Abstract
Two strategies were investigated to perform the controlled degradation of two grades of poly(butylene succinate) (PBS): the chemical and enzymatic degradation routes. PBS chemical degradation assays were carried out in alkaline and acidic media at the temperature range between 40 and 80 °C. Additionally, the effects of distinct enzymes (cutinase, lipase, cellulase, protease, and amylase) that exhibit distinct hydrolysis activities were investigated on the enzymatic degradation of PBS. Two grades of PBS were compared: PBS microparticles manufactured through suspension polycondensation (PBS1) and; commercial PBS (PBS2) manufactured in bulk processes. Polymer degradation was evaluated through high-performance liquid chromatography, gel permeation chromatography, weight loss analyses, and determination of the pH of the supernatant. It was observed that PBS1 was subject to high rates of chemical degradation at higher temperatures, especially when degradation was conducted in alkaline media. On the other hand, the analyzed operation conditions did not affect the degradability of PBS2 significantly, which fluctuated around 5% of weight loss after 4 weeks. The degradability of both polymer matrices was much more intense when enzymes were used. For PBS1, the enzyme degradation effect was significant (weight loss of 57%, 32%, 25% after 4 weeks of experimentation, using cutinase, lipase, and amylase, respectively); for PBS2, after 1 month of experimentation, the weight loss using cutinase and lipase approached respectively 100% and 81%. It can be concluded that the application of enzymes can be beneficial for the controlled degradation of PBS at mild process conditions, allowing the development of environmental-friendly strategies for the controlled degradation of this material, and that both chemical and enzymatic degradations are sensitive to the characteristics of the analyzed samples, which must be carefully considered during the process development stage.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The growing demand for new bio-based polymers obtained from renewable resources is expected to propel the renewable polymer market in the next years [1,2,3,4,5,6,7,8]. Particularly, researchers and industries have investigated and developed new technologies to explore and improve the properties of green polymer materials, including mechanical, thermal, rheological, and structural properties [9,10,11,12,13]. For this reason, according to Grand View Research, the global market for biodegradable plastics was estimated at approximately US$ 9.17 billion in 2020 and is expected to grow over US$ 15 billion by the end of 2027 [10]. In this context, considering modern circular economy concepts that assume that post-consumer polymer wastes must be reinserted into the chemical chain, many studies have also been conducted to evaluate the degradability of these green polymer materials through chemical, microbiological, enzymatic, and natural marine degradation routes [14,15,16,17,18,19]. The use of enzymes, in particular, can be very advantageous, enabling the controlled degradation of polymer materials under mild conditions [20].
Polyesters based on renewable monomers, such as furan-2,5-dicarboxylic acid, lactic acid, butane-1,4-diol, and succinic acid, have been widely studied due to their inherent biodegradability characteristics and excellent mechanical, thermal, rheological properties, when compared to standard oil-based polymers [8]. Among these monomers, succinic acid has been widely recognized as a fundamental bio-based building block monomer that can originate a large number of distinct chemical reactants, solvents, and polymer materials, including poly(butylene succinate) (PBS) [21, 22].
Over the recent years, the production of PBS has aroused enormous interest in both academia and industry, as shown in Fig. 1. As a consequence, based on the available data, it can be observed that the studies regarding biodegradable polymers and PBS materials exhibited similar trends. Particularly, the use of biodegradable PBS-based materials can be environmentally attractive, mitigating pollution impacts related to the release of microplastics in the environment and contributing to the reduction of marine contamination. As a matter of fact, when PBS resins are degraded, the monomers can be released in the environment. For this reason, it is important to notice that such compounds (mainly 1,4-butanediol and succinic acid) exhibit low toxicity for sea life and do not accumulate in marine organisms, as both molecules take part of distinct metabolic pathways [14].
Previous studies analyzed the degradability of different polymer materials and indicated that the rates of degradation depend not only on the polymer composition, but also on the polymer crystallinity (rates of both chemical and enzymatic degradations are usually higher in the amorphous phase due to the higher exposure and mobility of chemical groups that are subject to degradation), pH of the aqueous media, and presence of enzymes, when enzymatic biodegradation is considered [14, 22]. Particularly, the use of different hydrolases has been investigated, as these enzymes catalyze the cleavage of covalent bonds using water. This large group of enzymes comprises distinct enzymatic subgroups that include phosphatases (that act on ester bonds), proteases (that act on amide bonds), among others [23].
PBS and respective copolymers are well-known for their high rates of controlled degradability and high hydrolysis susceptibility. However, despite the efforts of different research groups to develop new strategies to efficiently degrade PBS, few works have compared the effects of enzymatic and chemical hydrolyses or evaluated comparatively the effects of distinct enzymes on the controlled degradation of PBS samples.
Ahn et al. [24] compared the degradation of PBS homopolymer and succinate-adipate copolymers. In this study, the effect of a specific enzyme on polymer degradation was not evaluated, but the effect of using activated sewage sludge to perform the degradation, which was compared to the chemical hydrolytic degradation under alkaline conditions (pH 10.6). The authors observed higher biodegradability for copolymers presenting higher adipate contents and smaller crystallinity. Regarding the controlled chemical degradation, the increase of the adipate content caused the decrease in the rate of degradation.
