Abstract
Poly(dl-lactic acid) or PLA is a biodegradable polymer. It has received much attention since it plays an important role in resolving the global warming problem. The protease produced by Actinomadura keratinilytica strain T16-1 was previously reported as having PLA depolymerase potential and being applicable to PLA biodegradation, which was used in this work. Therefore, this research demonstrates the important basic knowledge on the biological degradation process by the crude PLA-degrading enzyme from strain T16-1. Its re-polymerization was evaluated. The optimization of PLA degradation by statistical methods based on central composite design was determined. Approximately 6700 mg/l PLA powder was degraded by the crude enzyme under optimized conditions: an initial enzyme activity of 200 U/ml, incubated at 60 °C for 24 h released 6843 mg/l lactic acid with 82% conversion, which was similar to the commercial enzyme proteinase K (81%). The degradable products were re-polymerized repeatedly by using commercial lipase as a catalyst under a nitrogen atmosphere for 6 h. A PLA oligomer was achieved with a molecular weight of 378 Da (n = 5). This is the first report to demonstrate the high efficiency of the enzyme to degrade 100% of PLA powder and to show the biological recycling process of PLA, which is promising for the treatment and utilization of biodegradable plastic wastes in the future.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Poly(dl-lactic acid) (PLA) is a biodegradable polymer synthesized from lactic acid that is produced by microorganisms from cheap agricultural products. PLA has received much attention since it plays an important role in resolving the global warming problem. Since 1997, many researchers have isolated and identified PLA-degrading microorganisms. Several PLA-degrading microorganisms have been reported such as thermophilic bacteria, actinomycetes and fungi [1–6]. Furthermore, PLA-degrading actinomycetes were found to be distributed into various families such as Pseudonocardiaceae, Thermomonosporaceae, Micromonosporaceae, Streptosporangiaceae, Bacillaceae and Thermoactinomycetaceae [1–6]. Interestingly, our group previously described an actinomycetes, Actinomadura keratinilytica strain T16-1, that demonstrated a high potential to produce PLA-degrading enzymes [5, 7]. The maximum enzyme activity was obtained with 257 U/ml in a basal medium containing PLA under batch fermentation in a 3L airlift fermenter [8]. Therefore, the enzyme used in this study was produced by A. keratinilytica strain T16-1 and was used for optimization of the PLA degradation assay. The enzyme produced by strain T16-1 was previously purified and characterized. The optimum pH and temperature for the purified enzyme were 10 and 70 °C, respectively [5]. However, the optimum conditions for PLA degradation by crude PLA-degrading enzymes from A. keratinilytica strain T16-1 have not yet been investigated.
Since 1981, Williams [9] reported the degradation of L-PLA using a commercial enzyme proteinase K from Tritrirachium album. Afterwards, almost all studies of enzymatic degradation of PLAs were performed by using an alkaline protease at the optimal pH range of 6–10 and optimal temperature of 55–70 °C [1, 3, 6, 10–12]. Thus, the comparison of PLA degradation between the commercial protease and the crude enzyme produced by strain T16-1 should be evaluated.
In general, PLA is polymerized commercially by chemical processes such as poly condensation from lactic acid and/or ring opening polymerization from lactide [13, 14]. However, these processes require extreme conditions, such as high temperature, high solvent concentrations and high loading of catalysts. Recently, biological polymerization methods showed an alternative process that requires mild conditions, and that utilizes the lipase enzyme for bio-catalytic reaction such as poly lactic acid synthesis [15], multifunctional oligoester resins polymerization [16], enzymatic ring-opening polymerization by reactive extrusion (REX) [17] as well as polymerization of ε-caprolactone [18].
