Abstract
Due to the high mortality associated with heart disease, there is an urgent demand for advanced detection of abnormal heart beats. The use of dynamic electrocardiogram (DCG) provides a useful indicator of heart condition from long-term monitoring techniques commonly used in the clinic. However, accurately distinguishing sparse abnormal heart beats from large DCG data sets remains difficult. Herein, we propose an efficient fine solution based on 11 geometrical features of the DCG PQRST(P-T) waves and an improved hierarchical clustering method for arrhythmia detection. Data sets selected from MIT-BIH are used to validate the effectiveness of this approach. Experimental results show that the detection procedure of arrhythmia is fast and with accurate clustering.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Heart disease is a major cause of human morbidity and mortality [1]. The presence of abnormal heart beats indicates impaired cardiac function. Thus, detection of cardiac arrhythmia is important for predicting potential heart disease.
The dynamic electrocardiogram (DCG) analyzer is a widely used clinical method for recording the electro-activity of heart beats [2]. Because DCG records the heart state continuously, it provides more accurate information than the common short-term electrocardiogram. However, it is difficult for clinicians to detect sparse abnormal heart beats in large data sets [3]. ss than normal beats of a person in reality. Thus, 3 PVR are mistakenly clustered into normal group by our algorithms. However, the accuracy is still higher because of the full accounting of wave features.
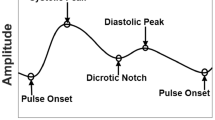
Since DCG signals are continuous sampling points, dividing data D={d 1,⋯ ,d n } into heart beats H={h 1,⋯ ,h m } is necessary, and is done by locating the R-peak of every cardiac [4]. As shown in Fig. 1, a full cardiac cycle mainly consists of a QRS complex, a P-wave and a T-wave. Thus, more attribute values from P-T waves must be extracted to describe H. DCG waveforms are various and our approach focuses far more on lesion waveforms than other waveforms which contain less-valued information. At the same time, we address to improve location accuracy of Q- and S-waves, and we have investigated a few algorithms about DCG feature extraction [8–14].
Given feature vectors X={x 1,⋯ ,x m }∈R p, where x i is an attribute vector of dimension p describing h i , it is infeasible to manually capture abnormal heart beats, and the clustering tool is urgently necessary. Clustering is the unsupervised tool of partitioning a set of data objects (data items) into multiple groups (clusters). Objects within a group have high similarities, but are very dissimilar to objects in other groups [5]. When processing a data set, different clustering methods lead to different clustering results. In our approach, agglomerative hierarchical clustering is suitable to the DCG data sets on accuracy. We create an adaptive model method to cooperate with hierarchical method for improving the speed of clustering. Some basic and advanced methods for clustering have been discussed [15–19].
The framework of arrhythmia detection consists of feature-vector extraction and clustering. First, we locate the R-wave to divide the cardiac cycles, and then extract position information to get P-T wave features, where p=11. Next, a template is selected to remove some heart beats, and the rest are clustered by hierarchical clustering method to capture arrhythmias.
The paper is organized as follows. Section “Related Work” presents a brief review on related work. Section “Feature extraction of P-T waves” details the process of feature extraction. Section “Improved Hierarchical Clustering” describes a clustering method of heart beats. Section “Experimental Results and Analysis” presents experimental results and their analysis. Section “Conclusion” concludes the paper.
Related Work
DCG waveforms.
DCG data contain at least 24 h of electrocardiogram waves, which can be divided into three categories: congenital waveforms, jamming waveforms, and lesion waveforms which can lead to some abnormal heart beats [6].
Congenital waveforms refer to some similar waveforms whose number can almost reach more than 90 percent of the total DCG waves. They are individual, vast, and maybe normal or abnormal. Congenital waveforms may cause severe congenital heart diseases [14] and common short-term electrocardiogram is a good tool for assessing pathological information of them.