Tsutsumi et al. [25] compared the chemical and enzymatic degradations of some commercial polyesters, such as PBS, poly(butylene succinate-co-adipate) (PBSA), poly(ethylene succinate) (PES), a blend of PBS and polycaprolactone (PBS/PCL), and poly(butylene adipate-co-terephthalate) (PBAT). The authors investigated the use of distinct lipases and the controlled chemical degradation was carried out in alkaline media. It was observed that the enzymatic degradation was more efficient than chemical degradation for PBS-based polymers, whilst PBSA was the most biodegraded material, reaching approximately 100% of weight loss after 10 days.
Bai et al. [26] used cutinase to perform the controlled degradation of three aliphatic polyesters: PBS; poly(butylene adipate) (PBA), and poly(butylene suberate) (PBSub). The authors reported higher rates of degradation for longer aliphatic polymer chains and that the action of cutinase was not very sensitive to the presence of amorphous and crystalline regions.
Qi et al. [27] compared the chemical and enzymatic degradations (using porcine pancreas lipase) of copolymers of poly(butylene-co-isosorbide succinate) (PBIS) and PBS homopolymer. The authors reported higher rates of chemical degradation for PBS under acidic conditions, when compared to neutral media, and that the enzymatic degradation of the polymer matrix was more efficient than the chemical hydrolysis.
Some additional studies compared the effects of hydrolases on PBS-based materials. Shi et al. [28], for example, compared the enzymatic degradation of PBS using cutinase and lipase. Results showed that biodegradation using cutinase was faster than using lipase, reaching 100% of degradation in less than two days. Besides, it was observed that cutinase acted preferentially on the surface of the test specimens, whilst lipase degraded mostly the bulk region of the polymer matrix.
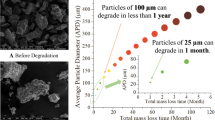
Most of the references previously discussed evaluated the degradation of films and test specimens of PBS, as this polymer is normally prepared through bulk polymerization processes. However, Dutra et al. [29] developed a new PBS suspension polycondensation process that allows the manufacture of PBS particles in a single reaction step. Dutra et al. [14] studied the chemical degradation of the obtained PBS particles in aqueous media under different conditions of salinity, pH, and temperature. As observed experimentally, the rates of PBS degradation are higher at higher temperatures and at acidic and alkaline conditions. However, at the analyzed degradation conditions and time, the polymer weight losses were smaller than 10%. Particularly, in saline conditions, the rates of polymer degradation were relatively smaller. It was also shown that the degradation rates depend on the particle morphology. According to the developed kinetic model, PBS microparticles showing 100 µm of diameter were expected to completely degrade in seawater in less than 1 year, whilst microparticles exhibiting 1 µm of diameter were expected to degrade completely in less than 1 day.
Based on the previous paragraphs, it can be said that few studies compared the controlled chemical and enzymatic degradations of PBS samples, and even fewer evaluated the effects of particle morphology on the degradation of PBS samples, although PBS is expected to be used in particulate form in cosmetic formulations. Besides, most of the works presented before used lipases and cutinases to perform the polyester degradation. However, considering the much wider group of hydrolases, the use of other enzymes, such as amylase, protease, and cellulase, should also be considered for evaluation of the most adequate conditions to perform the controlled degradation of PBS matrices. Nevertheless, to the best of our knowledge, no previous study investigated comparatively the effects of controlled chemical and enzymatic degradation of PBS, analyzing the effects of using distinct chemical media and different enzymes to perform the degradation assays. For these reasons, the present work investigates the controlled chemical and enzymatic degradation of distinct PBS grades at different conditions and in presence of different enzymes.
Materials and Methods
Materials
Two PBS grades were investigated in the present study. PBS1 was produced as described by Dutra et al. [29] using the monomers succinic acid (SA), provided by Vetec (Rio de Janeiro, Brazil), and 1,4-butanediol (BDO), supplied by Merck (Rio de Janeiro, Brazil). Span 80® (sorbitan monooleate) was provided by Fluka (Mexico City, Mexico) and used as surfactant. Soybean oil was supplied by Bunge Alimentos (Santa Catarina, Brazil) and used as the suspending medium [30, 31]. PBS2 was manufactured in bulk by Showa Denko (Tokyo, Japan) as pellets with an average diameter of 900 µm and was kindly provided by Braskem (Rio de Janeiro, Brazil), exhibiting Mn of 9.10 × 103 Da, Mw of 1.99 × 104 Da and crystallinity of 50% [32].
To perform the chemical degradation experiments, sodium hydroxide was provided by Caledon Laboratory Chemical (Ontario, Canada) and hydrochloric acid was provided by Vetec (Rio de Janeiro, Brazil). The buffer aqueous solutions were prepared using sodium phosphate (Vetec, Rio de Janeiro, Brazil) and sodium acetate (Vetec, Rio de Janeiro, Brazil).
Finally, all analyzed enzymes (lipase—Lipozyme® CALB, amylase, protease, cutinase, and cellulase) were supplied by Novozymes (Bagsvaerd, Denmark) and used without purifications steps.
