Abstract
In recent times, the use of computer aided diagnosis for detection of Glaucoma from fundus images has been prevalent. In the proposed work, a cloud based system is proposed for automatic and real-time screening of Glaucoma with the use of automatic image processing techniques. The proposed system offers scalability to the developers and easy accessibility to the consumers. The proposed system is device and location independent. The input digital image is analyzed and a comprehensive diagnostic report is generated consisting of detailed analysis of indicative medical parameters like optic-cup-to-disc ratio, optic neuro-retinal rim, ISNT rules making the report informative and clinically significant. With recent advances in the field of communication technologies, the internet facilities are available that make the proposed system an efficient and economical method for initial screening and offer preventive and diagnostic steps in early disease intervention and management. The proposed system can perform an initial screening test in an average time of 6 s on high resolution fundus images. The proposed system has been tested on a fundus database and an average sensitivity of 93.7% has been achieved for Glaucoma cases. In places where there is scarcity of trained ophthalmologists and lack of awareness of such diseases, the cloud based system can be used as an effective diagnostic assistive tool.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
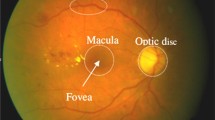
Eye is an important organ of the human body which allows vision. It is made up of 3 important layers: outer, middle and innermost layer. The outer layer consists of cornea and sclera which is responsible in controlling the amount of light entering the eye. The middle layer consists of iris, choroid and ciliary body. The innermost layer is called retina and consists mainly of retinal vessels and fovea and optic nerve head. The human eye is filled with some fluids, namely, aqueous and vitreous humor, which maintains a pressure inside the eye. This pressure inside the eye is called intra-ocular pressure (IOP). If IOP is not maintained, it may give rise to some aberrations in normal functioning of the tubular meshwork. The IOP may increase and create an inward force on the retina. This inward force pushes the optic nerve head and in turn leads in cupping of the nerve head. The more the cupping of the nerve head, the greater is the vision impairment. This condition is called Glaucoma and may need medical treatment. Glaucoma is an irreversible ocular disease which can be treated only if diagnosed at an early stage. If proper medical treatment is not given to the patient, then it may result into complete vision loss. Although, IOP is a valuable observation to examine the presence of Glaucoma, the appearance of the optic nerve head and the structural changes that develops during the progression of the disease, plays an important role in validating the Glaucoma presence [1]. The cup-to-disc ratio and ISNT rule has been used as two globally accepted clinical parameters used by ophthalmologists across the globe for Glaucoma diagnosis. These two parameters are used in the proposed method for Glaucoma detection in a fundus image using computer vision. A variation in the value of the CDR can be a detrimental factor to grade the severity of the disease progression [1].
In developing countries, there is a scarcity of trained ophthalmologists and lack of awareness regarding such diseases. So, there is a need to abridge this gap of trained ophthalmologists. Computer aided diagnosis (CAD) of diseases comes as a viable solution in scenarios where the ratio of doctors to patients is less. CAD is an efficient way to screen population for diseases by incorporating the medical feedback from the doctors in form of a computer program. The program is embedded into a computing hardware architecture and the combined embedded system may perform the real-time diagnosis automatically and generate a diagnosis report for the same which can be correlated with a medical practitioner.
The present work proposes a cloud computing based web application which can be used for screening patient with Glaucoma. The proposed system can be used as an assisting tool by primary health care centers in performing a screening test for population residing in rural areas. Also, such applications can be used at various eye hospitals, clinics and by non-profit organizations (NGOs) for upliftment of medicinal facilities in rural areas of countries.