Jamming waveforms generally occur at the beginning and the end of data acquisition or when the state of the subject changes, such as changing from sitting to walking. These waveforms are irregular, distinct, and complex. According to the difference of time-frequency characteristics of normal ECG signals and jamming signals, [14] uses FSWT (Frequency Slice Wavelet Transform) to deal with the ECG signal denoising. Although data cleaning is challenging, the proportion of pollution is small; we can analyze less-noisy waves to reduce negative effects.
Lesion waveforms are sudden cardiac waveforms relatively fewer in number, at most 5-10 % in DCG, and lead to some arrhythmia including atrial premature beats (APB), ventricular premature beats (VPB), and atrial fibrillation (AF). Lesion waveforms are sparse, and generally reflect impaired cardiac function, and can contribute to myocardial ischemia, myocardial infarction, and even sudden death [9]. Unfortunately, a common electrocardiogram cannot capture these subtle lesion waves, and it is difficult for a clinician to observe them visually from DCG. Nevertheless, lesion waveforms exhibit obvious differences in appearance to congenital waveform [10], and can be processed by clustering to detect arrhythmia.
Feature extraction.
Precise feature extraction of P-T waves is important prior to clustering. P-T waves are collected by DCG to reflect heart activities. In one cardiac cycle, the P-wave appears first and represents depolarization of the atrium. QRS complex and T-wave reflect the depolarization and re-polarization of the ventricle. For convenience, an interval between two adjacent R-waves is considered as a full cardiac cycle. Currently, several algorithms of DCG feature extraction have been proposed for abnormal heart beats, including time domain analysis, wavelet transformation, and neural networks. A combination of neural network and wavelet analysis [11], a simple autoregressive-moving average (ARMA) method [12], and a moving average filter [13] have been reported. In these studies, the width of P- and T-waves, which include important pathological information, has not been taken into account. In the present study, we specifically locate the starting and end points of P- and T-waves for more precise clustering.
When collected, DCG signals, especially low-amplitude waves, are polluted by serious noises, artifacts and baseline drift. The interference has severe influence on parameter measurement, waveform identification and disease diagnosing [8]. Literature [15, 16] remove DCG noises based on wavelet and literature [17] processes baseline drift by moving average algorithm. Pointing at Q- and S- waves, we raise curve fitting and threshold method to reduce the influence of noise.
Clustering.
The choice of clustering methods for DCG heart beats is also important. Basic clustering methods are including partitioning methods, density-based methods and hierarchical methods. Partitioning, which is generally distance-based such as k-means algorithm, is the simplest version and most effective for spherical clusters. Density-based methods can find arbitrarily shaped clusters but requires big internal storage and heavy computation than other methods when processing large data. Hierarchical methods can group objects into a hierarchical tree of clusters and suffer from the fact that it cannot correct erroneous split once a step is done; however, computation costs are smaller because of the rigidity. Agglomerative hierarchical clustering generally follows the following steps [18]:
-
1.
Representing each object as a cluster;
-
2.
Merging the two clusters as one cluster with the minimal distance;
-
3.
Repeating step 2 till only one cluster left.
Matrix D represents clustering objects in a pairwise dissimilarity distances. The pairwise Euclidean distance d i j is defined as:
Besides, a template-matching method for clustering is given in [19], an analytical method based on variable threshold is raised in [20], a kind of weighted hierarchical clustering algorithm is proposed in [21], and the application of genetic algorithm in electrocardiosignal is presented in [22] . Although accurate, they are complex for processing large DCG data sets. Herein, we improve hierarchical clustering to provide more rapid data analysis.
Feature extraction of P-T waves
The aim of extracting P-T waves is to process DCG data as appropriate inputs of clustering. Different methods are used to obtain the positions of the R-wave, Q- and S-waves, and the P- and T-waves. Eleven-dimension features are then extracted, standardized, and normalized.