All reactants and solvents used in the present study were provided as analytical grades. Unless stated otherwise, chemicals were used as received, without any further purification step.
Production of PBS Microparticles
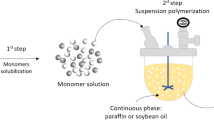
PBS microparticles (PBS1) were produced through suspension polycondensation, as described and detailed by Dutra et al. [33]. Polycondensations were performed in heterogeneous media for 5 h at 160 °C, using soybean oil as suspending phase, and Span 80® as a suspending agent. The obtained PBS microparticles presented Mn of 2.43 × 103 Da, Mw of 4.58 × 103 Da, crystallinity of 70% [34], and particle sizes ranging from 60 to 1000 µm, as shown in Fig. 2, with volume average particle size of 384 μm. It is important to point out that such microparticles have indeed been used for formulations of commercial cosmetic and personal care products [14, 33].
Degradation Experiments
Degradation Media
For chemical degradation, alkaline solutions (pH 10.0) and acid solutions (pH 4.0) were prepared using sodium hydroxide and hydrochloric acid. Two aqueous buffer solutions were synthesized and used to perform enzymatic degradation assays: sodium phosphate solution (0.1 M, pH 7.0), and sodium acetate solution (0.1 M, pH 5.5).
Chemical Degradation Assays
Chemical degradation experiments were performed with the two PBS grades under alkaline (NaOH solutions), acidic (HCl solutions), and neutral conditions (distilled water). The temperature and pH of each experiment are shown in Table 1. At this step, 9.5 mL of the corresponding solutions were added to 0.5 g of polymer samples at temperatures in the range between 40 and 80 °C. Samples of the polymer particles and the supernatant were withdrawn over the experimentation time for characterization: after 1 day (t1), 2 weeks (t2), and 4 weeks (t3). The experimental design was defined to verify the independent effects of the input variables (temperature and pH) on the output variables (weight loss and average molecular weights) as a complete 22 factorial experimental plan, comprising seven experiments with the replicates at the central point (22 + 3), as presented in Table 1.
Enzymatic Degradation Assays
The enzymatic degradation experiments were conducted with both PBS grades at the optimal reaction conditions (temperature and pH) of each enzyme, as reported by the supplier, using sodium phosphate or sodium acetate buffer aqueous media to control the pH values. In order to do that, 200 µL of solution of each enzyme were added to 0.5 g of each PBS grade (PBS1 or PBS2), previously dispersed in 9.5 mL of the buffer solution. Table 2 presents the experimental conditions of the enzymatic degradation step. Experiments were also conducted without enzymes for comparison. Assays were carried out in triplicates at 60 ºC using distilled water. Samples of the polymer particles and the supernatant were also withdrawn over the experimentation time for characterization: after 1 day (t1), 2 weeks (t2), and 4 weeks (t3).
Characterization
Determination of Weight Loss
After the specified experimentation time (1 day, 2 weeks, 4 weeks), polymer samples were filtrated, washed, and dried at 40 °C. The percentage of weight loss, \(\mathrm{\%}WL\), was calculated based on Eq. 1:
where \({m}_{0}\) corresponds to the initial mass of polymer and \({m}_{f}\) is the final collected polymer mass, after the drying step.
Gel Permeation Chromatography (GPC)
Gel permeation chromatography (GPC) was used to evaluate the average molar mass values of polymer samples during the degradation process. Analyzes were performed using a gel permeation chromatograph manufactured by VISCOTEK (model GPC Max VE2001, Malvern, Malvern, UK) equipped with a refractometer detector, also manufactured by Viscotek, model VE3580. During the analysis, the mobile phase flow rate was kept constant at 1.0 mL/min and 40 °C, using hexafluoroisopropanol (HFIP) as the mobile phase. Columns, manufactured by Shodex (Shodex GPC HFIP803, Shodex GPC HFIP-804, and Shodex GPC HFIP-805, Showa Denko, Tokyo, Japan), exhibited maximum pore sizes of 5 × 102, 1.5 × 103, and 5 × 103 Å and exclusion limits of 3 × 104, 1 × 105, and 1 × 106 Da. The calibration curves were built with poly(methyl methacrylate) standards provided by American Polymer Standards (Mentor, USA), with average molar masses ranging from 5 × 102 to 1 × 106 Da. Data acquisition and processing were performed with the proprietary OminiSEC program developed by Viscotek. The polymer samples were prepared using HFIP (3 mg/mL).
Scanning Electron Microscopy (SEM)
A scanning electron microscope (SEM) (Versa 3D, Fei Company, Eindhoven, The Netherlands) was used to analyze the morphology of the particles before and after the degradation experiments. For these analyses, samples were firstly coated with gold and the images were obtained in high-vacuum mode.