Some scientists have transformed their work into web applications or embedded systems through which a CAD for diseases can be performed and the society can benefit. Vilppu et al. [2] have developed a web application for quantitative image analysis of estrogen receptor which can be used for diagnosis of breast cancer. M. Ortega et al. [3] have developed a web-based screening system for retinal image analysis. Michael T. Krieger et al. [4] describe scalable open source cloud stack covering IaaS, PaaS and SaaS. Piotr Nowakowski et al. [5] describe a cloud computing platform for Virtual Physiological Human research community while discussing issues involved in porting existing desktop applications to cloud environment. Arnon Rosenthal et al. [6] examine how BMI community could take advantage of cloud computing for data sharing and application development. Kashish A. Shakil et al. [7] propose a model for efficient storage, biometric authentication and retrieval of e-healthcare data. Raul Alonso-Calvo et al. [8] propose a cloud computing service which provides load balancer for processing large image datasets. Khondaker Abdullah Al Mamun et al. [9] propose a cloud based framework for supporting Parkinson patients in a low resource setting. Pankaj Deep Kaur et al. [10] propose a cloud based intelligent system which gather user specific health data and store it in cloud based storage repositories for analysis. F. Yin et al. [11] propose a cloud-based system for automatic glaucoma screening using medical image-based patter classification technologies. Raw data including patient’s medical condition fundus image etc. are distributed through the public cloud tier. Glaucoma is a painless disease and can be a cause of irreversible blindness if not diagnosed at early stage. So, there arises a need for some system which can perform a screening test and generate a diagnostic report which can be clinically correlated with an ophthalmologist. The proposed work addresses this issue and proposes a cloud computing based application for Glaucoma screening. This application is economical, automated, real-time and easy-to-use.
Some work using image processing techniques has been proposed by researchers in the field of Glaucoma detection using fundus images. J Cheng et al. [12] proposed the use of sparse dissimilarity-constrained coding approach for optic disc segmentation. The reconstruction coefficients from SDC were used for cup-to-disc ratio calculation to determine if a fundus images is affected by Glaucoma. J. Cheng et al. [13] proposes the use of histograms and center surround statistics for segmentation of optic disc and cup from fundus images. The final decision regarding Glaucoma is made on the basis of segmented optic disc and cup. T. Khalil et al. [14] proposed a method using hybrid structural and textural features. The features extracted are fed to a support vector machine (SVM) based classification module to determine the symptoms of Glaucoma. G D Joshi et al. [15] has proposed a method for optic disc segmentation using local parameters in multi-dimensional space. The optic cup is segmented using bends in the blood vessels. The finally segmented disc and cup are used to perform a Glaucoma analysis. G D Joshi et al. [16] proposed a depth-discontinuity approach for estimation of cup boundary pixels in a fundus image. Best-fitting circles have been used for final boundary estimation and hence used for CDR detection for Glaucoma analysis. Although, a lot of work has been done in this area, yet there arises a need for an algorithm that can correctly segment the optic disc and analyze the structural changes in it caused due to progression of the disease. The present work proposes a cloud based system for Glaucoma detection using fundus images.
-
a)
The main contribution of the proposed work is the segmentation of the optic disc from a variety of non-uniformly illuminated images. The use of a statistical and geometrical framework provides robustness to the algorithm by correctly locating and segmenting the disc from the fundus images. Many challenges are faced by a computer vision scientist while performing a CAD for Glaucoma from fundus images. These challenges include presence of exudates, choroid vessels and reflections at the periphery of fundus images. These objects are present in images and can be like optic disc in intensity and colour. However, they may differ with respect to shape and size. Hence, the use of geometrical framework provides robustness by differentiating between objects on basis of geometrical properties like solidity, eccentricity, circularity and major-to-minor axis ratio. The statistical framework makes use of adaptive thresholds determined from local features and helps in making the algorithm image independent.
-
b)
Another contribution of the proposed work lies in the use of cloud computing which makes the proposed system location and device independent. With the recent advances made in the field of communication, the proposed cloud based system can be accessed with the help of web connection from anywhere and at any time. The proposed system can be used as an assisting tool for ophthalmologists and primary care centers to screen patients for Glaucoma. The use of a cloud computing environment makes the system scalable and easy to configure for any hardware or software upgradations in future.
-
c)
One more important contribution of the proposed work is the use of clinically significant parameters in detection of Glaucoma from fundus images. Cup-to-disc ratio and structural changes observed in the disc during the disease progression are some clinical parameters which are considered by ophthalmologists across the globe to identify the symptoms for Glaucoma. The use of image processing techniques to determine such important and detrimental features from the fundus images makes the proposed system clinically significant and provides strength to the system to be used for real-time eye check-ups of patients suffering from ocular disorders.
The remaining paper is structured as follows: Section 2 discusses various technologies which have been used in designing the architecture of the proposed system for Glaucoma detection from fundus images. Section 3 discusses the methodologies used for linking different technologies used in the proposed work. Section 4 discusses the web application, both front and backend. Image processing techniques involved to segment objects of interest from fundus images and use them to diagnose Glaucoma are covered under this section. Section 5 discusses the experimental results obtained while testing the developed algorithms on the fundus database available. Section 6 draws a conclusion to the work.