Compared with other methods, our algorithms are simple and stable. Further, both the peak point and the starting and end points of P- and T-waves are detected based on the importance of P- and T-wave width. We also use a combination of the curve fitting and threshold methods to avoid noise interference when locating the Q- and S-waves.
Location of R-waves
For convenience, researchers consider the signals between two adjacent R-waves as one complete cardiac cycle. Therefore, it is necessary to first locate the R-wave exactly to divide the heart beats.
Wavelet transform (WT) is a very promising technique for analyzing bio medical signals. The WT of f(t) is defined as [23]:
Where α is scale factor, and ψ(t) is basic wavelet. Herein, Mexican hat wavelet is used as ψ(t) to detect R-wave because of the similar appearance with QRS complex. Mexican hat wavelet is defined as:
Algorithm 1 shows the R-wave location method. As shown in Fig. 2, the DCG signals are relatively smooth and the singularities are more obvious after the wavelet transform in step 1. Because of the large computation involved in wavelet transform, we only use the method for initial R-wave location, which collects the R-waves and some error points.

Next, we find all singularities corresponding to the peaks of R-waves and other points that we wish to remove. The altitude threshold t h 1 is set in step 2 ∼3 to remove these points. However, altitude threshold occasionally identify a high T-wave as an R-wave by mistake. According to the R-R interval characteristics, we set t h 2 as the position threshold to improve accuracy. By repeated experiments, t h 2=200 is finally determined, and the intermediate result and final result of R-wave location are shown in Fig. 3.
Location of P- and T-waves
Several cardiac diseases can be identified by width and height features of freak P- and T-waves. Thus, it is important to locate the peaks, starting points, and end points of P- and T-waves.
Location of the peaks of T- and P-waves.
The R-R interval helps us to identify the peaks of P- and T-waves as follow steps:
-
1.
Search points on the band from the T T th point after the front R-wave to the midpoint of the R-R interval. The maximal absolute value on the band is the T-wave peak position, and TT is set as 40 according to the experience of cardiologists and results of repeated experiments.
-
2.
Search points on the band from the midpoint of the R-R interval to the P P th point before the latter R-wave. The maximal absolute value on the band is the P-wave peak position, and P P=40 is also empirical.
Location of starting and end points of T-waves.
we can use the same method to locate the starting and end points of P- and T- waves. Herein, we typically describe the processing of T-waves in algorithm 2 and show the location results in Fig. 4.

T-wave width is a key reference index used in Section “Improved Hierarchical Clustering”. It is impossible for the starting or end point of a normal T-wave to be right next to the peak point. Thus, steps 4 and 7 are necessary in our method to locate an abnormal T-wave according to its width.
The morphologies of P- and T-waves vary from person to person. Figure 5 shows twelve kinds of T-waves, including normal and abnormal shapes. The aim of our study is to distinguish sparse abnormal heart beats which include uncommon lesion waves, so that algorithm 2 does not account for all possible reported T-wave types [24] for efficiency. For example, assume that the inborn T-wave of person A is double-humped, as shown in Fig. 5f. It seems ill-behaved that only the first peak of this T-wave is located by algorithm 2. However, it does not contribute to bad clustering results after performing algorithm 4. This is expected for two reasons: (i) the similarities between double-humped T-waves and other congenital T-waves are higher than these between double-humped T-waves and unusual waves too tall, too wide, or disappeared; (ii) double-humped T-waves are similar to each other as all the first peaks are located.
The morphologies of T-waves [24]

Location of Q- and S-waves
Q- and S-waves can also be identified using the same method because of their similar characteristics. We have found that the wavelet transformation is not applicable in our experimental samples for Q- and S-wave location. Thus, algorithm 3 is proposed to locate Q-waves typically.
The amplitudes of the Q- and S-waves are very small and their directions are uncertain. Thus, it is difficult for location algorithms to differentiate the correct points when there is noise in the opposite direction. Figure 6 shows that the noise is easily mistaken for the Q-wave using some algorithms. The interference contributes to inaccurate feature values such as height difference between R-peak and Q-peak, and then has a negative impact on final result.