High-Performance Liquid Chromatography (HPLC)
High-Performance Liquid Chromatography (HPLC, Shimadzu, Kyoto, Japan) was used for quantification of succinic acid (SA) released in the supernatant during the experiment. In order to do that, a chromatograph (Shimadzu, Kyoto, Japan) equipped with: (i) an Aminex® HPX-87H column, coupled to a cation exchange column, with dimensions 300 mm × 7.8 mm (Bio-Rad Laboratories Ltd, California, USA); (ii) the RID-refractive index detector 10A (Shimadzu, Kyoto, Japan); and (iii) the LC-20ADSP pump (Shimadzu, Kyoto, Japan), was used for analyses. The proprietary LabSolutions chromatographic software (Shimadzu, Kyoto, Japan) was used for data acquisition and calculations. Samples (20 µL) were injected and eluted at a flow rate of 0.6 mL/min using aqueous sulfuric acid H2SO4 solution (5 mM) as the mobile phase. The column temperature was maintained at 55 °C. The samples were filtrated through a membrane (CHROMAFIL®) with pore diameters of 0.45 μm and injected in duplicates. The SA quantification was determined using a calibration curve built for the retention time of 11.5 min.
pH
The pH of the supernatant was monitored with help of pH strips. This step was conducted in order to evaluate the acidification of the supernatant caused by the release of succinic acid molecules during the polymer degradation process.
Results and Discussion
Chemical Degradation
Figure 3 shows the degradation of PBS1 and PBS2 samples under distinct chemical conditions. Analyzing particularly PBS1, it is possible to notice that under lower temperature values, the acidic degradation resulted in higher polymer weight loss. However, the largest degrees of polymer degradations were achieved when the polymer matrices were kept at the highest temperature value (80 °C) in an alkaline medium. Based on Fig. 3, temperature clearly exerts the most significant effect on the chemical degradation of both PBS grades, so that the hydrolysis reaction observed under neutral conditions at 60 °C was statistically similar to the one obtained when an alkaline medium was used at 40 °C. Besides, the existence of a synergetic effect can be observed graphically in Fig. 3, as the acidic condition led to the highest rate of degradation at 40 °C, while the alkaline condition led to the highest rate of degradation at 80 °C.
It must be noticed in Fig. 3 that the weight losses observed for grade PBS2 were always smaller than observed for grade PBS1 in all analyzed conditions, reinforcing the results obtained by Dutra et al. [14] who showed that weight losses were smaller for the PBS test pieces than for PBS microparticles. Therefore, one might already expect lower rates of degradation for grade PBS2 due to its larger diameter and higher average molar masses. The obtained results clearly indicate that the grade properties exert enormous influence on the course of the PBS chemical degradation.
It is important to point that Dutra et al. [14] also observed that the degradation of PBS samples was accelerated in both acidic and alkaline aqueous media, when compared to the rates of degradation in neutral aqueous environments. As highlighted by the authors, under an acidic degradation medium, PBS hydrolysis is catalyzed by protons, forming shorter chains and additional carboxylic groups, which may lead to autocatalytic degradation conditions [14, 35]. On the other hand, under alkaline conditions, cleavage of the ester bonds can also be catalyzed by the nucleophilic attack of the hydroxide ion [14, 36]. Thus, both chemical conditions can catalyze the hydrolysis of PBS [14, 37].
Figure 4 exhibits the molar mass distributions of the samples subjected to chemical degradation after 4 weeks of the assay. It must be noticed that the hydrolysis effect is present in all reaction conditions, even when degradation is performed under a neutral medium. Analyzing PBS1 and considering the initial time (t0) as a reference, the Mw presented a drop of 13% at 80 °C and pH 10; 14% at 80 °C and pH 4; 7% at 40 °C and pH 10; 2% at 40 °C and pH 4; and finally, 10% at 60 °C and pH 7. The obtained results corroborate the ones observed for the weight loss: higher weight losses and lower molar masses were obtained for samples submitted to higher temperatures. However, based on the molar mass distributions, no significant difference was observed between acidic and alkaline degradation conditions. For PBS2, as similar weight loss values were observed in all conditions, only the central point was analyzed, and no significant change of molar mass distributions was observed, as shown in Fig. 4B.
Based on Figs. 3 and 4, it becomes possible to conjecture that the degradation mechanism is probably related to cleavage of short molecular segments placed at vicinities of the chain ends, explaining why the weight losses were not accompanied by a significant reduction of average molar masses, as one might expect when chain cleavage occurs at random. This can possibly explain the effect of morphology too, as chain cleavage is expected to occur when the chain ends are placed near the particle surfaces and in close contact with the aggressive aqueous medium.
Table 3 summarizes some results obtained by different authors regarding the chemical degradation of PBS. It is possible to observe that different methodologies have been investigated in order to evaluate the most appropriate degradation condition. It is highlighted that the chemical conditions employed in the present work (high temperature of 80 °C, alkaline medium, and 4 weeks of the experiment) favor the chemical degradation of PBS matrices. Additionally, it becomes evident that the specific area of PBS significantly affected the degradation of the polymer matrices.
