure used CD3, the main marker on the T cell population, to engineer a bispecific T cell engager (TCEs) promoting cytolytic synapse with cancer cells [61]. The crosslinking of the different protagonists through CD3 bispecific targeting tumor antigen, P-cadherin (PF-06671008), has been investigated by a QSP model #40 to quantify the relationship of the tripartite partners (drugs-T cells-tumor cells) in vivo. The model predicted that the number of T cells and P-cadherin expression are crucial for clinical efficiency as the half-life of PF-06671008 only lasts one day. Therefore, such a model can predict and optimize the CD3 bispecific technology into the clinics for human PB/PK prediction.
Model #25 investigated molecular cancer therapy; this PK/PD model describes the efficiency of ORY‐1001, a small molecule inhibitor of LSD1—lysine-specific histone demethylase that acts as an epigenetic regulator in cancer. This predictive model examined the ORY-1001 pharmacodynamic response and response durability associated with tumor growth across multiple doses. The model was able to predict in vivo drug efficacy extrapolated exclusively from in vitro data. Such a mechanistic approach could reduce the use of animal models, the cost and time in drug development [45]. In another study that addresses drug therapies, Stroh et al. model the activatable antibody, Probody therapeutic (Pb-Tx), designed to keep the antigen-binding site of engineered antibody masks until local proteolytic activation in disease tissue. Model #39 integrated the in vitro and in vivo PK/PD effects of both prodrug CD166 and pharmacological properties for rational design and clinical translation. The QSP model predictions proposed a greater absorption of Pb-Tx than parental antibody and emphasized that the antibody masking strength can modulate the molecule’s absorption in desired sites, such as the tissue or the peripheral circulation. As a result, this study utilized interesting approaches to customize Pb‐Tx infiltration to desired sites of tumor niches instead of a healthy environment [62].
Another study presents a QSP model #18 of humanized mice [63]. Here, the authors modeled the interactions between tumor growth, T cells, cytokine secretion, immune checkpoint expression, and drug inoculation using experimental data from a xenograft mouse model. The critical aspect of such a model is that it can aid in extrapolating dose conversion between animals to humans, where often therapeutic dose translation from the rodent system to human fails [63].
The tumor microenvironment (TME) is an essential aspect of cancer development as it participates in survival needs, drug resistance and installs an auspicious immunosuppressive environment [8]. Several studies investigated the dynamic interplay between immune-mediated TME and immunotherapy treatment. Model #20 examines cellular communication and TME crosstalk by studying myelosuppression, a severe side-effect of anti-cancer therapies. To improve the understanding of drug-induced myelosuppression, Wilson et al. produced a QSP model of hematopoiesis in vitro to quantify the effects of anti-cancer agents on multiple hematopoietic cell lineages [40]. Model #21 is an open-source and expandable modeling IO platform that integrates tumor-T cell crosstalk in response to different combinatorial immunotherapy. The QSP tool integrates several critical modules of TME: a cancer module (tumor size and tumor antigen), dendritic cell as antigen-presenting cells, a T cell module (immunosuppressive regulatory T cell, cytotoxic and non-cytotoxic T cells), checkpoint module, and a pharmacokinetics module illustrating TME behavior upon therapeutic strategies [41]. Also, the model #3 utilized a QSP model to reproduce the main component of interaction between tumor and immune system to model TME response upon combination of radiation and immunotherapy [64]. These complex QSP frameworks can be utilized as clinical platforms to evaluate the dynamics of therapy responses at a larger scale than at the individual level.
Breast neoplasms
In breast cancer (BC), human epidermal growth factor receptor 2 (HER2) is a neoantigen protein that can promote the growth of cancer cells [65]. HER2-positive BC is an aggressive cancer subtype prevalent in 20% of cases. Despite improvements in anti-HER2 therapies, treatment resistance remains a clinical challenge [66].
Wang et al. proposed two different dynamical models (#24 and #2) of the TME to address the efficacy of existing therapies in different types of BC. For HER2-negative BC, they proposed a QSP model (#24) for a virtual clinical trial with immune checkpoint therapy in association with an epigenetic modulator. The authors integrated different modules describing immune activation, suppression, and trafficking into four separate compartments (lymph node, central, peripheral, and tumor site) and PK/PD of two therapeutic agents [44]. Their second (similarly constructed) model (#4) focuses on triple-negative breast cancer. This BC type is defined by the lack of three receptors (estrogen, progesterone receptors, and low HER2 expression). It classifies as highly invasive with limited treatment and poor outcomes [67]. The authors developed a virtual patient cohort of atezolizumab (anti-PD-L1) and nab-paclitaxel treatments for this cancer to identify immune biomarkers and optimal treatment for clinical trials [34].
The QSP model #2 addresses drug resistance by evaluating the efficacy of lapatinib (LAP), abemaciclib (ABE), and 5-fluorouracil individually and in combination using trastuzumab-resistant HER2-positive BC cell line. Their findings suggest synergistic effects between ABE and LAP while showing the impact of the triple combination therapy on tumor cell viability [68]. Overall, both models address the dynamics of tumor-immune-drug interaction for a virtual clinical trial to provide guidelines in drug development and clinical regiment design.
Colorectal neoplasms
Previous studies on bispecific TCE cited above led to the construction of a QSP model, combining TCE and immune checkpoint inhibitor, anti-PD-L1, with similar modules described in the Breast neoplasm section [34, 44]. They predict that the efficacy of treatment is dictated by the patient’s variability and unique characteristics. This model not only aids TCEs and immune checkpoint strategies but also is an interesting tool for precision medicine initiatives [39].
Lung neoplasms
In lung cancer, anti-PD-1 treatments show promising results in the survival rate of patients with advanced non-small-cell lung cancer [69]. The QSP model #37 integrated dynamic modules of tumor growth, antigen processing and presentation, T cell activation and trafficking, anti-PD-1, and antibody kinetic responses. The model predicted that the density of anti-tumoral effector T cells in the blood correlated with a better response to therapy than the density of pro-tumoral regulatory T cells [70]. Later Ma et al. extended this model to TCEs as a single therapy to explore the dynamic of inter-cellular interactions in the tumor microenvironment and identify immune biomarkers. This study predicted that indicators of responders versus non-responders to TCE therapies depend highly on the patient’s response and stage of disease (e.g., Non-responders, partial or complete response, stable or progressive disease conditions) [42].
Melanoma
This is a severe skin cancer derived from melanocytes, melanin-producing cells. Located in the bottom layer of the skin, cancerous melanocytes are likely to metastasize to any part of the body [71]. Two models address CPIs in melanoma cancer. Model #38 simulates CTLA-4, PD-1, and PD-L1 therapies with varying modes of treatment administration (single, dual, or sequential) to evaluate the optimal parameter for melanoma treatment. The dynamic response of their virtual patient model reproduced data from real clinical trials. The model also predicted the median response of each therapy and defined the physiological range of virtual responders for each combination [47]. Milberg et al. address the efficacy of a combination checkpoint therapy consisting of pembrolizumab (anti-PDL1) and ipilimumab (anti-CTLA4) in metastatic melanoma while taking into account lesions used for melanoma immunogenicity diagnosis. The model showed that combination therapy is significantly more efficient for intermediate lesions than non- or high metastatic lesions [72].
Prostatic neoplasms
The ODE-based model #23 explores castration-resistant prostate cancer, for which therapies are still non-conclusive [43]. This study presents a QSP model of prostate cancer immunotherapy, integrating different immune cells, tumor compartments, and seven treatments. Among numerous treatment combinations, the authors found that dual association of cancer vaccine and immune checkpoint blockade are the most effective combinatorial immunotherapy for subjects associated with androgen-deprivation therapy resistance.
Nutritional and metabolic diseases
Diabetes mellitus, type 2
Type 2 diabetes mellitus (T2DM), commonly known as type 2 diabetes, is a metabolic disorder translated by an aberrant accumulation of glucose in the blood due to a defect of insulin function and expression [73]. Different T2DM QSP models described below focused on drugs that could lower plasma glucose and filter it through other organs.