To improve the accuracy of location, mistakes (noise interference) like Fig. 6 must be avoided. After observing the features of these interferences, algorithm 3 firstly seeks out possible locations of Q-wave in step 1 ∼3. Each candidate is the highest singular point of its direction. One of the points is then chosen by step 4 ∼ 8. In step 4, curve-fitting is used to smooth the signals to get a line whose endpoint is one of the candidates. After that, four slopes of four lines can be calculated, f 1j p (t),f 2j p (t) represent candidate in positive direction and f 1j b (t), f 2j b (t) represent candidate in negative direction. In step 5 ∼ 8, the slopes describing candidates are compared with each other, all the situations are considered and corresponding judgements are given. In principle, the larger the corresponding slope value, the more likely the Q-wave position. To sum up, the whole algorithm 3 focuses on screening the right Q-wave from possible disturbance points (noise).
Figure 7a shows the location result of QRS complex, while Fig. 7b shows the location of all required DCG points. As demonstrated, this method is effective for locating normal signals. The location effects for freak waves are demonstrated in Section “Experimental Results and Analysis”.
Feature extraction
Representing H={h 1,⋯ ,h m }, vectors X={x 1,⋯ ,x m }∈R p are extracted to be the input of clustering. Experts have put forward consulting opinions about extracting features. For example, Aimei Zhai [25] believes that following items are useful:
-
(1) P-wave: time, amplitude, type, direction;
-
(2) P-R interval: time, amplitude;
-
(3) QRS wave group: time, amplitude, type, direction, Q-wave amplitude, Q- wave time, R-wave amplitude, R-wave peak, Q-wave amplitude, S-wave amplitude;
-
(4) S-T interval;
-
(5) Q-T interval: time;
-
(6) T-wave: time, amplitude, type, direction.
After repeated experiments and discussing with the cardiologists, the following characteristics are 11-dimensional most informative attributes describing x i ∈X, as shown in Fig. 8.
-
(1) Five time characteristics are used: the RR interval (RRL), the distance between the starting point and the end point of the T-wave (TLRL), the distance between the R-wave and the T-wave (RTL), the distance between the starting point and the end point of the P-wave (PLRL), and the distance between the R-wave and the P-wave (RPL).
-
(2) Six amplitude characteristics are also used: the amplitude difference between the starting point and the peak of the T-wave (TTLH), the amplitude difference between the end point and the peak of the T-wave (TTRH), the amplitude difference between the starting point and the peak of the P-wave (PPLH), the amplitude difference between the end point and the peak of the P-wave (PPRH), the amplitude difference between the P-wave and the Q-wave (PQHH), and the amplitude difference between the T-wave and the S-wave (TSHH).
Improved Hierarchical Clustering
As a type of classic algorithm, the hierarchical clustering provides highly accurate results. For DCG datasets, sparse abnormal heart beats are hidden in large heart beats; hierarchical clustering is very time consuming, thus reducing efficiency. Therefore, we developed an improved hierarchical clustering as algorithm 4.

A heart beat containing congenital P-T waves is selected as a template. Numerous heart beats are then eliminated by comparing them with the template. Finally, the remaining beats are clustered. This entire procedure is described in Algorithm 4.
Step 1 ∼5 selects the template from the current heart beats. Step 6 ∼8 rejects heart beats by template matching. Finally, the rest of beats are clustered by hierarchical algorithm. For step 1 ∼5, the whole procedure is faster than selecting the model from the template base, as different people have different congenital heart beats.