Currently, polymers recycling is necessary for utilizing materials efficiently, and this practice can help to reduce the plastics waste that affects global warming. The chemical methods for PLA recycling, such as pyrolysis [19] and chemical hydrolysis [20], have previously been reported. However, the pyrolysis of PLLA-Ca also leads to formation of dl lactide at high temperatures of approximately 250 °C [19]. In 2002, Tsuji and Nakahara [20] used high temperatures (140 °C) and acidic conditions for PLA polymerization. Biological processes of both microbial and enzymatic activities are currently considered as sustainable recycling methods for polyester formation. The process of PLA biological recycling by the enzyme activity occurs under mild conditions, is regarded as a clean process, and does not lead to any undesirable byproducts such as racemic mixtures of PLA after degradation [8–12, 15–18]. Poly(lactic acid) such as poly(dl-lactic acid), poly(d-lactic acid) and poly(l-lactic acid) were degraded by lipases in an organic solvent to produce cyclic oligomers by several enzymes such as lipase RM (Lipozyme RM IM) and lipase CA (Novozym 435) at 60–100 °C [21]. This report suggested that cyclic oligomers might be suitable for re-polymerization and recycling of PLA. The lipase catalysed transformation will open a novel route for sustainable chemical recycling of polymers. Jarerat et al. [22] reported the biological degradation of PLA using the stereospecific protease enzyme produced by Amycolatopsis orientalis in mild conditions (relatively low temperature, 40 °C) and in a clean process (without organic solvents), without the formation of a mixture of d- and l-lactic acids, which generally occurs in conventional hydrolysis in water at a temperature over 300 °C. In 2008, Lassalle and Ferreira [23] demonstrated optimal conditions for biological re-polymerization of PLA by lipase in hexane solvents. PLA was obtained at 55% (w/v) under optimal conditions (temperature 65 °C, reaction time for 96 h). Recently, Chuensangjun et al. [15] investigated the optimal conditions for the polymerization of PLA and showed that the low molecular weight of PLA is derived from commercial lactic acid by the commercial lipase, Lipozyme TL IM, at 50 °C for 5 h incubation.
Interestingly, the PLA degradation conditions and the recycling process have not been investigated so far. We aim to describe the optimization of PLA degradation conditions by using PLA-degrading enzymes produced by A. keratinilytica strain T16-1 and to determine the degradation products derived from enzymatic hydrolysis. Afterward, recycling of biopolymers, such as PLA, which uses microbial enzymes, should be described in more detail, especially the processes of biodegradation, bio-recycling and re-polymerization to develop new technologies for reducing plastic waste in the future.
Methods
Bacterial Strain and Growth Medium
A. keratinilytica strain T16-1 was reported to produce the highest PLA-degrading activity at 45 °C. A loopful of strain T16-1 grown on a yeast-malt extract agar slant was inoculated into 50 ml of yeast extract-malt extract broth at 50 °C, incubated at 150 rpm for 4 days. The cells were harvested by filtration through filter paper (Whatman No. 1), washed twice, and re-suspended in distilled water. The cell suspension was then added to the fermentation medium with 10% inoculum. The fermentation medium consisted of (w/v) 0.035% DL-PLA powder (80% L-PLA and 20% D-PLA with Mw of 43,000, Toyobo, Japan), 0.24% gelatin, 0.4% (NH4)2SO4, 0.4% K2HPO4, 0.2% KH2PO4, and 0.02% MgSO4·7H2O. The flasks were incubated at 45 °C for 3 days. PLA-degrading enzyme was then collected by filtration through filter paper and centrifuged at 8000 g. The supernatant was used for determination of enzyme activity.
Degradation of PLA
The crude enzyme produced by A. keratinilytica strain T16-1 (with an initial specific activity of 5.7 U/mg protein in 1000 U/ml crude extract) was diluted in 0.1 mM Tris–HCl buffer pH 9.0 according to the variable enzyme activity. Three millilitres of diluted enzyme were added to a test tube containing 0.02 g of PLA powder (6700 mg/l). The tubes were incubated at various time points and at various temperatures as described in the experimental design (Table 1). Degradation products were retrieved after centrifugation at 10,000g for 5 min. The supernatant was used for determination of lactic acid concentration. The negative control experiments were conducted using the diluted enzyme without PLA powder. The effect of the initial pH on PLA degradation was assayed under optimal conditions. A total of 0.02 g of PLA powder was suspended in 0.1 mM Tris–HCl at various pH values (8.0, 9.0 and 10.0). Three millilitres of diluted crude enzyme with an activity of 200 U/ml was added into the tube and incubated at 60 °C. Samples from this mixture were taken every 6 h for 30 h. The pH value was determined by pH meter (Benchtop pH/MV Meter-860,031, USA). The enzymatic hydrolysis of PLA by the commercial enzyme proteinase K (Amresco LLC, USA) was investigated. The enzyme was diluted in 0.1 mM Tris–HCl buffer pH 9.0 to a final activity of 100 and 200 U/ml. Three millilitres of diluted enzyme were added to a test tube containing 0.02 g of PLA powder (6700 mg/l). The tubes were incubated at 37 or 60 °C for 24 h. Degradation products were received after centrifugation at 10,000g for 5 min. The supernatant was then used for determination of lactic acid concentration by HPLC.