A recently approved class of antidiabetic medications includes gliflozins that target sodium‐glucose co‐transporter (SGLT), a class of receptors expressed in the kidney and small intestine and responsible for more than 80% of glucose reabsorption [74]. These drugs decrease glucose by increasing urine secretion and blocking renal re-consumption [8]. In the first model (#27), Mori-Anai et al. addressed the inhibitory action of three different SGLT2 inhibitors after food consumption with a model called human systemic glucose dynamics (HSGD) integrating glucose metabolism, intestinal uptake, and renal reabsorption. The model provided a quantitative estimation of drugs’ effect on dynamic glucose absorption after food consumption [75]. In another study, a QSP model (#47) investigated SGLT1 and SGLT2 activity in renal glucose circuits and estimated the PK/PD of SGLT2 inhibitors using clinical data of healthy and T2DM patients. Interestingly, the model showed that under SGLT2 inhibition, SGLT1 action increased, indicating compensatory relationships between SGLT receptors and an adverse effect of the drug selection [76]. Later, Sokolov et al. utilized this model (#29) to address SGLT1 inhibition in response to SGLT2 gliflozins inhibitors (dapagliflozin, empagliflozin, and canagliflozin). The QSP model indicated that only canagliflozin could inhibit renal SGLT1, resulting in identifying a critical therapy design to maximize the SGLT2 inhibitory effect [73]. Because glucose accumulation depends on insulin, model #10 considers glucose-insulin dynamics in the short and long-term under dapagliflozin treatment. According to this model, dapagliflozin is more beneficial to patients with more inadequate glycemic control by insulin [77]. Another model (#28) focuses on several protagonists in glucose levels after food consumption [78]. The incretin hormones, glucagon-like peptide-1 (GLP-1), glucose-dependent insulinotropic polypeptide (GIP) catalyzed by enzyme dipeptidyl-peptidase 4 (DPP4), and the neutral endopeptidase (NEP) stimulate insulin release to lower glucose. By modeling GLP-1 and GIP dynamics, and PK/PD of DPP4 inhibitors, model #28 showed that inhibition of DPP4 occurs in a dose-dependent manner. Still, the highest dose of DDP4 inhibitor stimulated a high GLP-1 secretion, suggesting the triggering of alternative pathways upon DPP4 inhibition [78].
Nervous system diseases
Alzheimer’s disease (AD) is a slow and irreversible degenerative disorder that leads to progressive neurocognitive dysfunction. One of the histological characteristics of AD is the formation of amyloid plaque due to the accumulation of insoluble extracellular amyloid-beta (Aβ) that causes inflammation and neurotoxicity [79]. Targeting the Aβ pathways is one of the main therapeutic strategies to slow down degenerescence; however, many clinical trials fail due to several reasons, including patient heterogeneity, disease stage, treatment timing, ineffective drug penetration, and mechanism of action. To explore Aβ therapy failures, Madrasi et al. constructed a QSP model of the Aβ pathways with three relevant drugs (elenbecestat, verubecestat, and semagacestat) and four anti-Aβ monoclonal antibodies (aducanumab, crenezumab, solanezumab, bapineuzumab). Their model (#11) predicted that among the different monoclonal therapies, aducanumab and bapineuzumab could induce the fastest plaque reduction, while drug molecules promote slow reduction and their efficiency depends on plaque turnover formation [80].
Model #33 was used to simulate a clinical trial using aducanumab combined with different genotypes of common variants affecting cognitive function (apolipoprotein E, Catechol -O -methyl Transferase, and 5-HT transporter genotypes). This study highlighted the variability of clinical response between phase II and III, determined mainly by the different variants and baseline Ab peptide accumulation [81]. Similarly, another study focused on the same variants under benzodiazepines, antidepressants, and antipsychotics drug treatments [82]. Model simulations indicated, once again, variability of response between baseline and mild stage of AD under different regiments.
In summary, we reviewed examples of recent QSP models across major disease areas. Notably, many of these studies focus on maximizing drug design, therapeutic strategies, understanding the dynamic of drug-target interaction at the system, finding optimal dosage, addressing toxicity and potential adverse side effects. Altogether, QSP is a growing platform in drug development with much potential as an integral approach to reconciling drug safety and clinical patients’ response to therapy.
Machine learning applications in QSP modeling
The staggering amount of data generated with recent technologies demands integrative approaches to address the pharmacological challenges more efficiently. The “big data” field aims to analyze information from datasets containing complex or extensive amounts of information [83]. An example of big data used for drug discovery is observational data such as Electronic Health Records (EHR), which encompasses patients’ unique medical characteristics such as laboratory results, comorbidities, treatments, and observed effects [84]. In drug development, machine learning has been used as part of automated pipelines to guide and accelerate preclinical wet-lab experiments, drug discovery, and clinical trials [83, 85]. In fact, there are opportunities to apply ML methods in nearly all stages of drug discovery and development [85]. For example, we can utilize ML to identify and validate novel targets [86, 87], predict treatment responses [88], discover biomarkers [89], predict disease progression [90] degeneration [91], and risk factors [92, 93], design and optimize small-molecule components [94], and improve analyses of high-throughput imaging in computational pathology [85] 89. ML can also optimize the drug candidate discovery field by predicting desirable physicochemical characteristics, pharmacokinetics, safety, and efficacy [20, 83, 95,96,97,98,99,100].
In this section, first, we briefly explain the basis of ML; we refer readers to the recent publications on ML methods [83, 101] for detailed information and additional relevant studies. Second, we review recent applications of ML in drug discovery and development. Finally, we provide some examples of recent QSP efforts that benefited from machine learning methods.
Machine learning
ML methods can be categorized into two groups: Supervised learning, which uses labeled data (the goal is to “predict”), and unsupervised learning, which deals with unlabeled data (the goal is to “explore”) [102].
Supervised ML algorithms require input data sets to be split into a “training” and a “test” data set. Model training fits the model to the training data set, and the trained ML model can then be validated using the test data set. The validated ML model can then be utilized to make predictions or decisions based on the new data set covariates [103]. Several algorithms have been developed in this field, such as linear and logistic regression, ridge regression, decision trees, random forest, gradient boosting, neural networks, and genetic algorithms [104,105,106]. Data sets that contain both covariates and outcomes are “labeled” and used in supervised ML.
Different studies approach drug discovery with supervised learning techniques such as regression analysis methods (e.g., disease and target druggability from multidimensional data [87], targets for Huntington disease [107], identify potential cancer biomarkers [108, 109], drug sensitivity prediction [110], image-based diagnosis [111]), and classifier methods (e.g., tissue-specific biomarkers from gene expression signatures [89], target druggability based on PK properties and protein structure [112, 113]).
Supervised learning methods also enable the modeling of response surfaces for estimating individualized patient outcomes. One way to accomplish this is to fit a single-output model with the treatment as an input feature, making it less flexible and providing the same outcome model for treated and untreated patients. Another approach is to fit two separate supervised models for different treatments, which provides more flexibility in estimating patient outcomes [114].
Unsupervised ML includes the covariates but not the outcomes. This technique is used to identify patterns and associations between data points. K‐means and hierarchical clustering are examples of the algorithms widely used in unsupervised ML [83]. Unsupervised clustering methods also have been used for drug discoveries such as de novo molecular design [115], deep feature selection for biomarkers [116], feature reduction in single-cell data to identify cell types [117], and biomarkers [118].
ML-facilitated causal inference can estimate the effects of single/multiple or time-dependent treatments on patient outcomes. Various types of data can be used for training ML models to evaluate treatment effects such as clinical data (e.g., age, sex, genetic information, laboratory measurement), type of treatment (e.g., binary treatment, single treatment, or multiple treatments), patient outcomes (e.g., survival probability, multiple outcomes), and treatment decisions (e.g., optimal single/combinatorial treatment, optimal dosage). As a result, causal inference methods can assist physicians in decisions about the treatment benefit, treatment options, and dosages [114].
Integration of QSP and ML
Developing methodologies to integrate clinical data such as EHR or biological data sets (e.g., human genetic information in large populations, omics profiling of healthy and not healthy individuals) with QSP models provide the opportunity for additional progress in the QSP field. Below, we provide examples of efforts that integrated QSP models with ML methods.
Recent studies illustrate the benefits of integrating ML approaches with mechanistic modeling in curation, optimization, parameter estimation, and simulations of QSP models that can be computationally costly [114, 119]. For example, Hartmann et al. presented a predictive ML model to assist in optimizing antithrombotic therapy [120]. For this study, routine clinical data were gathered from 479 patients during therapeutic antithrombotic drug monitoring. A QSP model of coagulation network was developed based on a humoral coagulation model [121] to observe the effect of rivaroxaban, warfarin, and enoxaparin treatment on clotting factors levels. The authors estimated the parameters (factor rate constants, and production rates of coagulation factors) using a nonlinear programming solver. A stiff ODE solver (a variable-step, variable-order solver based on the numerical differentiation formulas of orders 1 to 5) was utilized for model simulation. The QSP model predicted the steady‐state effects of the rivaroxaban, warfarin, and enoxaparin treatment on clotting factor levels. For example, the model predicted that rivaroxaban did not affect the inactivated coagulation factor levels (such as prothrombin, protein C, protein S). Due to the variability in individuals responding to drugs, estimating the interindividual variability is important [122]. ML methods were used to evaluate the importance of interindividual variability. Monte Carlo simulations [123] were performed for interindividual variability by adding 20% variability on estimated production rates. Sobol sensitivity analysis [124] was performed to recognize the parameters with a higher impact on the activation of clot‐dissolution under different treatments. The model-generated predictions suggest suppressing protein C and protein S (components that regulate blood clot formation) under treatment with warfarin compared to enoxaparin and rivaroxaban.