Figure 5 shows the pH values of the supernatant over the time of degradation. The pH of day 0 represents the pH of the solutions that were added to each polymer at the beginning of the assay. It must be observed that the pH of the supernatant decreased continuously under the distinct conditions over the experimentation time, with sharp and continuous reduction at 80 °C and alkaline condition for both PBS1 and PBS2 (condition P1), indicating that chemical degradation of the polymer matrices probably released succinic acid to the aqueous media, reinforcing the previously proposed interpretation of the degradation process. Particularly, by comparing results obtained at the conditions reported in Fig. 5, it becomes possible to observe that hydrolysis takes place in both caustic, neutral and acidic conditions for the analyzed resin in a broad range of temperatures. Furthermore, it is also important to point out that the pH values of the supernatants obtained after degradation under neutral reaction conditions (condition ACP) were not altered significantly after day 1, possibly indicating that succinic acid molecules released into the aqueous media remained adsorbed onto the polymer material because of the lower salinity and polarity of the aqueous system. This can also explain why pH values changed more significantly in the assay performed with PBS2 in neutral conditions, as this commercial resin is additivated and can release small amounts of additives in the aqueous phase, changing the solubility of acid residues in water. Finally, one must observe that the investigated hydrolytic degradation is an auto-catalytic reaction, as the release of protons due to hydrolysis of the ester groups catalyses the advancement of the hydrolytic reaction, which explains why the reaction can be performed effectively even when the initial reaction condition of the aqueous phase is near the neutral pH value.
The SEM micrographs of PBS1 and PBS2 before the degradation assays are presented in Fig. 6. As one can observe, the surface of both polymer matrices presented distinct features. Apparently, the surfaces of PBS2 pellets were more irregular than the surfaces of PBS1 particles produced in suspension, facilitating the chemical and enzymatic degradation of PBS2, as observed in most cases. To facilitate the comparison between the matrices before and after the degradation experiments, the micrographs showing higher magnifications (20000×) were selected and used as references (PBS1.2 and PBS2.2).
Figure 7 shows SEM micrographs of PBS samples before and after the chemical degradation assays. It is possible to observe for both polymer matrices PBS1 and PBS2 that more irregular surfaces were obtained after the alkaline and high-temperature degradation assays, once surface erosion can be observed in each micrograph and was not observed in matrices submitted to neutral degradation conditions (PBS1.A and PBS2.A).
The HPLC chromatograms of the supernatants obtained after degradation are shown in Fig. 8. The presence of the characteristic peaks of SA (11 min) and BDO (22 min) can be observed in the supernatants after degradation, reinforcing the previously proposed interpretation of the chemical degradation process. Observation of additional peaks (as the one located at 15 min) is probably related to the presence of dimers or trimers, indicating that the chemical degradation occurs primarily in the vicinities of the polymer chain end, as already explained. Based on the quantification of the released SA and considering that the average sizes of the polymer chains of grades PBS1 and PBS2 were different, it was possible to detect 1% of SA for PBS1 (condition P1), 0.07% of SA for PBS1 (condition P5), 5% of SA for PBS2 (condition P1) and 4% of SA for PBS2 (condition P5), which are very low values when compared to the weight loss values. Thus, this discrepancy indicates that other oligomeric byproducts with higher molar masses are also formed, as indicated by the HPLC chromatogram.
Enzymatic Degradation
The weight losses of PBS samples treated with enzymatic solutions are shown in Fig. 9. It is possible to observe that the addition of protease and cellulase did not modify the rates of polymer degradation significantly, resulting in weight loss values that were similar to the ones obtained when the degradation experiment was performed in absence of enzymes. On the other hand, the addition of amylase favored the degradation of grade PBS1, achieving almost 30% of degradation, but did not affect the degradation of grade PBS2 significantly. One must consider that many variables can affect the rates of enzymatic polymer degradation simultaneously, including the specific areas, the average polymer masses and molar mass distributions, the porosity of the polymer pieces, the reaction temperature, the intrinsic enzyme activity, among others. In the analyzed case, PBS1 presented higher specific area and lower average molar mass, which favor the adsorption of enzyme molecules on the particle surfaces and the chain unzipping mechanism (chain end degradation through sequential removal of chemical segments), which has been frequently assigned to enzymatic degradation processes.
Cutinase and lipase were the enzymes that degraded more effectively both analyzed grades of PBS. In both cases, the degradation caused by cutinase was higher than caused by lipase. This behavior had already been reported by Shi et al. [28] who showed that the degradation mechanisms for cutinase and lipase were different: for cutinase, the degradation occurs by surface erosion; whilst for lipase, the erosion occurs in the bulk. Consequently, the enzymatic degradation induced by cutinase tends to provide higher degradation rates, as observed in Fig. 9. Comparing grades PBS1 and PBS2, one can observe that grade PBS2 was more effectively degraded by both cutinase and lipase.
For PBS2, both cutinase, and lipase significantly affected the polymer degradation, leading to weight losses of 100% and 81%, respectively. For PBS1, cutinase, lipase, and amylase were the most effective enzymes, leading to weight losses of 57%, 32%, and 25%, respectively. It is important to highlight that the weight loss values achieved through the enzymatic degradation route for both PBS matrices were higher than the ones obtained through the chemical degradation route, encouraging the use of these enzymes for the controlled degradation of PBS grades. Given the much larger specific areas provided by grade PBS1, the lower rates of enzymatic degradation obtained for grade PBS1 were somewhat surprising and possibly indicates that the rate of enzymatic degradation diminishes with the average molar mass of the polymer chains. This result can indicate the existence of important geometrical effects that can affect the interaction of the enzymes with the polymer chains and that should be investigated more thoroughly in future works.