Methods
Technologies used
Docker and components
Docker is an open platform which is used to develop and run applications. It can be used to interlink applications which are developed on hybrid platforms. The use of docker in the proposed work is to design a processing environment which can diagnose a fundus image for Glaucoma detection using image processing techniques. This is achieved by creating a docker container and installing various technologies and libraries like OpenCV, Python, Matplotlib, Numpy, Django on the Heroku server. These applications are installed and can be used on different machines with similar host OS. The architecture of a docker is explained in the later section.
Docker registry
It is a stateless, highly scalable server-side application that stores and facilitates distribution of docker images, controls the image distribution pipeline and is an open source application operating under Apache license. It consists of image repositories which can be managed with high customizability. Docker provides a zero maintenance Docker Registry called Docker Hub where all the images, officially supported Docker images (such as python images, ubuntu images, etc) as well as 3rd party images, are stored. In the proposed work, a diagnostic tool has been proposed for identification of eye disease like Glaucoma from fundus images. The computations and processing involved in the proposed work are based on imaging and mathematical techniques. The programming environment used in proposed work has been achieved using open-source Python language and OpenCV libraries. An automatic-build image has been created based on the official Python image along with some supporting libraries installed, like OpenCV, Matplotlib and Numpy, Pillow and Virtualenv. The Docker Image build for the proposed work is completely open-source and is available for pull requests publicly (https://hub.docker.com/r/hulkinbrain/docker-opencv2/).
Docker engine
It is a client-server application which is responsible for fetching images from Docker Registry. The programming environment image created in previous section consists of programming tools and libraries which are capable of detecting Glaucoma from fundus images. Before a user uploads the fundus image for testing, an application and system responsible for performing the assigned task must get live. This task is achieved with the help of Docker engine. The container consisting of Python, OpenCV and depending libraries is executed and brought running and live to actually develop the algorithms for segmentation of objects from fundus images and perform diagnostic test.
-
Docker Daemon: A server which is a type of long-running program. A docker daemon is the main component of a docker engine which communicates with the kernel regarding the creation, operation and management of containers. The container contains images of Python, OpenCV and supporting libraries like numpy, matplotlib, etc.
-
Docker Client: The client represents a command line interface which is used by programmers/developers to run docker commands. Once the images of libraries are stored in the containers, the developer needs to install them, and this is achieved by giving some instructions to the kernel. These instructions are coded on client program and the execution of these instructions takes place using REST API.
-
REST API: It specifies an interface that programs can use to talk to daemon and instruct it what to do. It acts as an interface between the client and engine and transmits all commands from clients to engine.
Figure 1 shows a graphical representation of a docker engine specifying important components and management of various protocols and data.
Docker architecture
The CLI uses the Docker REST API to control or interact with the Docker daemon through scripting or direct CLI commands. Many other Docker applications use the underlying API and CLI. Figure 2 shows the docker architecture and describes the interlinking of various executable files with the architecture.
-
Docker daemon (dockerd): Manages Docker objects such as images, containers, networks, and volumes, listens for REST API requests and can also communicate with other daemons to manage Docker services.
-
Docker client: Primary way through which users interact with Docker components. All commands executed by users in the Docker client are relayed to daemon which carries out the execution. Commands like docker run are sent to daemon for execution.
-
Docker registry: Docker registry is a server which stores all Docker images. Whenever commands like docker push and docker run are run, configured docker images are fetched, pulled or pushed to the registry. By default, docker hub is used as the registry service. The python based OpenCV image created for this paper is also stored in docker hub.
-
Docker objects:
-
Images: These are read-only templates containing instructions for creating a Docker container. These images can be based on other images or can be made from scratch by creating a Dockerfile which is nothing a file containing bash scripts with instructions for installing libraries. After each command, a layer is created so, only those layers are rebuilt/added/deleted which have been changed. This makes Docker very lightweight, small and fast since whole image is not being rebuilt when update is made to the Dockerfile. In this paper, an automated-build Docker image is built. Dockerfile contains necessary scripts to install OpenCV, Matplotlib, Numpy which get executed when docker run command is executed in the cmd window or a bash shell in order to create a docker image automatically.
-
Containers: Container is a runnable instance of an image. A container can be created, ran, stopped, moved or deleted using Docker API or CLI. By default, a container is isolated from other containers and the host machine, but they can be connected to one another and deployed on the same host machine. For this paper, only one container was created since all the libraries were bundled in a single image.