Illustrating the benefits of using ML to analyze information from databases and predict drug targets, Pei et al. utilized QSP methods to provide a comprehensive study of cellular pathways involved in 50 drugs of abuse [125]. For this study, 50 drugs of abuse and their relative pharmacological actions were gathered. Utilizing the DrugBank [126], the STITCH database [127] (drug/ligand-target interaction databases), 142 known targets of these drugs were identified. Probabilistic matrix factorization (PMF) [128, 129] based machine learning methodology was subsequently applied to identify 48 new targets. Studies show that the PMF model, which scales linearly with the number of observations, can perform well on large, sparse, and imbalanced datasets [128]. The PMF models were trained on 11,681 drug-target interactions and 8,579,843 chemical-target interactions. The study evaluated and associated a confidence score to each predicted drug-target interaction and selected high confidence predictions, leading to the identification of 161 novel interactions between 27 out of the 50 input drugs and 89 targets. The authors also identified and categorized 173 human molecular pathways associated with the drug targets from the KEGG database. Finally, the authors examined the involvement of these targets and pathways in predicting drug addiction. Using ML methods, this study provided novel target predictions and detected critical signaling modules sensing the effects of drugs of abuse.
Another study focused on the modulation of autophagy, an important process with cellular functions such as cell death/survival [130]. The authors used QSP models to investigate the mechanism of action of autophagy modulators by predicting novel drug-target reactions and studying the drug effects using pathway/network analysis tools. Two hundred twenty-five autophagy modulators were collected, including various drugs such as fostamatinib, olanzapine, melatonin, and artenimol. Data collection was performed using the DrugBank database, and the selected modulators were manually classified into inhibitors, activators, and dual-modulators. ML was subsequently used to predict the drug-target interaction applying the PMF algorithm [129]. Using the DrugBank database, the PMF model was trained by 14,983 interactions between 5,494 drugs and 2,807 targets. A confidence score was evaluated for each predicted interaction, and the predicted interactions with high scores were selected for each drug. This ML approach led to 368 novel drug-target interactions. Functional analysis was performed using the predicted targets to present the enriched pathways involved in the regulation of autophagy. The study assists in new investigations related to the mechanism of action of autophagy modulators [130].
Coletti et al. developed the QSP model #23 of prostate cancer immunotherapy to identify the effective drug combinations for prostate cancer treatment [43]. The model was calibrated, and the numerical optimization method [131] was used for parameter estimation. The model was used to compute the synergistic effects and predict the percentage of tumor inhibition. A decision tree was built to integrate the results for making predictions about potential causality that facilitate obtaining a more comprehensive view of the system’s behavior. They set the androgen deprivation therapy as the root of the decision tree to identify efficacious treatments for castration-resistant prostate cancer. The decision tree edges were annotated with Bliss Combination Index value, a commonly used correlation measure for evaluating the synergistic effects of the therapies. The position of the nodes along the decision tree indicated the efficacy of the possible combined therapies. The results suggest that adding immune checkpoint blockade to cancer vaccines is the most effective combinatorial immunotherapy to inhibit tumor growth in castration-resistant prostate cancer.
Gaweda et al. presented QSP model #17 for chronic kidney disease mineral bone disorder (CKD-MBD) [20], where ML methods were utilized to estimate model parameters of the differential equations representing the CKD-MBD compartments. A better understanding of CKD-MBD and the variability of individuals’ CKD-MBD indications can facilitate achieving the therapeutic intentions for reducing mortality and morbidity [132]. The CKD-MBD model was constructed by applying modifications to a previously published model [133]. The modifications include adding new components to the model and using ML methods to estimate the parameters related to CKD-MBD model (such as parameters in the parathyroid gland compartment, renal phosphate reabsorption, and smooth muscle cell compartments of model #17). The CKD-MD model contains individual functions with parameters that require to be estimated. Utilizing data from 5496 CKD patients, they estimated 23 parameters associated with components of the modified model. The model fitting was performed using nonlinear least-squares regression with the trust-region reflective algorithm. The resulting model was validated by ten-fold cross-validation (each fold included 30,106 training vectors and 3345 testing vectors).
Another study integrated mechanistic models with ML to predict treatment response [134]. The authors developed a QSP model (#12) for blood pressure regulation. High blood pressure enhances the risk for various cardiovascular diseases [135]. Studying the treatment response in hypertensive patients is essential since about half of the patients do not reach adequate blood pressure control after treatment [136]. The QSP model of blood pressure regulation was constructed to provide insight into utilizing precision medicine in hypertension. A sex-specific virtual population was built to consider the heterogeneity between the sexes and within hypertension physiopathology. After constructing the sex-specific QSP model and creating the virtual population, ML methods integrated with the mechanistic model evaluated the response to antihypertensive therapies. The authors constructed a decision tree to identify the optimal drug class. This decision tree was trained to predict which drug class causes the optimal reduction in mean arterial pressure across the virtual population. Several variables can influence hypertension physiopathology and the mean arterial pressure. The features of the virtual individuals include antidiuretic hormone secretion rate, arterial resistance, renin secretion rate, the strength of the myogenic response, aldosterone secretion rate, renal sympathetic nerve activity, afferent arteriolar resistance, and venous resistance, which are pathophysiological variables. The model was validated using five-fold cross-validation [134].
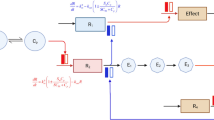
Mathematical modeling can be helpful in order to estimate risks versus potential benefits when quick decision-making is required. For example, several proposed drugs for coronavirus disease 2019 (COVID‐19) patients were associated with cardiac adverse events [137]. Model #16 presented cardiac risks of COVID‐19 therapies using a combination of PK and QSP modeling [38]. For this purpose, the authors investigated the potential effects of azithromycin, lopinavir, chloroquine, and ritonavir on cardiac electrophysiology. In order to predict cardiac adverse events, PK with the QSP model of ventricular myocytes has been utilized. A QSP model developed by O’Hara et al. was applied to simulate the effects of the drugs on ventricular action potentials [138]. Then, the QSP simulations’ drug concentrations were linked with patients’ free plasma drug concentrations using PK models to simulate drug disposition. This study predicted a greater action potential prolongation by using the combination therapy involving these drugs compared with drugs given in isolation. In order to study the influence of sex and pre-existing heart failure, models for different patient groups were developed, and virtual populations were generated to simulate the individual’s physiological variability. A logistic regression analysis was performed on population outcomes to evaluate why individual cells were resistant or susceptible to arrhythmias. Modeled ventricular myocytes were labeled as 1 (arrhythmic dynamics) and 0 (no arrhythmic dynamics) in the simulated population. The developed logistic model predicted the probability of arrhythmia from the parameter values in each cell. The simulations of patient groups suggest that women with pre‐existing heart failure are particularly susceptible to drug‐induced arrhythmias.
In this section, we presented examples of applying ML methods in different steps of QSP modeling, such as predicting treatment response, evaluating risks versus potential benefits for clinical decision-making, estimating the model parameters, model simulation, analyzing the information from databases, and predicting drug targets. Figure 2 summarizes the potential areas in which QSP modeling can benefit from ML methods. However, we note that substantial opportunities exist for the integration of QSP models with ML to further fuel pharmacometrics and drug development in general. Hence, there is a need for collaboration between statisticians, clinical pharmacologists, QSP modelers, and ML engineers to benefit from the full potential of using integrative approaches and extensive data resources.
Discussion
In this review, we present recent efforts utilizing mechanistic QSP models in drug development and clinical strategies across various pathologies. Given the multidisciplinary potential of QSP methodologies, we considered ML as a complementary tool to be used soon along with QSP to improve empiric simulation and predictions for drug development.
QSP is a quantitative framework that mimics the mechanistic knowledge of biological systems. QSP approaches can provide several advantages. First, QSP provides a platform to assess preclinical and clinical outcomes during drug development. Second, QSP can parametrize complex molecular and cellular interactions to evaluate the overall behavior of drug-target and drug-drug interaction under any biological system. For example, some I-O trials faced an increased therapy failure because of the complex dynamic interaction between TME, drug, and cancer cells, indicating the relevance of the development of virtual systems to comprehend such complex cross-interaction [8]. Third, QSP can model patient cohorts using individual patient clinical data and optimize the clinical trial design. Fourth, the personalization of QSP models can maximize the clinical trial design calibrated to patients’ background variability. Finally, QSP models can reduce the time and the cost during the drug development during the decision-making process.