Experimental Design for Optimization of PLA-Degradation Conditions
Three factors (temperature, time and enzyme activity) and five levels, including two- star point and factorial point values, were designed according to a central composite design (CCD). The star points represent new extreme values (low and high) for each factor in this design. The value of the α variable was calculated depending on the number of variables to define the rotatability of this model. In this study, considering that k = 3 factors (temperature, time and enzyme activity), the star point code (α) could be written as
Seventeen experimental runs were calculated by Eq. (2) when k = number of variables, and n0 = number of centre points. Tables 1 and 2 show the factors and their values and the experimental design, respectively. This design includes the lactic acid concentrations as response variable.
A quadratic regression model was achieved by Eq. (3) using the method of least squares as follows:
where Y is the predicted response (lactic acid concentration); X1, X2, and X3 are the coded forms of the input variables (temperature, time and enzyme activity, respectively); b0 is a constant; b1, b2, and b3 are the linear coefficients; b12, b13, and b23 are interactional coefficients; and b11, b22, and b33 are the quadratic coefficients. The data from the experimental design were subjected to multiple regression analysis for the parameter estimation of the statistical model.
Evaluation of Enzymatic Re-polymerization of PLA by the Enzyme Lipase
Polymerization experiments were performed using the condenser system. The PLA degraded products were dissolved in toluene at a concentration of 250 mg/ml and heated to 70 °C, and then 15% (w/v) of the immobilized enzyme (Lipozyme TL IM Novo Nordisk, Denmark) was added to the solution. The reaction mixture was then continuously stirred at 70 °C under a nitrogen atmosphere with a flow rate of 0.1 vvm for 6 h. After the reaction, the enzyme particles were removed by filtration, the PLA products were isolated by precipitation, and the re-polymerization of PLA was used to determine the structure and molecular weight of the polymer as show below.
Analytical Methods
The PLA-degrading enzyme activity was measured by the method previously reported by Sukkhum et al. [5]. A 0.1% (w/v) mixture of PLA was emulsified in 0.1 mM Tris–HCl buffer at pH 9.0 using an ultrasonic processor, and this mixture was used as a substrate for our experiments. The reaction mixture was incubated at 60 °C for 30 min. One unit of the enzyme activity was defined as a 1unit decrease in optical density at 630 nm under the assay conditions described. Protein concentrations were assayed by the Lowry method using bovine serum albumin as a standard. Lactic acid concentrations in the reaction mixture were analysed by the HPLC method (InertSustain C18 column, with a mobile phase of 10 mM NH4H2PO4 (pH 2.6), a flow rate of 1.0 ml/min, and a temperature of 40 °C). The percentage of PLA converted to liberate lactic acid (% conversion efficiency) was calculated according to the following equation.
The structure of re-polymerized PLA was measured by 1H Nuclear Magnetic Resonance (NMR) data analysis (Bruker Avance 300 FT-NMR spectrometer, operating at 300 MHz in CDCl3). The molecular weight of PLA was obtained by LC/MS (Thermo Finnigan LC-Q mass spectrometer).
Statistical Analysis
The data from the experimental designs were analyzed a second-order multiple regression analysis using least squares regression methodology to obtain the parameter estimators of the mathematical model. SPSS Statistics 20.0 and Statistica 5.0 software (Statsoft, USA) were used for regression analysis and graphical analysis of the data, respectively.