Methodologies
Building automated docker container image
The proposed work deals with the detection of Glaucoma from fundus images using image processing techniques. The web-app and the image processing algorithm requires OpenCV, Numpy, Matplotlib, Python and Django to work on the cloud. So, a container consisting of all the above-mentioned libraries is created using Dockerfile. The Dockerfile contains bash script commands which install all the libraries on a Linux machine. The Dockerfile is stored on GitHub and publicly available (https://hub.docker.com/r/hulkinbrain/docker-opencv2/). Figure 3 shows the container structure for the proposed web-app.
Bash commands for installing OpenCV on Linux, available on the official OpenCV website, were written into the Dockerfile. Numpy, Django and other libraries were installed by using requirements.txt along with pip.
Using Heroku container registry
Heroku Container Registry allows the user to deploy Docker based app to Heroku. Heroku runs a container registry on registry.heroku.com . Traditionally Heroku runs on an Ubuntu based stack, but with Container Registry, any stack can be deployed to Heroku using Docker. Since Heroku dynos are basically virtual machines running Ubuntu and since Docker operates above the OS level, it enables users to install all the dependencies using Dockerfile onto the dynos. After logging into the container registry service from Heroku CLI, running heroku container: push web in the project’s root folder registers the Docker container name after Heroku app name in the Heroku Container Registry, runs the Dockerfile to build the container image and then finally deploys the app online. Figure 4 shows the stack of Docker running on a Heroku dyno.
Django web application
A basic block diagram of the proposed system is shown in Fig. 5. A patient with vision problem visits an eye clinic, hospital or primary care center. A concerned person attends the patient and captures the fundus image using a fundus camera. This fundus image is fed to the proposed system using a web browser which can be accessed on any device with net connected facility. The attendant enters the URL in the browser and it directs the user to the webpage consisting the diagnostic tool (http://glaucoma.herokuapp.com/tool). Some details like name and age are entered on the webpage which are later used for report generation. The fundus image is uploaded, and some automatic cloud based diagnostics is performed and finally a report is generated regarding the pathologies and structural changes observed in the image which can be correlated with the disease progression.
Django is a python based web framework, this facilitates variable passing from a python based image processing script to a Django web-app conveniently. Following figure shows the process flow which happens in a Django web-app:
Django is a Model Template View (MTV) web framework, but the user uploaded images are not stored on the server so, Model is not used since its purpose is to validate and store data. User uploaded fundus image of JPEG extension (only JPEG images are supported right now) is converted to a base64 string which is passed to the fundus image processing script. The base64 string is then converted to a Numpy array format so that it can be processed using OpenCV. To run the web-app, a server must be started which is mapped to a port (for ex 8080). This is demonstrated in Fig. 6.
Additionally, the web-app is fully responsive i.e. it scales perfectly according to the device’s viewport size. Figure 7 shows the web-app display on devices with different screen sizes.
Upload fundus image
Base64 encoding
The proposed method uses Base64 encoding while uploading the fundus image for processing. It is used to convert any binary data file to a Base64 string. During HTML5 image upload, image’s base64 string can be accessed directly and then be directly displayed in the browser since HTML5 offers out of the box support for viewing base64 encoded images after appending a header to it. In the proposed work, only jpg images are supported. Jpg images are stored in hex format and then get automatically converted to base64 while being uploaded using HTML5 image upload. In other words, the jpg image stored in hex format is converted to a base64 string which finally is converted to a data URI. This URI can be used as a src attribute value of an <img > tag to be displayed directly in the browser. Base64 encoded image string ensure that the image is not in a readable format providing a basic format of encryption. Also, since the character set being used in this is used everywhere else, there is surety factor that the data will reach the server without being corrupted. Although there is a minor caveat, Base64 encoded string has a 33.3% overhead, i.e. the string’s size would be larger than the size of the binary data file.
Given an input of n bytes, the output will be \( \frac{4n}{3} \) bytes long, including padding characters. The padding character is not needed for decoding, since the number of missing bytes can be calculated from the number of Base64 digits. In the above example, n = 3 since the first 3 bytes of the hexadecimal values are considered, are encoded to 4 bytes of base64 characters. But since in the proposed work, user’s privacy concern was of utmost priority, Base64 encoding of images provided as a viable method to transfer image from user end to the server without storing it on the server-end.
Base64 encoding acts as a bridge between the gap of two different technologies used in present work. The browser and python script (after converting base64 encoded image to Numpy image) can parse the image for display (and processing in the case of python).