Challenges and opportunities
Despite the wide range of applications, QSP approaches also exhibit certain limitations and challenges. With the expansion of omics technologies, QSP rarely integrates omics data into the framework, possibly due to the amount of information needed to combine during model construction. Notably, the confidence of the QSP model largely depends on experimental data available at the biological scale of interest to parametrize the model. However, quantitative data are often missing. The gaps can be filled using various experimental resources that may or may not be fully compatible with each other (e.g., by simultaneously considering data from in vitro and in vivo studies or different animal models). Thus, integrating omics data with QSP models will provide additional biological knowledge to fill mechanistic and clinical data gaps during model construction [13].
As indicated in Fig. 1, QSP approaches have focused on a limited number of diseases. While I-O appears to leverage QSP models recently most widely, other areas of medicine, such as transplantation management and rare diseases, have also begun applying QSP-informed approaches in drug discovery, clinical strategies, characterization of side effects, and set up a virtual patient cohort to support more personalized design therapies.
Another challenge is that QSP mainly focuses on the prominent cells or molecules and does not integrate all biological interactions of the environment. For example, TME supports cancer cells through direct and indirect effects that influence drug infiltration and resistance [8]. Nevertheless, this limitation can be addressed by using computational platforms such as Cell Collective to analyze large-scale biological systems to predict biomarkers of biological interactions. In addition, the Systems Biology Markup Language (SBML) is the most adopted standardized file format that is developed for storing computational models in a form that various modeling platforms can exchange [139]. However, based on a survey published in 2019, using SBML has not become a dominant approach in QSP modeling [140]. Utilizing a standard format for QSP models would enable the exchange and reusability of QSP models and their collection in online repositories and modeling platforms of mathematical models such as Cell Collective [31] and BioModels [141]. For example, Balbas-Martinez et al. illustrated the benefits of such model-sharing cyberinfrastructure by constructing and sharing their model for inflammatory bowel diseases in the Cell Collective platform [33].
Given their mechanistic nature, QSP models can also be utilized to understand the normal physiological behavior of molecules and cells in non-disease conditions. For example, Puniya et al. developed a mechanistic logical model of T cell plasticity and discovered the potential of a hybrid T cell population upon external cytokine stimulation [142]. Knowing the complexity of T cell biology, a recent study integrated four different modeling approaches to build three different scales (e.g. signaling, metabolism and cellular) capturing the essential biological phenomena of T cell biology [143]. Understanding the importance of different T cell populations in disease, QSP models can address the dynamic of cell development to predict biomarkers and conditions where the cellular balance is disrupted.
Notably, the QSP approach is often mentioned by many modeling approaches such as PBPK and PK/PD models, creating disparate communication between models and corporations. Therefore, defining a clear consensus for QSP among academia and pharmaceutical companies will facilitate interaction between fields and accelerate drug development strategies.
ML benefits in QSP methodologies
Mechanistic QSP models can integrate multi-layered data and characterize mechanisms that explain the emergence of biological function, phenomenon, or disease. However, as QSP models grow in scope and depth, their simulations and analyses become computationally too expensive [144]. ML models can be highly predictive and integrate multi-modal, multi-fidelity data relatively easily to reveal correlations between intertwined phenomena. However, ML models alone ignore the fundamental mechanisms behind their predictions. The integration of these approaches can result in a computationally efficient approach that can generate predictions with high accuracy and identify the underlying mechanism of the disease or its treatment [145]. With the expansion of multi-dimensional biomedical data, integrative strategies are developed to exploit and process these data to sustain a rich platform of information and support research and development. ML provides a powerful computational approach to handle and leverage big data to make intelligent decisions. The potential of ML in processing big data can transform the QSP platform into larger complex modeling systems. For example, QSP models are built as “horizontal integration” systems including structural networks (e.g., receptors, signaling pathways, metabolic pathways, or cell types); however, vertical integrations such as multiscale modeling (e.g., molecule, cells, tissue, and organs) are more challenging to conceptualize [7]. In addition, QSP modeling requires a human intervention to curate biological networks and literature review manually; therefore, adding ML in QSP can reduce bias in manual curation and allow automated data mining. Another benefit of ML to consider is parameter estimation during the development of the QSP model. The lack of prior knowledge and heterogeneity of data used during QSP model development can cause modeling uncertainty beyond the scope of biological knowledge. Therefore, ML algorithms can calibrate an interval for parameter estimation using robust statistical analysis to minimize prediction uncertainties [26, 49]. ML is broadly utilized in several fields; however, it is only now being considered as a companion to QSP models.
Conclusion
Quantitative systems pharmacology is increasingly solicited in drug development and clinical areas. QSP has demonstrated a positive impact in modern medicine through understanding mechanistic pathways of drug-target interactions, absorption, trafficking, metabolism, and side effects. QSP can help decrease the time and cost of drug development by systematically evaluating drug targets’ safety and efficacy. QSP can further increase its impact by integrating with ML and expanding to many other diseases.
References
Sever R, Brugge JS (2015) Signal transduction in cancer. Cold Spring Harb Perspect Med. https://doi.org/10.1101/cshperspect.a006098
Yildirim O, Gottwald M, Schüler P, Michel MC (2016) Opportunities and challenges for drug development: public-private partnerships, adaptive designs and big data. Front Pharmacol 7:461. https://doi.org/10.3389/fphar.2016.00461
Kitano H (2002) Systems biology: a brief overview. Science 295:1662–1664. https://doi.org/10.1126/science.1069492
Quantitative and Systems Pharmacology in the Post-genomic Era: New
Leil TA, Ermakov S (2015) Editorial: The emerging discipline of quantitative systems pharmacology. Front Pharmacol 6:129. https://doi.org/10.3389/fphar.2015.00129
Azer K, Kaddi CD, Barrett JS et al (2021) History and future perspectives on the discipline of quantitative systems pharmacology modeling and its applications. Front Physiol 12:637999. https://doi.org/10.3389/fphys.2021.637999
Quantitative and systems pharmacology in the post-genomic era: new approaches to discovering drugs and understanding therapeutic mechanisms | pharmaceutical. Report. https://pharmaceutical.report/whitepapers/quantitative-and-systems-pharmacology-in-the-post-genomic-era-new-approaches-to-discovering-drugs-and-understanding-therapeutic-mechanisms/2440. Accessed 29 Apr 2021
Roma-Rodrigues C, Mendes R, Baptista PV, Fernandes AR (2019) Targeting tumor microenvironment for cancer therapy. Int J Mol Sci. https://doi.org/10.3390/ijms20040840
Gershell LJ, Atkins JH (2003) A brief history of novel drug discovery technologies. Nat Rev Drug Discov 2:321–327. https://doi.org/10.1038/nrd1064