The molar mass distributions of samples of PBS1 treated with lipase and cutinase are shown in Fig. 10. It is possible to observe the displacement of the molar mass distributions towards lower values using both enzymes, lipase (Fig. 10A) and cutinase (Fig. 10B). The increase of the low molar mass chains was even more significant when cutinase was added to the reaction medium, reinforcing the great potential of such enzyme for PBS degradation. Nevertheless, the persistent observation of long chains in the polymer residue, even after very significant losses of mass, probably indicates once more that degradation involves the release of short molecular segments placed in the vicinities of the polymer chain ends, which can justify the better performances of the enzymes for degradation of longer PBS molecular chains.
Figure 11 exhibits the molar mass distributions of PBS2 chains after degradation treatments using lipase and cutinase. It must be noticed that significant narrowing of the molar mass distributions was observed with both enzymes, suggesting once more the existence of significant synergetic effects between the chain size and the degradation efficiency of the enzymes. Despite the significant variation of the weight loss values, the Mw values of the polymer residues diminished only 8% using lipase and 2% using cutinase. This suggests that the enzymatic degradation action possibly occurs repeatedly through chain unzipping (chain end degradation) along the same polymer chain to which the enzyme molecule is attached, mimicking the action of a zipper that cleaves repeatedly the bonds of long chains, preserving the characteristics of the residual chains.
The pH values of the supernatants for both PBS1 and PBS2 are shown in Fig. 12. The supernatant of the enzymatic solutions significantly decreased when lipase and cutinase were added to the media, indicating the increase of the succinic acid molecules over time and indicating the occurrence of polymer degradation. The effect of amylase on PBS1 was not detected, probably because the weight losses were small in this case, which means that the concentration of released succinic acid was not sufficient to significantly affect the pH of the medium.
Figure 13 shows SEM micrographs of the samples before and after the enzymatic degradation assays. It is possible to observe for both polymer matrices PBS1 and PBS2 that irregular surfaces were also obtained after the degradation assays conducted in enzymatic media, as surface erosion could be observed in each micrograph and was not observed on the matrices submitted to degradation under neutral conditions (PBS1.A and PBS2.A). According to the SEM analyses, it can be noticed that the surfaces of the enzyme-treated PBS samples (C and D) were degraded more intensively than observed previously with the chemical treatment (B).
Figure 14 shows the HPLC chromatograms of the supernatants obtained after the enzymatic degradation experiments. It can be observed that these chromatograms are very different from the ones obtained after the chemical degradation experiments. Particularly, the much larger number of peaks in Fig. 14 suggests that the mechanisms of chemical and enzymatic degradations are different and that the oligomeric species released by the enzymatic degradation are larger. Based on the quantification of the released SA, 12% of SA was detected for PBS1_lipase, 12% for PBS1_cutinase, 7% for PBS2_lipase, and 28% for PBS2_cutinase. Thus, it seems evident that the enzymes can play important roles in the PBS degradation process, making the degradation process faster even under milder reaction conditions. The obtained results encourage the deeper study of the enzymatic degradation process as a potential polymer degradation platform based on circular economy principles.
Conclusions
The chemical and enzymatic degradations of two grades of poly(butylene succinate) (PBS), presenting different morphological aspects and average molar masses, were successfully conducted. The PBS grade that presented lower average molar masses and average diameters (PBS1 microparticles) provided higher rates of chemical degradation, which increased with temperature (from 40 to 80 °C), especially when placed in alkaline conditions (pH = 10). The distinct reaction conditions did not affect the rates of degradation of the PBS grade that presented higher average molar masses and diameters (PBS2 pellets). Based on the obtained results, it was conjectured that the degradation mechanism is probably related to cleavage of short molecular segments placed at vicinities of the chain ends, explaining why the weight losses were not accompanied by a significant reduction of average molar masses and why the particle morphology affected degradation, as chain cleavage is expected to occur when the chain ends are placed near the particle surfaces and in close contact with the aggressive aqueous medium. These conjectures were confirmed by independent HPLC characterizations, which detected significant amounts of short chains in the supernatants of the aqueous solutions.
Additionally, it was shown that the degradability of the polymer matrices was much more intense when enzymes were added to the reaction media. For PBS2, the cutinase and lipase enzymes significantly affected the polymer degradation, leading to weight losses of 100% and 81% after 4 weeks of experimentation, respectively. For PBS1, cutinase, lipase, and amylase were the most effective enzymes and caused weight losses of 57%, 32%, and 25% after 4 weeks of experimentation, respectively. Particularly, the much larger number of peaks detected in HPLC characterizations suggested that the mechanisms of chemical and enzymatic degradations are different and that the oligomeric species released by the enzymatic degradation were larger.
Finally, it can be concluded that the application of enzymes for the degradation of PBS under mild process conditions clearly encourages the use of the enzymatic route as a promising environmental-friendly pathway for controlled PBS degradation and monomer recycling.