Workflow between view, fundus processing script (FPS) and template
As mentioned above, View processes the data which is sent to the server when user tries to access an URL. Since the View is a python file, other scripts can be imported into it. The Fundus processing script used in this paper is imported into Views.py for fundus image processing. The fundus image, which is in base64 format, gets converted into Numpy array format. Many resultant images, consisting of segmented objects having clinical relevance, are generated by the FPS script and sent back to Views.py. All the images are converted back to base64 format and are packed into a JSON object. These base64 image strings are then parsed in the template and set as src attribute for appropriate <img > tags. Figures 8 and 9 shows the variable passing between views.py, FPS.py and template:
Image processing techniques
Once the image in form of a Numpy array is sent to the server for processing, a script is executed using to process the fundus image on the web interface. This step involves execution of various image processing methods which involves the following core steps:
-
a.
Optic disc segmentation
-
b.
Blood vessel segmentation and inpainting
-
c.
Optic cup segmentation
-
d.
Feature determination
-
e.
Decision making
The entire process involved in determining if an image shows signs of Glaucoma or is unaffected, is represented graphically in Fig. 10 below.
Optic disc segmentation
The fundus images are captured as colour images in RGB format. The red channel provides highest contrast between the optic disc and boundary pixels [17, 18]. Hence, it has been chosen for optic disc segmentation. The procedure for candidate pixels which are segmented using the intensity based segmentation is summarized in Algorithm 1 below.

The candidate pixels consist of various noisy pixels which might occur due to presence of exudates, reflections, choroid vessels, non-uniform illumination (low or high contrast). The objects like reflections, exudates and choroid vessels are like optic disc in intensity and colour. However, these may be differentiated based on some geometrical features. These features include solidity, circularity, eccentricity, major-to-minor axis ratio and area. The process to differentiate between noisy and optic disc pixels is proposed in [19] and explained in Algorithm 2 below.

Blood vessel segmentation and inpainting and optic cup segmentation
Once the optic disc is segmented, next important object which plays a key role in Glaucoma detection is blood vessel. The blood vessels exit the eye through optic disc and hence, many vessels spread over it. During the segmentation of optic cup, there vessels cause hinderance and on basis of intensity, the segmented cup becomes fragmented. However, sometimes morphological operations may come to rescue in joining the fragmented portions, but the actual shape of segmented cup may change. So, in the proposed work, an imaging technique of blood vessel inpainting [20], has been used.
In the proposed work, the blood vessels are segmented using morphological operation, namely, bottom – hat transformation. In this transformation, a disk-shaped structuring element is used, and the luminance image is filtered using this filter. The size of the structuring element is chosen to be 25. This value is selected empirically. In the proposed work, the blood vessels on the optic disc are of interest for inpainting process, so the segmented vessels are multiplied with binary mask of optic disc. The blood vessels on the optic disc are inpainted using a statistical variance maximization approach [21, 22]. In this approach, the blood vessels are piece-wise linear and uniform intensity regions, so a window of specific size is created and a linear region with maximum variance is selected and the vessel pixels inside the window are replaced by the maximum variance value. The process of blood vessel segmentation and inpainting is discussed below in Algorithm 3:

The optic cup is determined from the blood vessel inpainted green channel image. It has been observed that the pixels belonging to the cup region are the pixels having high intensities in the green channel image. To segment the optic cup from the inpainted image, the image is subjected to an intensity based thresholding operation. Empirically, an intensity range greater than 70% of the difference between the maximum and minimum pixel intensity value has been considered as threshold for cup segmentation. Mathematically, the threshold can be represented as:
Where,
- Th2:
-
threshold for optic cup segmentation.
- Max:
-
maximum intensity of pixels in disc region of blood vessels inpainted green channel.
- Min:
-
minimum intensity of pixels in disc region of blood vessels inpainted green channel.
Feature determination
Once, the optic disc and cup are segmented, both these objects are utilized to determine the features to assess if an image shows signs of Glaucoma or is normal. 3 distinctive features are used in the proposed work which are listed below:
-
a.
Area cup-to-disc ratio
-
b.
Vertical cup-to-disc ratio
-
c.