DiMasi JA, Grabowski HG, Hansen RW (2016) Innovation in the pharmaceutical industry: new estimates of R&D costs. J Health Econ 47:20–33. https://doi.org/10.1016/j.jhealeco.2016.01.012
Scannell JW, Blanckley A, Boldon H, Warrington B (2012) Diagnosing the decline in pharmaceutical R&D efficiency. Nat Rev Drug Discov 11:191–200. https://doi.org/10.1038/nrd3681
Rogers M, Lyster P, Okita R (2013) NIH support for the emergence of quantitative and systems pharmacology. CPT Pharmacometrics Syst Pharmacol 2:e37. https://doi.org/10.1038/psp.2013.13
Nijsen MJMA, Wu F, Bansal L et al (2018) Preclinical QSP modeling in the pharmaceutical industry: an IQ consortium survey examining the current landscape. CPT Pharmacometrics Syst Pharmacol 7:135–146. https://doi.org/10.1002/psp4.12282
Van Der Graaf PH, Gabrielsson J (2009) Pharmacokinetic-pharmacodynamic reasoning in drug discovery and early development. Future Med Chem 1:1371–1374. https://doi.org/10.4155/fmc.09.124
Lees P, Cunningham FM, Elliott J (2004) Principles of pharmacodynamics and their applications in veterinary pharmacology. J Vet Pharmacol Ther 27:397–414. https://doi.org/10.1111/j.1365-2885.2004.00620.x
Huang XH, Zheng QS (2010) Pharmacokinetic and pharmacodynamic data analysis: concepts and applications. Am J Pharmac Educ 74:53
Derendorf H, Meibohm B (1999) Modeling of pharmacokinetic/pharmacodynamic (PK/PD) relationships: concepts and perspectives. Pharm Res 16:176–185. https://doi.org/10.1023/a:1011907920641
Gabrielsson J, Dolgos H, Gillberg P-G et al (2009) Early integration of pharmacokinetic and dynamic reasoning is essential for optimal development of lead compounds: strategic considerations. Drug Discov Today 14:358–372. https://doi.org/10.1016/j.drudis.2008.12.011
Paul SM, Mytelka DS, Dunwiddie CT et al (2010) How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nat Rev Drug Discov 9:203–214. https://doi.org/10.1038/nrd3078
Gaweda AE, McBride DE, Lederer ED, Brier ME (2021) Development of a quantitative systems pharmacology model of chronic kidney disease: metabolic bone disorder. Am J Physiol Renal Physiol 320:F203–F211. https://doi.org/10.1152/ajprenal.00159.2020
Brown RP, Delp MD, Lindstedt SL et al (1997) Physiological parameter values for physiologically based pharmacokinetic models. Toxicol Ind Health 13:407–484. https://doi.org/10.1177/074823379701300401
Kuepfer L, Niederalt C, Wendl T et al (2016) Applied concepts in PBPK modeling: how to build a PBPK/PD model. CPT Pharmacometrics Syst Pharmacol 5:516–531. https://doi.org/10.1002/psp4.12134
Fidler M, Wilkins JJ, Hooijmaijers R et al (2019) Nonlinear mixed-effects model development and simulation using nlmixr and related R open-source packages. CPT Pharmacometrics Syst Pharmacol 8:621–633. https://doi.org/10.1002/psp4.12445
Geerts H, Wikswo J, van der Graaf PH et al (2020) Quantitative systems pharmacology for neuroscience drug discovery and development: current status, opportunities, and challenges. CPT Pharmacometr Syst Pharmacol 9:5–20. https://doi.org/10.1002/psp4.12478
Cheng Y, Thalhauser CJ, Smithline S et al (2017) QSP toolbox: computational implementation of integrated workflow components for deploying multi-scale mechanistic models. AAPS J 19:1002–1016. https://doi.org/10.1208/s12248-017-0100-x
Helmlinger G, Sokolov V, Peskov K et al (2019) Quantitative systems pharmacology: an exemplar model-building workflow with applications in cardiovascular, metabolic, and oncology drug development. CPT Pharmacometr Syst Pharmacol 8:380–395. https://doi.org/10.1002/psp4.12426
Quantitative Systems Pharmacology (QSP) - MATLAB & Simulink. https://www.mathworks.com/discovery/quantitative-systems-pharmacology.html. Accessed 8 Jun 2021
Elmokadem A, Riggs MM, Baron KT (2019) Quantitative systems pharmacology and physiologically-based pharmacokinetic modeling with mrgsolve: a hands-on tutorial. CPT Pharmacometr Syst Pharmacol 8:883–893. https://doi.org/10.1002/psp4.12467
Wang W, Hallow KM, James DA (2016) A tutorial on rxode: simulating differential equation pharmacometric models in R. CPT Pharmacometrics Syst Pharmacol 5:3–10. https://doi.org/10.1002/psp4.12052
Pinheiro J, Bates D, DebRoy S, Sarkar D (2006) nlme: Linear and nonlinear mixed effects models. R package …
Helikar T, Kowal B, McClenathan S et al (2012) The Cell Collective: toward an open and collaborative approach to systems biology. BMC Syst Biol 6:96. https://doi.org/10.1186/1752-0509-6-96
Bloomingdale P, Nguyen VA, Niu J, Mager DE (2018) Boolean network modeling in systems pharmacology. J Pharmacokinet Pharmacodyn 45:159–180. https://doi.org/10.1007/s10928-017-9567-4
Balbas-Martinez V, Ruiz-Cerdá L, Irurzun-Arana I et al (2018) A systems pharmacology model for inflammatory bowel disease. PLoS ONE 13:e0192949. https://doi.org/10.1371/journal.pone.0192949
Wang H, Ma H, Sové RJ et al (2021) Quantitative systems pharmacology model predictions for efficacy of atezolizumab and nab-paclitaxel in triple-negative breast cancer. J Immunother Cancer. https://doi.org/10.1136/jitc-2020-002100
Shimizu R, Katsube T, Wajima T (2021) Quantitative systems pharmacology model of thrombopoiesis and platelet life-cycle, and its application to thrombocytopenia based on chronic liver disease. CPT Pharmacometr Syst Pharmacol 10:489–499. https://doi.org/10.1002/psp4.12623
Rogers KV, Martin SW, Bhattacharya I et al (2021) A dynamic quantitative systems pharmacology model of inflammatory bowel disease: part 1 - model framework. Clin Transl Sci 14:239–248. https://doi.org/10.1111/cts.12849
Rogers KV, Martin SW, Bhattacharya I et al (2021) A dynamic quantitative systems pharmacology model of inflammatory bowel disease: part 2 - application to current therapies in Crohn’s disease. Clin Transl Sci 14:249–259. https://doi.org/10.1111/cts.12850
Varshneya M, Irurzun-Arana I, Campana C et al (2021) Investigational treatments for COVID-19 may increase ventricular arrhythmia risk through drug interactions. CPT Pharmacometr Syst Pharmacol 10:100–107. https://doi.org/10.1002/psp4.12573
Ma H, Wang H, Sové RJ et al (2020) Combination therapy with T cell engager and PD-L1 blockade enhances the antitumor potency of T cells as predicted by a QSP model. J Immunother Cancer. https://doi.org/10.1136/jitc-2020-001141
Wilson JL, Lu D, Corr N et al (2020) An in vitro quantitative systems pharmacology approach for deconvolving mechanisms of drug-induced, multilineage cytopenias. PLoS Comput Biol 16:e1007620. https://doi.org/10.1371/journal.pcbi.1007620
Sové RJ, Jafarnejad M, Zhao C et al (2020) QSP-IO: A Quantitative Systems Pharmacology Toolbox For Mechanistic Multiscale Modeling For Immuno-Oncology Applications. CPT Pharmacometr Syst Pharmacol 9:484–497. https://doi.org/10.1002/psp4.12546
Ma H, Wang H, Sove RJ et al (2020) A quantitative systems pharmacology model of T cell engager applied to solid tumor. AAPS J 22:85. https://doi.org/10.1208/s12248-020-00450-3
Coletti R, Leonardelli L, Parolo S, Marchetti L (2020) A QSP model of prostate cancer immunotherapy to identify effective combination therapies. Sci Rep 10:9063. https://doi.org/10.1038/s41598-020-65590-0
Wang H, Sové RJ, Jafarnejad M et al (2020) Conducting a virtual clinical trial in HER2-negative breast cancer using a quantitative systems pharmacology model with an epigenetic modulator and immune checkpoint inhibitors. Front Bioeng Biotechnol 8:141. https://doi.org/10.3389/fbioe.2020.00141
Bouhaddou M, Yu LJ, Lunardi S et al (2020) Predicting in vivo efficacy from in vitro data: quantitative systems pharmacology modeling for an epigenetic modifier drug in cancer. Clin Transl Sci 13:419–429. https://doi.org/10.1111/cts.12727
Balbas-Martinez V, Asin-Prieto E, Parra-Guillen ZP, Troconiz IF (2020) A quantitative systems pharmacology model for the key interleukins involved in Crohn’s disease. J Pharmacol Exp Ther 372:299–307. https://doi.org/10.1124/jpet.119.260539
Milberg O, Gong C, Jafarnejad M et al (2019) A QSP model for predicting clinical responses to monotherapy, combination and sequential therapy following CTLA-4, PD-1, and PD-L1 checkpoint blockade. Sci Rep 9:11286. https://doi.org/10.1038/s41598-019-47802-4
Cucurull-Sanchez L, Chappell MJ, Chelliah V et al (2019) Best practices to maximize the use and reuse of quantitative and systems pharmacology models: recommendations from the united kingdom quantitative and systems pharmacology network. CPT Pharmacometr Syst Pharmacol 8:259–272. https://doi.org/10.1002/psp4.12381