Results and Discussion
Optimization of PLA-Degradation Condition
In this study, three variables were selected for optimization of PLA-degradation conditions, namely temperature (X 1), time (X 2), and activity of the enzyme (X 3). The effects of these controlled variables were determined by the response surface methodology using a central composite design. The coded levels and the actual levels of these factors are shown in Table 1. The experimental design and the predicted and observed results according to the second order model achieved are shown in Table 2. The treatment numbers, 1–8, 9–14 and 15–17, were considered a full factorial point, a star point and a centre point, respectively. The number of repetitions at the centre point was used as an experimental control and shows the determination of standard error. Multiple linear regressions analysis was used for construction of the second order equation with the lactic acid concentration as the dependent variable in relation to the controlled variation of the three factors, regardless of the significance of the coefficients:
where Y is the predicted response (lactic acid concentration); X 1, X 2, X 3 are coded values of temperature, time and activity of the enzyme, respectively.
The statistical significance of Eq. (6) was tested with an F-test, and the results from analysis of variance (ANOVA) for the response surface quadratic model is shown in Table 3. The model was significant, p value (0.000). The results from analysis of variance (F-test) shows that the second order model obtained is well adjusted to the experimental data. The precision of the model can be determined by the termination coefficient (R 2) and the correlation coefficient (R). The response variations for PLA degradation were attributed to a level of 80.4% compared to the independent variables. The closer the value of R (correlation coefficient) is to 1, the better the correlation between the experimental and predicted values. Here, the value of R (0.896) for Eq. (6) indicates a satisfactory agreement between the experimental results and the theoretical values predicted by the model equation.
Student’s t-distribution and the corresponding p value, with the parameter estimates are given in Table 4. The p values are used as a tool to verify the significance of each of the coefficients, which, in turn, are necessary to understand the pattern of the mutual interactions between the best variables. The smaller the p values are, the larger the significance of the corresponding coefficient is. The quadric term of these three variables had a significant effect at the 95% confidence level. In this case, X 2 1 , X 2 2 and X 2 3 were significant model terms. In general, the velocity of the enzyme reaction is increased by various factors such as substrate concentration, amount of enzyme, temperature and time. As shown in Table 4, the negative regression coefficient of variables, X 1 , X 1 X 2 , X 1 X 3 , X 2 X 3 , X 2 1 , X 2 2 and X 2 3 in from Eq. (6) suggested a decrease in the lactic acid concentration with an increase in these variables, while a positive regression coefficient of variables X 2 and X 3 in from Eq. (6) indicated an increase in time and enzyme activity with an increase lactic acid concentration.
From the 3-dimension response surface plots and the corresponding contour plots, the optimal values of the independent variables could be observed. Figure 1 depicts the 3-dimensional plots and their corresponding contour plots. Figure 1a shows the effect of temperature and time on lactic acid concentration, while enzyme activity is fixed at its middle level (200 U/ml). Figure 1b demonstrates the combination of temperature and enzyme activity, and Fig. 1c shows the effect of enzyme activity and time. The optimal ranges of temperature, time and enzyme activity for lactic acid concentration were 58–62 °C, 23–26.6 h and 150–250 U/ml, respectively. The maximum predicted lactic concentration of 6970 mg/l was obtained. In this situation, the optimal conditions of temperature, time and enzyme activity in the un-coded unit were 60 °C, 24 h and 200 U/ml, respectively. These results suggested that the enzyme produced by strain T16-1 showed higher degrading activity at lower temperatures than previously reported (70 °C) [5]. The enzyme was expected to be stable and have sustained activity on degradation for a long time. In this case, the highest degradation product was obtained at temperature of 60 °C for 24 h. Moreover, the velocity of the enzyme may be decreased and constant, as