Neuro-retinal rim in ISNT Quadrants
Area cup-to-disc ratio (ACDR)
This cup-to-disc ratio is the ratio of cup area to the disc area. The area in terms of image processing can be determined by finding the number of white pixels belonging to the region. Mathematically, this can be expressed as:
Vertical Cup-to-disc ratio (VCDR)
This is defined as the ratio of the vertical diameter of the cup to that of the disc. The cupping of the disc usually spreads along the vertical direction and this ratio can be very helpful in identifying the vertical expansion of cup inside the disc region. Mathematically, this can be expressed as:
Neuro-retinal rim in ISNT Quadrants (NRR)
The neuro-retinal rim can be obtained by removing the cup from the disc region. It represents the region between the cup and disc boundaries. This gives information regarding the structural changes in the disc. Using the center of the disc, the complete rim structure is divided into 4 quadrants, namely, inferior, superior, nasal and temporal. As per clinical terms, the rim must follow the ISNT rule to belong to normal class. The rule states that for an eye to be normal, the area of the rim must be largest in inferior, followed by superior, nasal and temporal quadrants. Mathematically, this value is determined using the following expression [18]:
Decision making
In the proposed work, the images are divided into 3 categories, namely, Glaucoma, Suspect Glaucoma and Normal. The criteria to decide the class, an image belongs to, is explained in Table 1.
Based on the features discussed previously, a decision algorithm is designed to evaluate the medical condition of a fundus image. The algorithm is represented in form of a pseudo code as follows:

Viewing diagnostic report
Figures 11 and 12 shows the results which are displayed by default after selecting “Process Image” button.
The web interface has an option to view a more comprehensive report. By selecting “View Full Report” button, more detailed results can be viewed which contains Name and age of the user (If supplied otherwise displays NaN), Date of image processing (system generated), Area CDR, Vertical CDR, ISNT NRR value and Result analysis in form of some intermediate results obtained during segmentation of important objects from fundus images. Green channel image of the input image, Segmented blood vessels, Inpainted Blood vessels inside the OD, images containing contours of the Segmented optic disc and optic cup, ISNT masks, ISNT NRR images and the total execution time of the algorithm are provided in results section which can be used by ophthalmologist while performing a correlation. Figure 6 shows the comprehensive report (https://github.com/hulkinBrain/docker-opencv2/blob/master/app/Dockerfile/).
Experimental results
The proposed algorithm has been tested on a database [23] consisting of a total of 364 fundus images, classified into 3 classes, namely, Glaucoma, Suspect Glaucoma and Normal. The images have been labelled by the ophthalmologists into the mentioned classes. The decision criteria used by ophthalmologist in labelling the images is same as mentioned in Section 4.3.5. Out of 364 images, 32 images are labelled as Glaucoma, 107 images as Suspect Glaucoma and 225 images as Normal. The images were captured using a high resolution digital fundus camera which produced images of a resolution of 1696*2544 pixel. The proposed algorithm is developed and implemented on a Heroku cloud server. The hardware configurations are mentioned in Table 2.
Table 3 displays the computation time elapsed by the proposed algorithm working on a server with mentioned specifications. The execution time of the fundus processing script (FPS) is mentioned in seconds for a few samples from the database. An average time of 5.4 s is needed for data access and transfer on cloud and an average time of 6 s is needed by the proposed algorithm to test the image for Glaucoma screening. The computation time to process an image of high resolution seems encouraging and can be used by any organization working in this field to arrange screening camps and provide a feedback regarding the medical condition of their eyes. The diagnostic report generated can be correlated with a trained ophthalmologist.
Figure 13 shows the experimental results for some samples from the database. Figure 13(a) shows the input fundus image in RGB format which is fed to the system for diagnostic test. Figure 13(b) shows the segmented optic disc and cup marked on the fundus image with black and blue ink respectively. It can be observed from the figure that the database consists of images of varied illumination and various aberrations. The developed image processing framework is able to correctly reject the false positives and correctly segment the optic disc and cup in the fundus image. The figure contains samples from all 3 classes discussed above. The boundaries of optic disc and cup obtained using the proposed algorithm are encouraging and suggests that the proposed thresholds used for the segmentation of the objects are adaptive and can work on images with non-uniform illumination and aberrations.
Table 4 shows the confusion matrix obtained for the images used for experimentation. It can be observed that a sensitivity of 100% is achieved for Glaucoma cases. Although, there are some cases in case of Normal and Suspect Glaucoma class which have been predicted as member of different class. A sensitivity of 95.1% and 86% has been achieved for Normal and Suspect Glaucoma classes, respectively, using the proposed algorithm. The results obtained seems encouraging and indicates that the proposed system can be used for real time screening of the patients.