Experimental Results and Analysis
Using MATLAB2010b and DCG data sets from the MIT-BIH arrhythmia database (http://www.physionet.org/cgi-bin/atm/ATM), we quantitatively evaluate the performance of proposed detection algorithms. Our experimental data sets were collected through lead MLII, and were mainly from patients No. 100, 105, and 108 in MIT-BIH database. All collected heart beats have been labeled manually by prestigious doctors. For ease of viewing, we mainly investigated 1-min DCG data of above three patients. Each 1-min DCG record includes 60 ∼80 heart beats, but only 1 ∼4 abnormal beats. Besides, we also performed comparison of our approach to methods proposed in [12] using the data sets from [12].
Extraction of P-T features
-
1)
According to the MIT-BIH records, the 7th heart beat of patient No. 100 is an atrial premature beat (APB). As shown in Fig. 9a, the R-R interval of this abnormal heart beat is shorter than others, and the T-wave changes its position from where it should have been. Our results show that the abnormal beat is detected accurately by our algorithm. Based on the exact location of the lesion wave, we can extract its width, relative position, and other information to help us distinguish sparse abnormal heart beats by the clustering technique.
-
2)
According to the MIT-BIH records, the 21st, 30th, 48th, and 81st heart beats of patient No. 105 are ventricular premature beats (VPB). Abnormal heart beats shown in Fig. 9b only include the 21st and the 30th heart beats, marked as 3 and 12, respectively. As shown, the R-wave heights in these two abnormal heart beats are worlds away from other beats, and the T-waves are freak.
-
3)
According to the MIT-BIH records, the 19th and 30th heart beats of patient No. 108 are ventricular premature beats (VPB), while the 20th and 31st heart beats are blocked atrial premature beats (blocked APB). Abnormal heart beats shown in Fig. 9c only include the 19th and 20th heart beats, marked as 1 and 2, respectively. As shown, the P-wave in the 1st beat disappears. In the 2nd beat, the P-wave is copied to replace the T-wave. All of the abnormal beats were detected exactly. With respect to the absent P-wave, our algorithm did not report that it was abnormal, but marked the fictitious start, end, and peak points of this wave, despite the ”peak” possibly being lower than the other two points. The feature vector extracted in the next step illustrates the abnormalities and clustering information are used to make a correct judgement.
Analysis of clustering results
1) We take patient No.105 as an example to show the differences between our clustering algorithm and the hierarchical clustering algorithm [21]. Compared with the ground truth (abnormal heart beats: 21t h,30t h,48t h,81t h), hierarchical clustering [] is accurate, but performs unnecessary clustering, as seen in Fig. 10. For instance, both the 75 th and the 18 th heart beats are normal, and should not have been distinguished from each other. Our algorithm eliminated the majority of these beats prior to hierarchical clustering, with only 23 heart beats clustered by algorithm 4 in Fig. 11. The 23 beats are stored in an array n e w c l[] in the originally specified sequence, and the 21 st, 30 th, 48 th and 81 st original heart beats are stored in n e w c l[2], n e w c l[5], n e w c l[12], n e w c l[23]. From Table 1, the 21 st, 30 th, 48 th, and 81 st original heart beats are VPB, and the correct findings are shown in Fig. 11. Thus, our algorithm exhibits high accuracy owing to the hierarchical clustering, and improves the clustering efficiency.
Running time is an effective index for evaluating algorithm complexity. Figure 12 describes the comparison of running time between the weighted hierarchical clustering method [13] and the algorithm proposed in this study. It is evident from Fig. 12 that the running time of our algorithm, fluctuating from 0.217 ms to 0.277 ms among five data sets, is almost half as much as that of hierarchical clustering method [13].
2) Next, samples selected from MIT-BIH were used to test and verify the clustering accuracy. Table 1 lists the comparison of grond truth and our clustering results to show the accuracy of the approach. In Table 1, A represents APB, V is VPB, N is normal beat, X is blocked AP, and U is unknown abnormal heart beat. It can be seen that there are only 2 different labels between the ground truth and our result using data sets from No.100, No. 105 and No.108. Among them, several normal heart beats were not mismatched although they contain double-humped P-waves which are located by algorithm 2 inaccurately. In contrast, a few normal heart beats are separated from the others due to serious noises.