described by Michaelis–Menten kinetics, by increasing the amount of enzyme to excess and limiting the substrate concentration. Afterwards, we investigated the statistically optimal values of the variables and carefully considered the major and minor axis of the response and contour plots at the centre point that yielded the maximum lactic acid concentration. A repeated degradation of PLA using a crude enzyme produced by A. keratinilytica strain T16-1 under optimal conditions was carried out for verification of the optimization procedure. The maximal lactic acid level that was obtained was 6843 mg/l (predicted as 6970 mg/l) at a temperature of 60 °C, for 24 h at an ezyme activity of 200 U/ml (Fig. 1). The percentage of PLA converted to lactic acid was 82%. Jarerat et al. [22] demonstrated the biological recycling of PLA using enzyme activity by a process that was conducted at low temperature, 40 °C and without organic solvents. However, the optimization conditions were not described. Lomthong et al. [24] reported biodegradation conditions of a 2% poly l-lactic acid thermoplastic starch (PLLA/TPS) blend (50:50) film using the enzyme produced by Laceyella sacchari LP175 with an initial pH of 9.0, a degradation time of 4 h and a 99.7% weight loss. Nevertheless, this is the first study of 100% PLA powder degradation with a high conversion efficiency using a crude enzyme produced by strain T16-1. In contrast, Piemonte and Gironi [25] reported chemical recycling of PLA at 160–180 °C with 95% hydrolysed PLA and showed that the temperature used in the chemical method was higher than in the actual biological process.
Response surface and contour plots described by the model, representing lactic acid concentration (mg/l) as a function of temperature (°C), time (h) and enzyme activity (U/ml). a The combined effects of temperature (°C) and time (h) with constant enzyme activity (U/ml) of 200. b The combined effects of temperature (°C) and enzyme activity (U/ml) with constant time of 24 h. c The combined effects of time (h) and enzyme activity (U/ml) with constant temperature of 60 °C
The effect of pH on the PLA degradation rate was determined to enhance the degradation efficiency by varying pH values of 8.0, 9.0 and 10.0. The degradation of 0.02 g of PLA powder suspended in 0.1 mM Tris–HCl at various pH values is shown in Fig. 2. At the beginning stages of PLA degradation at pH 9.0 and 10.0, the conversion efficiency rapidly increased during the first 6 h of reaction time while at an initial pH of 8.0, the efficiency was quite slow (Fig. 2a). Then, the enzyme degradation efficiency at all three pH values reached the optimal point. The highest conversion (82% at 24 h) was achieved at the initial pH of 9.0. These results mean that PLA-degrading enzymes produced by strain T16-1 showed high activity at a wide range of alkaline pH values [5]. Figure 2b depicts the pH profile of PLA degradation at various pH values. During PLA degradation, pH values were decreased with increasing conversion efficiency up to the maximal point used for enzyme degradation. In any event, the degradation ability of the crude PLA-degrading enzyme was unchanged and seemed to decline 24 h after incubation. The result indicated that raising the lactic acid concentration in parallel with decreasing the pH values to 5–6 inhibited the enzyme activity and stability. Product inhibition can be a form of a negative feedback which an important enzyme limiting factor. In this case, the acidic conditions after 24 h of degradation were not at the optimal pH for the enzyme activity which reduced the percentage of conversion efficiency at all pH values.
The Comparison of PLA Degradation Between the Crude Enzyme Produced by Strain T16-1 and Commercial Proteinase K
According to the optimal conditions for PLA degradation, the crude enzyme produced by strain T16-1 showed high (82%) conversion. Commercial proteinase K with enzyme activity of 100 and 200 U/ml was used to evaluate PLA degradation efficiency at 37 and 60 °C for 24 h. The maximum lactic acid concentration of 5590 and 6779 mg/l was observed by the enzyme activity at 100 and 200 U/ml, respectively (Table 5). The highest conversion efficiency of 81% was achieved when using commercial proteinase K at 200 U/ml at 60 °C for 24 h. The results indicated that the crude PLA-degrading enzyme produced by A. keratinilytica strain T16-1 exhibited a similar efficiency of PLA conversion to a commercial enzyme and could be used for eliminating PLA waste-related degradation in the future.