Leil TA, Bertz R (2014) Quantitative systems pharmacology can reduce attrition and improve productivity in pharmaceutical research and development. Front Pharmacol 5:247. https://doi.org/10.3389/fphar.2014.00247
Niarakis A, Helikar T (2021) A practical guide to mechanistic systems modeling in biology using a logic-based approach. Brief Bioinform. https://doi.org/10.1093/bib/bbaa236
Lipscomb CE (2000) Medical subject headings (mesh). Bull Med Libr Assoc 88:265–266
Bejarano L, Jordāo MJC, Joyce JA (2021) Therapeutic targeting of the tumor microenvironment. Cancer Discov 11:933–959. https://doi.org/10.1158/2159-8290.CD-20-1808
Waldman AD, Fritz JM, Lenardo MJ (2020) A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat Rev Immunol 20:651–668. https://doi.org/10.1038/s41577-020-0306-5
Abril-Rodriguez G, Ribas A (2017) Snapshot: immune checkpoint inhibitors. Cancer Cell 31:848-848.e1. https://doi.org/10.1016/j.ccell.2017.05.010
Topalian SL, Drake CG, Pardoll DM (2015) Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell 27:450–461. https://doi.org/10.1016/j.ccell.2015.03.001
Menon S, Shin S, Dy G (2016) Advances in cancer immunotherapy in solid tumors. Cancers (Basel). https://doi.org/10.3390/cancers8120106
Hegde PS, Chen DS (2020) Top 10 challenges in cancer immunotherapy. Immunity 52:17–35. https://doi.org/10.1016/j.immuni.2019.12.011
Schubert ML, Schmitt M, Wang L et al (2021) Side-effect management of chimeric antigen receptor (CAR) T-cell therapy. Ann Oncol 32:34–48. https://doi.org/10.1016/j.annonc.2020.10.478
Hardiansyah D, Ng CM (2019) Quantitative systems pharmacology model of chimeric antigen receptor T-cell therapy. Clin Transl Sci 12:343–349. https://doi.org/10.1111/cts.12636
Labrijn AF, Janmaat ML, Reichert JM, Parren PWHI (2019) Bispecific antibodies: a mechanistic review of the pipeline. Nat Rev Drug Discov 18:585–608. https://doi.org/10.1038/s41573-019-0028-1
Betts A, Haddish-Berhane N, Shah DK et al (2019) Correction to: a translational quantitative systems pharmacology model for CD3 bispecific molecules: application to quantify T cell-mediated tumor cell killing by P-cadherin LP DART®. AAPS J 21:73. https://doi.org/10.1208/s12248-019-0348-4
Stroh M, Sagert J, Burke JM et al (2019) Quantitative systems pharmacology model of a masked, tumor-activated antibody. CPT Pharmacometr Syst Pharmacol 8:676–684. https://doi.org/10.1002/psp4.12448
Ma H, Pilvankar M, Wang J et al (2021) Quantitative systems pharmacology modeling of PBMC-humanized mouse to facilitate preclinical immuno-oncology drug development. ACS Pharmacol Transl Sci 4:213–225. https://doi.org/10.1021/acsptsci.0c00178
Balti A, Zugaj D, Fenneteau F et al (2021) Dynamical systems analysis as an additional tool to inform treatment outcomes: the case study of a quantitative systems pharmacology model of immuno-oncology. Chaos 31:023124. https://doi.org/10.1063/5.0022238
Burstein HJ (2005) The distinctive nature of HER2-positive breast cancers. N Engl J Med 353:1652–1654. https://doi.org/10.1056/NEJMp058197
Nagaraj G, Ma CX (2021) Clinical challenges in the management of hormone receptor-positive, human epidermal growth factor receptor 2-negative metastatic breast cancer: a literature review. Adv Ther 38:109–136. https://doi.org/10.1007/s12325-020-01552-2
Garrido-Castro AC, Lin NU, Polyak K (2019) Insights into molecular classifications of triple-negative breast cancer: improving patient selection for treatment. Cancer Discov 9:176–198. https://doi.org/10.1158/2159-8290.CD-18-1177
Franco YL, Ramakrishnan V, Vaidya TR et al (2021) A quantitative systems pharmacological approach identified activation of JNK signaling pathway as a promising treatment strategy for refractory HER2 positive breast cancer. J Pharmacokinet Pharmacodyn 48:273–293. https://doi.org/10.1007/s10928-020-09732-x
Sui H, Ma N, Wang Y et al (2018) Anti-PD-1/PD-L1 therapy for non-small-cell lung cancer: toward personalized medicine and combination strategies. J Immunol Res 2018:6984948. https://doi.org/10.1155/2018/6984948
Jafarnejad M, Gong C, Gabrielson E et al (2019) A computational model of neoadjuvant PD-1 inhibition in non-small cell lung cancer. AAPS J 21:79. https://doi.org/10.1208/s12248-019-0350-x
Haass NK, Smalley KSM, Li L, Herlyn M (2005) Adhesion, migration and communication in melanocytes and melanoma. Pigment Cell Res 18:150–159. https://doi.org/10.1111/j.1600-0749.2005.00235.x
Kumar R, Thiagarajan K, Jagannathan L et al (2021) Beyond the single average tumor: understanding IO combinations using a clinical QSP model that incorporates heterogeneity in patient response. CPT Pharmacometr Syst Pharmacol. https://doi.org/10.1002/psp4.12637
Sokolov V, Yakovleva T, Chu L et al (2020) Differentiating the sodium-glucose cotransporter 1 inhibition capacity of canagliflozin vs. dapagliflozin and empagliflozin using quantitative systems pharmacology modeling. CPT Pharmacometr Syst Pharmacol 9:222–229. https://doi.org/10.1002/psp4.12498
DeFronzo RA, Davidson JA, Del Prato S (2012) The role of the kidneys in glucose homeostasis: a new path towards normalizing glycaemia. Diabetes Obes Metab 14:5–14. https://doi.org/10.1111/j.1463-1326.2011.01511.x
Mori-Anai K, Tashima Y, Nakada T et al (2020) Mechanistic evaluation of the effect of sodium-dependent glucose transporter 2 inhibitors on delayed glucose absorption in patients with type 2 diabetes mellitus using a quantitative systems pharmacology model of human systemic glucose dynamics. Biopharm Drug Dispos 41:352–366. https://doi.org/10.1002/bdd.2253
Yakovleva T, Sokolov V, Chu L et al (2019) Comparison of the urinary glucose excretion contributions of SGLT2 and SGLT1: a quantitative systems pharmacology analysis in healthy individuals and patients with type 2 diabetes treated with SGLT2 inhibitors. Diabetes Obes Metab 21:2684–2693. https://doi.org/10.1111/dom.13858
Shah M, Stolbov L, Yakovleva T et al (2021) A model-based approach to investigating the relationship between glucose-insulin dynamics and dapagliflozin treatment effect in patients with type 2 diabetes. Diabetes Obes Metab 23:991–1000. https://doi.org/10.1111/dom.14305
Balazki P, Schaller S, Eissing T, Lehr T (2020) A physiologically-based quantitative systems pharmacology model of the incretin hormones GLP-1 and GIP and the DPP4 inhibitor sitagliptin. CPT Pharmacometr Syst Pharmacol 9:353–362. https://doi.org/10.1002/psp4.12520
Tiwari S, Atluri V, Kaushik A et al (2019) Alzheimer’s disease: pathogenesis, diagnostics, and therapeutics. Int J Nanomed 14:5541–5554. https://doi.org/10.2147/IJN.S200490
Madrasi K, Das R, Mohmmadabdul H et al (2021) Systematic in silico analysis of clinically tested drugs for reducing amyloid-beta plaque accumulation in Alzheimer’s disease. Alzheimers Dement. https://doi.org/10.1002/alz.12312
Geerts H, Spiros A (2020) Learning from amyloid trials in Alzheimer’s disease. A virtual patient analysis using a quantitative systems pharmacology approach. Alzheimers Dement 16:862–872. https://doi.org/10.1002/alz.12082
Geerts H, Spiros A (2020) Simulating the effects of common comedications and genotypes on Alzheimer’s cognitive trajectory using a quantitative systems pharmacology approach. J Alzheimers Dis 78:413–424. https://doi.org/10.3233/JAD-200688
Talevi A, Morales JF, Hather G et al (2020) Machine learning in drug discovery and development part 1: a primer. CPT Pharmacometr Syst Pharmacol 9:129–142. https://doi.org/10.1002/psp4.12491
Adler-Milstein J, Holmgren AJ, Kralovec P et al (2017) Electronic health record adoption in US hospitals: the emergence of a digital “advanced use” divide. J Am Med Inform Assoc 24:1142–1148. https://doi.org/10.1093/jamia/ocx080
Vamathevan J, Clark D, Czodrowski P et al (2019) Applications of machine learning in drug discovery and development. Nat Rev Drug Discov 18:463–477. https://doi.org/10.1038/s41573-019-0024-5
Jeon J, Nim S, Teyra J et al (2014) A systematic approach to identify novel cancer drug targets using machine learning, inhibitor design and high-throughput screening. Genome Med 6:57. https://doi.org/10.1186/s13073-014-0057-7