In addition to the local database, the proposed method is also tested on publicly available DRISHTI database. The algorithm has been used without parameter tuning and has achieved an accuracy of 85%.
Table 5 shows the comparison of the performance parameters such as accuracy, sensitivity, specificity and accuracy with some of the existing literature in the field of Glaucoma detection using computer vision. The proposed method has achieved good values for all three parameters used for comparison. The results show that the proposed method is capable enough of handling all three classes based on the features reported in the text.
Discussions
The proposed glaucoma screening system is a cloud based architecture, built on a Heroku server with a free tier subscription, which can be used by a person anytime, anywhere and possess capabilities to act as an assisting tool in urban areas and primary health care center in rural areas in screening of patients suffering from Glaucoma. It is an open source architecture with Python Django framework as backend and HTML frontend. The proposed system is location and device independent. The application can be accessed using a web browser from any device, such as laptops, tablets, smartphones, etc. and can generate a diagnostic report instantaneous round the clock. The clinical features such as CDR and structural changes in NRR due to progression of Glaucoma have been considered and involved in the proposed system. So, the diagnosis performed is clinically significant and can be correlated with an ophthalmologist. The system uses industry standards which empowers it to be easily scalable in future, if required.
References
Tsai, J. C., How to evaluate the suspicious optic disc. Rev. Ophthalmol. 2005.
Tuominen, V. J., Ruotoistenmäki, S., Viitanen, A., Jumppanen, M. and Isola, J., ImmunoRatio: a publicly available web application for quantitative image analysis of estrogen receptor (ER), progesterone receptor (PR), and Ki-67”, in 12:R56.
Ortega, M., Barreira, N., Novo, J., Penedo, M. G., Pose-Reino, A., and Gomez-Ulla, F., Sirius: A web-based system for retinal image analysis. Int. J. Med. Inform. 79(10):722–732, 2010 ISSN 1386-5056.
Krieger, M. T., Torreno, O., Trelles, O., and Kranzlmuller, D., Building an open source cloud environment with auto-scaling resources for executing bioinformatics and biomedical workflows. Fut. Gen. Comput. Syst. 67:329–340, 2017 ISSN 0167-739X.
Nowakowski, P., Bubak, M., Tomasz Bartyński, Tomasz Gubała, Harezlak, D., Kasztelnik, M., Malawski, M., and Meizner, J., Cloud computing infrastructure for the VPH community, J. Comput. Sci., 2017. ISSN 1877-7503.
Rosenthal, A., Mork, P., Li, M. H., Stanford, J., Koester, D., and Reynolds, P., Cloud computing: A new business paradigm for biomedical information sharing. J. Biomed. Inform. 43(2):342–353, 2010 ISSN 1532-0464.
Shakil, K. A., Zareen, F. J., Alam, M., and Jabin, S., BAMHealthCloud: A biometric authentication and data management system for healthcare data in cloud. J. King Saud University - Computer and Information Sciences, 2017, ISSN 1319-1578.
Alonso-Calvo, R., Crespo, J., Garc'ia-Remesal, M., Anguita, A., and Maojo, V., On distributing load in cloud computing: A real application for very-large image datasets. Proc. Comput. Sci. 1(1):2669–2677, 2010 ISSN 1877-0509.
Al Mamun, K. A., Alhussein, M., Sailunaz, K., and Islam, M. S., Cloud based framework for Parkinson’s disease diagnosis and monitoring system for remote healthcare applications. Fut. Gen. Comput. Syst. 66:36–47, 2017 ISSN 0167-739X.
Kaur, P. D., and Chana, I., Cloud based intelligent system for delivering health care as a service. Comput. Methods Programs Biomed. 113(1):346–359, 2014 ISSN 0169-2607.
Yin, F. et al., A cloud-based system for automatic glaucoma screening. 37th Ann. Int. Conf. IEEE Eng. Med. Biol. Soc. (EMBC), Milan 2015:1596–1599, 2015.
Cheng, J., Yin, F., Wong, D. W. K., Tao, D., and Liu, J., Sparse dissimilarity-constrained coding for Glaucoma screening. IEEE Trans. Biomed. Eng. 62(5):1395–1403, 2015. https://doi.org/10.1109/TBME.2015.2389234.
Cheng, J. et al., Superpixel classification based optic disc and optic cup segmentation for Glaucoma screening. IEEE Trans. Med. Imag. 32(6):1019–1032, 2013. https://doi.org/10.1109/TMI.2013.2247770.