Evaluation of Enzymatic Re-polymerized of PLA by Lipase
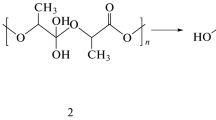
According to the optimized condition for PLA degradation, the degraded lactic acid dissolved in toluene can be used for the biological process of re-polymerization by commercial lipase. The reaction was loaded with immobilized lipase and further incubated at 70 °C under a nitrogen atmosphere with a flow rate of 0.1 vvm for 6 h. The structure of the re-polymerized product was obtained by 1H NMR data analysis. In the 1H NMR spectrum in CDCl3, the most intense signals were assigned to CH (δ 4.19 ppm, q, 2H, J = 6.8 Hz) and CH 3 (δ 1.35 ppm, d, 3H, J = 6.8 Hz) from the monomer lactic acid, while the signals with very much lower intensity could be attributed to a series of mixed lactic acid oligomers. Therefore, a set of resonances at δ 5.15 (q like, J ca 7.0 Hz, ca 0.09H, H a), 5.06 (q, J ca 7.1 Hz, ca 0.27H, H b), 4.28 (q, obscured with other oligomer signal, H c), 1.52 (d, J = 7.1 Hz, 0.31H, CH a 3 ), 1.47 (d, J = 7.1 Hz, 0.80H, CH b 3 ) and 1.40 (d, J = 7.0 Hz, obscured with other oligomer signal, CH c 3 ) was allowed for the proton assignments in the pentamer structure. The integration ratio of 2.6 for the two CH3 doublets at δ 1.47 and δ 1.52 in its 1H NMR data together with the observation of a pseudo molecular ion peak at m/z 413 [M + K]+in the ESIMS also suggested the number of the repeated lactic acid monomer of 5 with molecular weight of 378. The structure of the oligomers with molecular weights of 378 Da (n = 5) is shown in Fig. 3. Kondabagil et al. [26] reported that the maximum molecular weight of PLA using lipase-catalysed preparation of poly (lactic acid) was approximately 3300 Da using many solvent applications. Chuensangjun et al. [15] demonstrated that PLA could be obtained through a lipase-catalysed polymerization of commercial lactic acid using commercial lipase Lipozyme TL IM in a 5 h reaction time at 50 °C under a nitrogen atmosphere. An increase in the concentration of the immobilized enzyme in the reaction mixture increased the average molecular weight (Mn) of the PLA products. Products with molecular weights in the range of 2600–4500 Da were produced. From our results, the molecular mass of the oligomers was lower than in previous reports. This result might be due to the impurity of lactic acid derived from the degradation reaction (without the purification step) such as a residual protein or enzyme, some nutrient derived from the crude extract production step as well as a D/L enantiomer of lactic acid, while another report used pure lactic acid [27, 28]. In addition, the effect of various factors on the re-polymerization conditions should be investigated to obtain high Mw of PLA such as the type of enzyme, incubation time, temperature, organic solvent and flow rate of nitrogen gas [26]. However, these short chain oligomers of lactic acid were obtained after passing through the biological recycling process using microbial enzymes that can be used for many applications. Xiao et al. [29] and Jamshidian et al. [30] reported that the short oligomer of PLA can be applied for the synthesis of co-polymer and nano drug delivery devices.
Conclusions
This work demonstrates the important basic research on the biological degradation process of PLA by the crude enzyme PLA-degrading enzyme from strain T16-1 and its re-polymerization which is promising for the treatment and utilization of biodegradable plastic wastes in the future. Approximately 6700 mg/l PLA powder was degraded and released 6843 mg/l lactic acid with 82% conversion which was similar to the commercial enzyme proteinase K (81%). The degradable products were re-polymerized repeatedly by using commercial lipase as a catalyst. The oligomers derived from degradable products was achieved with molecular weight of 378 Da (n = 5).