3) We also input 18 normal sinus rhythms (NRS) and 40 premature ventricular contractions (PVC) from MIT-BIH database [12] and get the clustering results in Table 2. As we can see, the results of our algorithm are very little different from that of algorithm 2 in [12], but much superior to that of algorithm 1 in [12]. There are more PVR than NRS in this data set, whereas our approach respects the objective law that abnormal PVR heart beats are far less than normal heart beats.
Results Discussion
To sum up, the experimental part of this paper made two levels of work. In Section “Extraction of P-T features”, based on the MIT-BIH open data, experiments on extracting P-T features are carried out. Although lots of abnormal features, the location results are accurate (Fig. 9: No. 100 and 105). To save time, our algorithm did not deal with the absent P-wave, but marked three obvious wrong positions (Fig. 9: No. 108). In Section “Analysis of clustering results”, the clustering efficiency (Figs. 10 and 11) and running time (Fig. 12) are firstly compared with those of other hierarchical clustering methods and our algorithm speed is distinctly fast. Then, the clustering accuracy is tested by using the same data sets as those used in Section “Extraction of P-T features”; and ideal results are obtained even on dataset No.108 whose abnormal beats contain absent P-wave (Table 1). Finally, based on the dataset in [12], our clustering algorithm is compared with the methods proposed in [12], and the result is better but not excellent (Table 2). The reason is that our algorithms apply to hunt sparse abnormal heart beats, whereas PVR beats are more than normal beats in dataset [12].
Conclusion
Based on DCG PT features, we proposed a noise-tolerant detection system for capturing rare ectopic heart beats. D={d 1,⋯ ,d n } are firstly divided into H={h 1,⋯ ,h m } by retrieving the location information of R-waves. Then, algorithms are designed to locate P- and T- waves, and Q- and S-waves of h i . After that, 11 attributes are extracted based on above points to form feature vectors X={x 1,⋯ ,x m }∈R p,p=11 describing H. Finally, feature vectors X are processed by our clustering algorithm to detect sparse abnormal heart beats. In this system, we locate more feature points including start and end points of P- and T-waves to generate more geometrical pieces of information. Additionally, we balance the accuracy and the time complexity when extracting P- and T-waves. Furthermore, the consideration of noise with characteristics similar to Q- and S-waves also provides increased accuracy. The extraction algorithms are simple but stable, and the clustering procedure is fast but effective.
Future studies are required to improve the clustering method to automatically obtain clearer boundaries between normal and abnormal heart beats. Because some contaminated normal DCG signals can be mistaken as arrhythmias, further improvement in de-noising is also required. What’s more, it is valuable to help diagnosis a kind of specific heart disease, and further research on Coronary Heart Disease (CHD) may be feasible in the future.
References
Tu, Y.W., Chen, H., Fu, X.Q., et al., Beats clustering based algorithm for fast recognition of motion artifact sections in Holter system[J]. J. Zhejiang Univ. Eng. Sci. 46(6):1148–1156, 2012.
Tu, Y.W., Yu, X.M., Chen, H., and et al., A novel method for the detection of sleep apnea syndrome based on single-lead ECG signal[C]. Appl. Mech. Mater. Trans. Technol. Publ. 239:1079–1083, 2012.
Gang, Z., Yalou, H., and Pengtao, W., Study of Classficication Rules on Weighted Coronary Heart Disease Data[C], pp. 1845–1850. Guangzhou: Proceedings of the Fourth International conference on Machine Learning and Cybernetics, 2005.
Ghongade, R., and Ghatol, A.A., brief performance evaluation of ECG feature extraction techniques for artificial neural network based classification[C]. In: TENCON 2007-2007 IEEE Region 10 Conference, pp. 1–4. IEEE (2007)
Jiawei Han, M.K., and Pei, J., Data mining: Concepts and Techniques, Concepts and Techniques[J] (2011)
Yue, Q., Research of clustering strategies for dynamic electrocardiogram waveform Tianjin University of Technology (2008)
Desai, U., Martis, R.J., NAYAK C, G., and et al., Decision support system for arrhythmia Beats using ECG signals with dct, dwt and Emd methods: a comparative study[J], p. 1640012: Journal of Mechanics in Medicine and Biology, 2016.