Costa PR, Acencio ML, Lemke N (2010) A machine learning approach for genome-wide prediction of morbid and druggable human genes based on systems-level data. BMC Genomics 11(Suppl 5):S9. https://doi.org/10.1186/1471-2164-11-S5-S9
Athreya AP, Neavin D, Carrillo-Roa T et al (2019) Pharmacogenomics-driven prediction of antidepressant treatment outcomes: a machine-learning approach with multi-trial replication. Clin Pharmacol Ther 106:855–865. https://doi.org/10.1002/cpt.1482
Mamoshina P, Volosnikova M, Ozerov IV et al (2018) Machine learning on human muscle transcriptomic data for biomarker discovery and tissue-specific drug target identification. Front Genet 9:242. https://doi.org/10.3389/fgene.2018.00242
Lim B, van der Schaar M (2018) Disease-atlas: navigating disease trajectories with deep learning. Proc Mach Learn Res 85:1–23
Tomašev N, Glorot X, Rae JW et al (2019) A clinically applicable approach to continuous prediction of future acute kidney injury. Nature 572:116–119. https://doi.org/10.1038/s41586-019-1390-1
McCauley MD, Darbar D (2016) A new paradigm for predicting risk of Torsades de Pointes during drug development: Commentary on: “Improved prediction of drug-induced Torsades de Pointes through simulations of dynamics and machine learning algorithms.” Clin Pharmacol Ther 100:324–326. https://doi.org/10.1002/cpt.408
Alaa AM, van der Schaar M (2018) Prognostication and risk factors for cystic fibrosis via automated machine learning. Sci Rep 8:11242. https://doi.org/10.1038/s41598-018-29523-2
Riniker S, Wang Y, Jenkins JL, Landrum GA (2014) Using information from historical high-throughput screens to predict active compounds. J Chem Inf Model 54:1880–1891. https://doi.org/10.1021/ci500190p
Yamashita F, Fujita A, Sasa Y et al (2017) An evolutionary search algorithm for covariate models in population pharmacokinetic analysis. J Pharm Sci 106:2407–2411. https://doi.org/10.1016/j.xphs.2017.04.029
Gayvert KM, Madhukar NS, Elemento O (2016) A data-driven approach to predicting successes and failures of clinical trials. Cell Chem Biol 23:1294–1301. https://doi.org/10.1016/j.chembiol.2016.07.023
Sun R, Limkin EJ, Vakalopoulou M et al (2018) A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol 19:1180–1191. https://doi.org/10.1016/S1470-2045(18)30413-3
Zhao K, So H-C (2019) Drug repositioning for schizophrenia and depression/anxiety disorders: a machine learning approach leveraging expression data. IEEE J Biomed Health Inform 23:1304–1315. https://doi.org/10.1109/JBHI.2018.2856535
Korolev D, Balakin KV, Nikolsky Y et al (2003) Modeling of human cytochrome p450-mediated drug metabolism using unsupervised machine learning approach. J Med Chem 46:3631–3643. https://doi.org/10.1021/jm030102a
Wang Y-H, Li Y, Yang S-L, Yang L (2005) Classification of substrates and inhibitors of P-glycoprotein using unsupervised machine learning approach. J Chem Inf Model 45:750–757. https://doi.org/10.1021/ci050041k
Badillo S, Banfai B, Birzele F et al (2020) An introduction to machine learning. Clin Pharmacol Ther 107:871–885. https://doi.org/10.1002/cpt.1796
Maglogiannis IG (2007) Emerging artificial intelligence applications in computer engineering: real word AI systems with applications in EHealth, HCI, information retrieval and pervasive technologies, illustrated. IOS Press, New York
Kotsiantis SB, Zaharakis I (2007) Supervised machine learning: a review of classification techniques. … intelligence applications in …
Bies RR, Muldoon MF, Pollock BG et al (2006) A genetic algorithm-based, hybrid machine learning approach to model selection. J Pharmacokinet Pharmacodyn 33:195–221. https://doi.org/10.1007/s10928-006-9004-6
Ma J, Sheridan RP, Liaw A et al (2015) Deep neural nets as a method for quantitative structure-activity relationships. J Chem Inf Model 55:263–274. https://doi.org/10.1021/ci500747n
Friedman JH (2001) Greedy function approximation: a gradient boosting machine. Ann Stat 29:1189–1232. https://doi.org/10.1214/aos/1013203451
Ament SA, Pearl JR, Cantle JP et al (2018) Transcriptional regulatory networks underlying gene expression changes in Huntington’s disease. Mol Syst Biol 14:e7435
Mohammed A, Biegert G, Adamec J, Helikar T (2017) Identification of potential tissue-specific cancer biomarkers and development of cancer versus normal genomic classifiers. Oncotarget 8:85692–85715
Mohammed A, Biegert G, Adamec J, Helikar T (2018) CancerDiscover: an integrative pipeline for cancer biomarker and cancer class prediction from high-throughput sequencing data. Oncotarget 9:2565–2573
Costello JC, Heiser LM, Georgii E et al (2014) A community effort to assess and improve drug sensitivity prediction algorithms. Nat Biotechnol 32:1202–1212. https://doi.org/10.1038/nbt.2877
Sharma H, Zerbe N, Klempert I et al (2017) Deep convolutional neural networks for automatic classification of gastric carcinoma using whole slide images in digital histopathology. Comput Med Imaging Graph 61:2–13. https://doi.org/10.1016/j.compmedimag.2017.06.001
Bakheet TM, Doig AJ (2009) Properties and identification of human protein drug targets. Bioinformatics 25:451–457. https://doi.org/10.1093/bioinformatics/btp002
Wang Q, Feng Y, Huang J et al (2017) A novel framework for the identification of drug target proteins: combining stacked auto-encoders with a biased support vector machine. PLoS ONE 12:e0176486. https://doi.org/10.1371/journal.pone.0176486
Bica I, Alaa AM, Lambert C, van der Schaar M (2021) From real-world patient data to individualized treatment effects using machine learning: current and future methods to address underlying challenges. Clin Pharmacol Ther 109:87–100. https://doi.org/10.1002/cpt.1907
Olivecrona M, Blaschke T, Engkvist O, Chen H (2017) Molecular de-novo design through deep reinforcement learning. J Cheminform 9:48. https://doi.org/10.1186/s13321-017-0235-x
Wang B, Zhu J, Pierson E et al (2017) Visualization and analysis of single-cell RNA-seq data by kernel-based similarity learning. Nat Methods 14:414–416. https://doi.org/10.1038/nmeth.4207
Ding J, Condon A, Shah SP (2018) Interpretable dimensionality reduction of single cell transcriptome data with deep generative models. Nat Commun 9:2002. https://doi.org/10.1038/s41467-018-04368-5
Rashid S, Shah S, Bar-Joseph Z, Pandya R (2019) Dhaka: variational autoencoder for unmasking tumor heterogeneity from single cell genomic data. Bioinformatics. https://doi.org/10.1093/bioinformatics/btz095
Rieger TR, Allen RJ, Bystricky L et al (2018) Improving the generation and selection of virtual populations in quantitative systems pharmacology models. Prog Biophys Mol Biol 139:15–22. https://doi.org/10.1016/j.pbiomolbio.2018.06.002
Hartmann S, Biliouris K, Lesko LJ et al (2016) Quantitative systems pharmacology model to predict the effects of commonly used anticoagulants on the human coagulation network. CPT Pharmacometr Syst Pharmacol 5:554–564. https://doi.org/10.1002/psp4.12111
Wajima T, Isbister GK, Duffull SB (2009) A comprehensive model for the humoral coagulation network in humans. Clin Pharmacol Ther 86:290–298. https://doi.org/10.1038/clpt.2009.87
Board on Life Sciences, Division on Earth and Life Studies, National Academies of Sciences, Engineering, and Medicine (2016) Interindividual variability: new ways to study and implications for decision making: workshop in brief. National Academies Press (US), Washington (DC)
Goodfellow I, Bengio Y, Courville A (2016) Deep learning, illustrated. MIT Press
Nossent J, Elsen P, Bauwens W (2011) Sobol’ sensitivity analysis of a complex environmental model. Environ Model Softw 26:1515–1525. https://doi.org/10.1016/j.envsoft.2011.08.010
Pei F, Li H, Liu B, Bahar I (2019) Quantitative systems pharmacological analysis of drugs of abuse reveals the pleiotropy of their targets and the effector role of mTORC1. Front Pharmacol 10:191. https://doi.org/10.3389/fphar.2019.00191
Wishart DS, Feunang YD, Guo AC et al (2018) DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 46:D1074–D1082. https://doi.org/10.1093/nar/gkx1037
Szklarczyk D, Santos A, von Mering C et al (2016) STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res 44:D380–D384. https://doi.org/10.1093/nar/gkv1277