Khalil, T., Usman Akram, M., Khalid, S., and Jameel, A., Improved automated detection of glaucoma from fundus image using hybrid structural and textural features. IET Imag. Process. 11(9):693–700, 2017. https://doi.org/10.1049/iet-ipr.2016.0812.
Joshi, G. D., Sivaswamy, J., and Krishnadas, S. R., Optic disk and cup segmentation from monocular color retinal images for Glaucoma assessment. IEEE Trans. Med. Imag. 30(6):1192–1205, 2011. https://doi.org/10.1109/TMI.2011.2106509.
Joshi, G. D., Sivaswamy, J., and Krishnadas, S. R., Depth discontinuity-based cup segmentation from Multiview color retinal images. IEEE Trans. Biomed. Eng. 59(6):1523–1531, 2012. https://doi.org/10.1109/TBME.2012.2187293.
Issac, A., Parthasarthi, M., and Dutta, M. K., An adaptive threshold based algorithm for optic disc and cup segmentation in fundus images. 2nd Int. Conf. Sign. Process. Integ. Netw. (SPIN), Noida 2015:143–147, 2015.
Issac, A., Partha Sarathi, M., and Dutta, M. K., An adaptive threshold based image processing technique for improved glaucoma detection and classification. Comput. Methods Prog. Biomed. 122(2):229–244, 2015 ISSN 0169-2607, https://doi.org/10.1016/j.cmpb.2015.08.002.
Issac, A., Sengar, N., Singh, A., Sarathi, M. P., and Dutta, M. K., Automated computer vision method for optic disc detection from non-uniform illuminated digital fundus images. 2nd Int. Conf. Commun. Control Intell. Syst. (CCIS), Mathura 2016:76–80, 2016. https://doi.org/10.1109/CCIntelS.2016.7878204.
Partha Sarathi, M., Dutta, M. K., Singh, A., and Travieso, C. M., Blood vessel inpainting based technique for efficient localization and segmentation of optic disc in digital fundus images. Biomed. Sign. Process. Contrl 25:108–117, ISSN 1746-8094, 2016. https://doi.org/10.1016/j.bspc.2015.10.012.
Heneghan, C., Flynn, J., O’Keefe, M., and Cahill, M., Characterization of changes in blood vessel width and tortuosity in retinopathy of prematurity using image analysis. Med. Image Anal. 6:407–429, 2002.
Aquino, A., Gegundez-Arias, M. E., and Marin, D., Detecting the optic disc boundary in digital fundus images using morphological, edge detection, and feature extraction techniques. IEEE Trans. Med. Imag. 29(11):1860–1869, 2010. https://doi.org/10.1109/TMI.2010.2053042.
Soorya, M., Issac, A., and Dutta, M. K., An automated and robust image processing algorithm for glaucoma diagnosis from fundus images using novel blood vessel tracking and bend point detection. Int. J. Med. Inform. 110:52–70.
Salam, A. A., Khalil, T., Akram, M. U., Jameel, A., and Basit, I., Automated detection of glaucoma using structural and non structural features. Springerplus. 5(1):1519, 2016.
Noronha, K. P., Acharya, U. R., Nayak, K. P., Martis, R. J., and Bhandary, S. V., Automated classification of glaucoma stages using higher order cumulant features. Biomed. Sign. Process. Contrl. 10:174–183, 2014.
Acharya, U. R., Ng, E. Y. K., Eugene, L. W. J., Noronha, K. P., Min, L. C., Nayak, K. P., and Bhandary, S. V., Decision support system for the glaucoma using Gabor transformation. Biomed. Sign. Process. Contrl. 15:18–26, 2015.
Divya, L., and Jacob, J., Performance analysis of Glaucoma detection approaches from fundus images. Proc. Comput. Sci. 143:544–551, 2018.
Acknowledgements
This work was supported in part by the Grants from Department of Science and Technology, No. DST/TSG/ICT/2013/37. Also, the authors express their thankfulness to Dr. S.C. Gupta, Medical Director of Venu Eye Research Centre for his contribution in classification of the images.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Image & Signal Processing
Rights and permissions
About this article
Cite this article
Soorya, M., Issac, A. & Dutta, M.K. Automated Framework for Screening of Glaucoma Through Cloud Computing. J Med Syst 43, 136 (2019). https://doi.org/10.1007/s10916-019-1260-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10916-019-1260-2