References
Pranamuda H, Tokiwa Y, Tanaka H (1997) Appl Environ Microbiol 63:1637–1640
Tomita K, Kuroki Y, Nakai K (1999) J Biosci Bioeng 87:752–755
Tomita K, Nakajima T, Kikuchi Y, Miwa N (2004) Polym Degrad Stab 84:433–438
Tomita K, Tsuji H, Nakajima T, Kikuchi Y, Ikarashi K, Ikeda N (2003) Polym Degrad Stab 81:167–171
Sukkhum S, Tokuyama S, Tamura T, Kitpreechavanich V (2009) J Gen Appl Microbiol 55:459–467
Hanphakphoom S, Maneewong N, Sukkhum S, Tokuyama S, Kitpreechavanich V (2014) J Gen Appl Microbiol 60:13–22
Sukkhum S, Tokuyama S, Kitpreechavanich V (2009) Biotechnol Bioprocess Eng 14:302–306
Sukkhum S, Tokuyama S, Kitpreechavanich V (2012) J Microbiol Biotechnol 22(1):92–99
Williams DF (1981) Eng Med Biol Soc 10:5–7
Pranamuda H, Tsuchii A, Tokiwa Y (2001) Macromol Biosci 1:25–29
Jarerat A, Tokiwa Y, Tanaka H (2003) Biotechnol Lett 25:2035–2038
Jarerat A, Tokiwa Y (2001) Macromol Biosci 1:136–140
Cui Y, Li D, Gao B, Zhou Y, Chen L, Qiu B, Li Y, Duan Q, Hu N (2015) J Coord Chem 69(4):656–667
Dai Z, Cui Y, Chena C, Wu J (2016) Chem Commun 52:8826–8829
Chuensangjun C, Pechyen C, Chisti Y, Sirisansaneeyakul S (2012) Adv Mater Res 506:154–157
Semlitsch S, Torron S, Johansson M, Martinelle M (2016) Green Chem 18(7):1923–1929
Spinella S, Ganesh M, Re GL, Zhang S, Raquez JM, Dubois P, Gross RA (2015) Green Chem 17(8):4146–4150
He W, Fang Z, Zhu N, Ji D, Li Z, Guo K (2015) Biocatal Biotransformation 33(3):150–155
Fan Y, Nishida H, Hoshihara S, Shirai Y, Tokiwa Y, Endo T (2003) Polym Degrad Stab 79:547–562
Tsuji H, Nakahara K (2002) J Appl Polym Sci 86:186–194
Takahashi Y, Okajima S, Toshima K, Matsumura S (2004) Macromol Biosci 4:346–353
Jarerat A, Tokiwa Y, Tanaka H (2006) Appl Microbiol Biotechnol 72:726–731
Lassalle VL, Ferreira ML (2008) J Chem Technol Biotechnol 83(11):1493–1502
Lomthong T, Hanphakphoom S, Yoksan R, Kitpreechavanich V (2015) Int Biodeterior Biodegrad 104:401–410
Piemonte V, Gironi F (2013) J Polym Environ 21(2):313–318
Kondabagil R, Kiran D (2003) Soundar. World J Microbiol Biotechnol 19:859–865
Kumar S, Bhatnagar N, Ghosh AK (2016) Polym Bull 73:2087–2104
Kobayashi S (2010) Proc Jpn Acad Ser 86:338–365
Xiao RZX, Zhao WZ, Guang LZ, Jun JW, Fan ZL (2010) Int J Nanomed 5:1057–1065
Jamshidian M, Arab T, Imran E, Jacquot M, Desobry S (2010) Compr Rev Food Sci Food Saf 9:552–572
Acknowledgements
This work received financial support from the matching fund between Thailand Research Fund through and Srinakharinwirot University (Grant No. TRG5680026) and under the Core-to-Core Program, which was financially supported by Japan Society for the Promotion of Science (JSPS), National Research Council of Thailand (NRCT), Vietnam Ministry of Science and Technology (MOST), the National University of Laos, Beuth University of Applied Sciences and Brawijaya University as well as, Thailand Toray science foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Youngpreda, A., Panyachanakul, T., Kitpreechavanich, V. et al. Optimization of Poly(dl-Lactic Acid) Degradation and Evaluation of Biological Re-polymerization. J Polym Environ 25, 1131–1139 (2017). https://doi.org/10.1007/s10924-016-0885-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10924-016-0885-1