Mnih A, Salakhutdinov RR (2007) Probabilistic matrix factorization. Advances in neural information …
Cobanoglu MC, Liu C, Hu F et al (2013) Predicting drug-target interactions using probabilistic matrix factorization. J Chem Inf Model 53:3399–3409. https://doi.org/10.1021/ci400219z
Shi Q, Pei F, Silverman GA et al (2020) Mechanisms of action of autophagy modulators dissected by quantitative systems pharmacology analysis. Int J Mol Sci. https://doi.org/10.3390/ijms21082855
Nocedal J, Wright SJ (2006) Numerical optimization. Springer, New York
Jovanovich A, Kendrick J (2018) Personalized management of bone and mineral disorders and precision medicine in end-stage kidney disease. Semin Nephrol 38:397–409. https://doi.org/10.1016/j.semnephrol.2018.05.009
Peterson MC, Riggs MM (2010) A physiologically based mathematical model of integrated calcium homeostasis and bone remodeling. Bone 46:49–63. https://doi.org/10.1016/j.bone.2009.08.053
Ahmed S, Sullivan JC, Layton AT (2021) Impact of sex and pathophysiology on optimal drug choice in hypertensive rats: quantitative insights for precision medicine. iScience 24:102341. https://doi.org/10.1016/j.isci.2021.102341
Virani SS, Alonso A, Benjamin EJ et al (2020) Heart disease and stroke statistics-2020 update: a report from the American Heart Association. Circulation 141:e139–e596. https://doi.org/10.1161/CIR.0000000000000757
Ritchey MD, Gillespie C, Wozniak G et al (2018) Potential need for expanded pharmacologic treatment and lifestyle modification services under the 2017 ACC/AHA hypertension guideline. J Clin Hypertens (Greenwich) 20:1377–1391. https://doi.org/10.1111/jch.13364
Liu D, Zeng X, Ding Z et al (2021) Adverse cardiovascular effects of anti-COVID-19 drugs. Front Pharmacol 12:699949. https://doi.org/10.3389/fphar.2021.699949
O’Hara T, Virág L, Varró A, Rudy Y (2011) Simulation of the undiseased human cardiac ventricular action potential: model formulation and experimental validation. PLoS Comput Biol 7:e1002061. https://doi.org/10.1371/journal.pcbi.1002061
Hucka M, Finney A, Sauro HM et al (2003) The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics 19:524–531. https://doi.org/10.1093/bioinformatics/btg015
Ermakov S, Schmidt BJ, Musante CJ, Thalhauser CJ (2019) A survey of software tool utilization and capabilities for quantitative systems pharmacology: what we have and what we need. CPT Pharmacometr Syst Pharmacol 8:62–76. https://doi.org/10.1002/psp4.12373
Malik-Sheriff RS, Glont M, Nguyen TVN et al (2020) BioModels-15 years of sharing computational models in life science. Nucleic Acids Res 48:D407–D415. https://doi.org/10.1093/nar/gkz1055
Puniya BL, Todd RG, Mohammed A et al (2018) A mechanistic computational model reveals that plasticity of CD4+ T cell differentiation is a function of cytokine composition and dosage. Front Physiol 9:878. https://doi.org/10.3389/fphys.2018.00878
Wertheim KY, Puniya BL, La Fleur A et al (2021) A multi-approach and multi-scale platform to model CD4+ T cells responding to infections. PLoS Comput Biol 17:e1009209. https://doi.org/10.1371/journal.pcbi.1009209
Dada JO, Mendes P (2011) Multi-scale modelling and simulation in systems biology. Integr Biol (Camb) 3:86–96. https://doi.org/10.1039/c0ib00075b
Alber M, Buganza Tepole A, Cannon WR et al (2019) Integrating machine learning and multiscale modeling-perspectives, challenges, and opportunities in the biological, biomedical, and behavioral sciences. NPJ Digital Med 2:115. https://doi.org/10.1038/s41746-019-0193-y
Stroh M, Green M, Millard BL et al (2021) Model-informed drug development of the masked anti-PD-L1 antibody CX-072. Clin Pharmacol Ther 109:383–393. https://doi.org/10.1002/cpt.1985
Mackey MC, Glisovic S, Leclerc J-M et al (2021) The timing of cyclic cytotoxic chemotherapy can worsen neutropenia and neutrophilia. Br J Clin Pharmacol 87:687–693. https://doi.org/10.1111/bcp.14424
Derbalah A, Duffull S, Moynihan K, Al-Sallami H (2021) the influence of haemostatic system maturation on the dose-response relationship of unfractionated heparin. Clin Pharmacokinet 60:491–499. https://doi.org/10.1007/s40262-020-00949-0
Yu H, Tang W, Greasley PJ et al (2021) Predicted cardiac hemodynamic consequences of the renal actions of SGLT2i in the DAPA-HF study population: a mathematical modeling analysis. J Clin Pharmacol 61:636–648. https://doi.org/10.1002/jcph.1769
Miyano T, Irvine AD, Tanaka RJ (2021) A mathematical model to identify optimal combinations of drug targets for dupilumab poor responders in atopic dermatitis. Allergy. https://doi.org/10.1111/all.14870
Nguyen HQ, Iskenderian A, Ehmann D et al (2020) Leveraging quantitative systems pharmacology approach into development of human recombinant Follistatin fusion protein for Duchenne muscular dystrophy. CPT Pharmacometr Syst Pharmacol 9:342–352. https://doi.org/10.1002/psp4.12518
Abrams R, Kaddi CD, Tao M et al (2020) A quantitative systems pharmacology model of Gaucher disease type 1 provides mechanistic insight into the response to substrate reduction therapy with eliglustat. CPT Pharmacometr Syst Pharmacol 9:374–383. https://doi.org/10.1002/psp4.12506
Hartmann S, Biliouris K, Lesko LJ et al (2020) Quantitative systems pharmacology model-based predictions of clinical endpoints to optimize warfarin and rivaroxaban anti-thrombosis therapy. Front Pharmacol 11:1041. https://doi.org/10.3389/fphar.2020.01041
PLOS One Staff (2020) Correction: higher naloxone dosing in a quantitative systems pharmacology model that predicts naloxone-fentanyl competition at the opioid mu receptor level. PLoS ONE 15:e0240148. https://doi.org/10.1371/journal.pone.0240148
Varshneya M, Irurzun-Arana I, Campana C et al (2020) Investigational treatments for COVID-19 may increase ventricular arrhythmia risk through drug interactions. medRxiv. https://doi.org/10.1101/2020.05.21.20109397
Li M, Nguyen L, Subramaniyan B et al (2019) PBPK modeling-based optimization of site-specific chemo-photodynamic therapy with far-red light-activatable paclitaxel prodrug. J Control Release 308:86–97. https://doi.org/10.1016/j.jconrel.2019.07.010
Wang H, Milberg O, Bartelink IH et al (2019) In silico simulation of a clinical trial with anti-CTLA-4 and anti-PD-L1 immunotherapies in metastatic breast cancer using a systems pharmacology model. R Soc Open Sci 6:190366. https://doi.org/10.1098/rsos.190366
Vaidya TR, Ande A, Ait-Oudhia S (2019) Combining multiscale experimental and computational systems pharmacological approaches to overcome resistance to HER2-targeted therapy in breast cancer. J Pharmacol Exp Ther 369:531–545. https://doi.org/10.1124/jpet.118.255752
Kalra P, Brandl J, Gaub T et al (2019) Quantitative systems pharmacology of interferon alpha administration: a multi-scale approach. PLoS ONE 14:e0209587. https://doi.org/10.1371/journal.pone.0209587
Sokolov V, Helmlinger G, Nilsson C et al (2019) Comparative quantitative systems pharmacology modeling of anti-PCSK9 therapeutic modalities in hypercholesterolemia. J Lipid Res 60:1610–1621. https://doi.org/10.1194/jlr.M092486
Gebremichael Y, Lahu G, Vakilynejad M, Hallow KM (2019) Benchmarking renin suppression and blood pressure reduction of direct renin inhibitor imarikiren through quantitative systems pharmacology modeling. J Pharmacokinet Pharmacodyn 46:15–25. https://doi.org/10.1007/s10928-018-9612-y
Serrano Castillo F, Bertrand CA, Myerburg MM et al (2019) A physiologically-motivated model of cystic fibrosis liquid and solute transport dynamics across primary human nasal epithelia. J Pharmacokinet Pharmacodyn 46:457–472. https://doi.org/10.1007/s10928-019-09649-0
Maass C, Sorensen NB, Himmelfarb J et al (2019) Translational assessment of drug-induced proximal tubule injury using a kidney microphysiological system. CPT Pharmacometr Syst Pharmacol 8:316–325. https://doi.org/10.1002/psp4.12400
Acknowledgements
The work was supported by NIH grant 1R35GM119770 to T.H.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Aghamiri, S.S., Amin, R. & Helikar, T. Recent applications of quantitative systems pharmacology and machine learning models across diseases. J Pharmacokinet Pharmacodyn 49, 19–37 (2022). https://doi.org/10.1007/s10928-021-09790-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10928-021-09790-9