References
Aasberg-Petersen K, Christensen TS, Dybkjaer I, Sehested J, Østberg M, Coertzen RM, Keyser MJ, Steynberg AP (2004). In: Steynberg A, Dry M (eds) Studies in surface science and catalysis. Elsevier, Amsterdam
Gomes FW, Lima RC, Piombini CR, Sinfitele JF Jr, Souza FG Jr, Coutinho PLA, Pinto JC (2018) Comparative analyses of poly(ethylene 2,5-furandicarboxylate) − PEF − and poly(ethylene terephthalate) − PET − resins and production processes. Macromol Symp 381(1):1800129. https://doi.org/10.1002/masy.201800129
Kaushal J, Khatri M, Arya SK (2021) Recent insight into enzymatic degradation of plastics prevalent in the environment: a mini—review. Cleaner Eng Technol 2:100083. https://doi.org/10.1016/j.clet.2021.100083
Lomelí-Rodríguez M, Corpas-Martínez JR, Willis S, Mulholland R, Lopez-Sanchez JA (2018) Synthesis and characterization of renewable polyester coil coatings from biomass-derived isosorbide, FDCA, 1,5-pentanediol, succinic acid, and 1,3-propanediol. Polymers 10:600. https://doi.org/10.3390/polym10060600
Nakajima H, Dijkstra P, Loos K (2017) The recent developments in biobased polymers toward general and engineering applications: polymers that are upgraded from biodegradable polymers, analogous to petroleum-derived polymers, and newly developed. Polymers 9:523. https://doi.org/10.3390/polym9100523
Othman MH (2020). In: Hashmi S, Choudhury IA (eds) Encyclopedia of renewable and sustainable materials. Elsevier, Oxford
Pellis A, Malinconico M, Guarneri A, Gardossi L (2021) Renewable polymers and plastics: performance beyond the green. New Biotechnol 60:146–158. https://doi.org/10.1016/j.nbt.2020.10.003
Zhu Y, Romain C, Williams CK (2016) Sustainable polymers from renewable resources. Nature 540:354. https://doi.org/10.1038/nature21001
Bagheri E, Mosaddegh P, Behzad T (2020) Mechanical, thermal, and structural properties of uniaxially drawn polylactic acid/halloysite nanocomposites. J Appl Polym Sci 137:49134. https://doi.org/10.1002/app.49134
Grand View Research (2021) Bioplastics market size, share & trends analysis report by product (biodegradable, non-biodegradable), by application (packaging, automotive & transportation, textile), by region, and segment forecasts, 2021–2028. https://www.grandviewresearch.com/industry-analysis/bioplastics-industry. Accessed 27 Oct 2021
Song J, Zhou H, Wang X, Zhang Y, Mi J (2019) Role of chain extension in the rheological properties, crystallization behaviors, and microcellular foaming performances of poly(butylene adipate-co-terephthalate). J Appl Polym Sci 136:47322. https://doi.org/10.1002/app.47322
Tábi T, Wacha AF, Hajba S (2019) Effect of d-lactide content of annealed poly(lactic acid) on its thermal, mechanical, heat deflection temperature, and creep properties. J Appl Polym Sci 136:47103. https://doi.org/10.1002/app.47103
Verbeek CJR, Smith MJ, Cozens WC (2019) Rheology and sheet extrusion of novatein thermoplastic protein/poly(butylene adipate-co-terephthalate) blends. J Appl Polym Sci 136:47977. https://doi.org/10.1002/app.47977
Dutra LS, Costa TSB, Lobo VTV, Paiva TF, Nele M, Pinto JC (2019) Preparation of polymer microparticles through non-aqueous suspension polycondensations: part III—degradation of PBS microparticles in different aqueous environments. J Polym Environ 27:176–188. https://doi.org/10.1007/s10924-018-1329-x
Jia Z, Wang J, Sun L, Zhu J, Liu X (2018) Fully bio-based polyesters derived from 2,5-furandicarboxylic acid (2,5-FDCA) and dodecanedioic acid (DDCA): from semicrystalline thermoplastic to amorphous elastomer. J Appl Polym Sci 135:46076. https://doi.org/10.1002/app.46076
Nakayama A, Yamano N, Kawasaki N (2019) Biodegradation in seawater of aliphatic polyesters. Polym Degrad Stabil 166:290–299. https://doi.org/10.1016/j.polymdegradstab.2019.06.006
Noordzij GJ, Roy M, Bos N, Reinartz V, Wilsens CHRM (2019) Improving the hydrolysis rate of the renewable poly(hexamethylene sebacate) through copolymerization of a bis(pyrrolidone)-based dicarboxylic acid. Polymers 11:1654. https://doi.org/10.3390/polym11101654
Rivas MV, Petroselli G, Erra-Balsells R, Varela O, Kolender AA (2019) Synthesis, characterization and chemical degradation of poly(ester-triazole)s derived from d-galactose. RSC Adv 9:9860–9869. https://doi.org/10.1039/C9RA00398C
Tokiwa Y, Calabia BP (2007) Biodegradability and biodegradation of polyesters. J Polym Environ 15:259–267. https://doi.org/10.1007/s10924-007-0066-3
Kundys A, Białecka-Florjańczyk E, Fabiszewska A, Małajowicz J (2018) Candida antarctica lipase B as catalyst for cyclic esters synthesis, their polymerization and degradation of aliphatic polyesters. J Polym Environ 26:396–407. https://doi.org/10.1007/s10924-017-0945-1
Ferone M, Raganati F, Olivieri G, Marzocchella A (2019) Bioreactors for succinic acid production processes. Crit Rev Biotechnol 39:571–586. https://doi.org/10.1080/07388551.2019.1592105
Xu J, Guo B-H (2010) Poly(butylene succinate) and its copolymers: research, development and industrialization. Biotechnol J 5:1149–1163. https://doi.org/10.1002/biot.201000136
Nothling MD, Xiao Z, Bhaskaran A, Blyth MT, Bennett CW, Coote ML, Connal LA (2019) Synthetic catalysts inspired by hydrolytic enzymes. ACS Catal 9:168–187. https://doi.org/10.1021/acscatal.8b03326
Ahn BD, Kim SH, Kim YH, Yang JS (2001) Synthesis and characterization of the biodegradable copolymers from succinic acid and adipic acid with 1,4-butanediol. J Appl Polym Sci 82:2808–2826. https://doi.org/10.1002/app.2135
Tsutsumi C, Hayase N, Nakagawa K, Tanaka S, Miyahara Y (2003) The enzymatic degradation of commercial biodegradable polymers by some lipases and chemical degradation of them. Macromol Symp 197:431–442. https://doi.org/10.1002/masy.200350737
Bai Z, Shi K, Su T, Wang Z (2018) Correlation between the chemical structure and enzymatic hydrolysis of poly(butylene succinate), poly(butylene adipate), and poly(butylene suberate). Polym Degrad Stabil 158:111–118. https://doi.org/10.1016/j.polymdegradstab.2018.10.024
Qi J, Wu J, Chen J, Wang H (2019) An investigation of the thermal and (bio)degradability of PBS copolyesters based on isosorbide. Polym Degrad Stabil 160:229–241. https://doi.org/10.1016/j.polymdegradstab.2018.12.031
Shi K, Su T, Wang Z (2019) Comparison of poly(butylene succinate) biodegradation by fusarium solani cutinase and candida antarctica lipase. Polym Degrad Stabil 164:55–60. https://doi.org/10.1016/j.polymdegradstab.2019.04.005
Dutra L, Nele M, Pinto JC (2018) A novel approach for the preparation of poly(butylene succinate) microparticles. Macromol Symp 381:1800118. https://doi.org/10.1002/masy.201800118
ANVISA - Agência Nacional de Vigilância Sanitária (2004) Brazil. https://bvsms.saude.gov.br/bvs/saudelegis/anvisa/2005/rdc0270_22_09_2005.html. Accessed 28 Mar 2020
Bunge (2012) Soya. http://www.soya.com.br/produtos/oleos/oleo-de-soja-soya. Accessed 28 Mar 2020
Poussard L, Mecheri A, Mariage J, Barakat I, Bonnaud L, Raquez JM, Dubois P (2014) Synthesis of oligo(butylene succinate)-based polyurethanes. J Renew Mater 2:13–22. https://doi.org/10.7569/JRM.2013.634132
Dutra LS, Nele M, Pinto JC (2021) Preparation of polymer microparticles through non-aqueous suspension polycondensations: part IV—effect of the continuous phase on the characteristics of final poly(butylene succinate) particles. J Polym Environ 29:219–229. https://doi.org/10.1007/s10924-020-01869-7
Dutra LS (2019) Desenvolvimento de um Processo de Policondensação em Suspensão para Produção de Micropartículas Biodegradáveis. PhD thesis in Portuguese- Graduated Program in Engineering of Chemical and Biochemical Processes, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil
Antheunis H, van der Meer J-C, de Geus M, Kingma W, Koning CE (2009) Improved mathematical model for the hydrolytic degradation of aliphatic polyesters. Macromolecules 42:2462–2471. https://doi.org/10.1021/ma802222m
Solomons TWG, Fryhle CB (2007) Organic chemistry, 9th edn. John Wiley and Sons, Hoboken, NJ
Lucas N, Bienaime C, Belloy C, Queneudec M, Silvestre F, Nava-Saucedo J-E (2008) Polymer biodegradation: mechanisms and estimation techniques – a review. Chemosphere 73:429–442. https://doi.org/10.1016/j.chemosphere.2008.06.064
Cho K, Lee J, Kwon K (2001) Hydrolytic degradation behavior of poly(butylene succinate)s with different crystalline morphologies. J Appl Polym Sci 79:1025–1033. https://doi.org/10.1002/1097-4628(20010207)79:6%3c1025::AID-APP50%3e3.0.CO;2-7
Acknowledgements
Authors thank CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) and FAPERJ (Fundação Carlos Chagas Filho de Apoio à Pesquisa do Estado do Rio de Janeiro) for scholarships and financial support.
Funding
This study were funded by Conselho Nacional de Desenvolvimento Científico e Tecnológico, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior and Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Dutra, L., Pinto, M.C.C., Lima, R.C. et al. Preparation of Polymer Microparticles Through Non-aqueous Suspension Polycondensations: Part VI—Analyses of Chemical and Enzymatic Degradation of Poly(Butylene Succinate) (PBS). J Polym Environ 30, 1893–1907 (2022). https://doi.org/10.1007/s10924-021-02313-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10924-021-02313-0