Li, N., Yang, Z., Sun, L., and Wei, R., Extraction method based on Q wavelet transform of ECG signal characteristic. Comput. Sci. 41:61?-64, 2014.
Pang, Z., The comparison of dynamic electrocardiogram and common electrocardiogram in diagnosis coronary heart disease. China Foreign Med. Treat. 28(13):1–1, 2009.
Perren, F., Gschwind, M., Landis, T., and et al., Multiple sclerosis relapse mimicking a transient ischemic attack[J]. Mult. Scler. Relat. Disord. 5:27–28, 2016.
Banerjee, S., and Mitra, M., ECG feature extraction and classification of anteroseptal myocardial infarction and normal subjects using discrete wavelet transform[C]. In: 2010 International Conference on Systems in Medicine and Biology (ICSMB), pp. 55–60. IEEE (2010)
Mao X, M., Zhang T, T., and LI, Q., ECG Clustering algorithm based on ARMA[j]. Chin. J. Biomed. Eng. 6:004, 2012.
Chen, H.C., and Chen, S.W., A moving average based filtering system with its application to real-time QRS detection[C]. In: Computers in Cardiology, 2003, pp. 585–588. IEEE (2003)
Li, N., and Yang, Z., The application of frequency slice wavelet transform in ECG signal feature Extraction[J]. J. Fiber Bioeng. Inf. 8(3):461–472, 2015.
Xu, J., A-ming, W., and Zheng, X., Analysis of ECG Signals Based on Wavelet Threshold De-noising[J]. Comput. Simul. 12:068, 2011.
Wang, Y., and Zeng, D., Processing of ECG signal based on compared with wavelet denoising and the EMD de-noising in quantitative. J. Numer. Methods Comput. Appl. 32:274–282 , 2011.
Yuan, F., Research about noise pocessing of ECG signals. Electron. Tecnol. 5:130–131, 2014.
Zhang, X., and Xu, Z., Hesitant fuzzy agglomerative hierarchical clustering algorithms[J]. Int. J. Syst. Sci. 46(3):562–576, 2015.
Jing, Y., Zhou, Q., and Chen, W., An improved template matching algorithm and its application in ECG waveform recognition. Chin. J. Biomed. Eng. 31(5):775–780, 2012.
Ge, D., and Weng, J., Discrimination of myocardial infarction based on 2d-LDA and high frequency electrocardiogram. Space Med. Med. Eng. 26(2):125–130, 2013.
Yuan, Z., Li, J., and Zhang, B., A weighting system clustering method and application. J. Shenyang Univ. Nat. Sci. 26(3):201–207, 2014.
Ma, Y.J., and Yun, W.X., Research progress of genetic algorithm[J]. Jisuanji Yingyong Yanjiu 29(4), 2012.
Bushra J., Olivier L., Eric F., et al.: Detection of QRS complex in ECG signal based on classification approach[C]. In: 2010 17th IEEE International Conference on Image Processing (ICIP), pp. 345–348. IEEE (2010)
Liu, J., Analysis of ECG signal and software implement beijing jiaotong university (2010)
Liu, S., Research on clustering in typical character of ECG by fuzzy C-Means Zhejiang University (2006)
Acknowledgments
This research is sponsored by National Natural Science Foundation of China (No.61171014, 61371185, 61401029, 61472044, 61472403, 61571049), Beijing Advanced Innovation Center for Future Education (BJAICFE2016IR-004), the BNU Graduate Students’ Platform for Innovation & Entrepreneurship Training Program (No.1601121E1), the Fundamental Research Funds for the Central Universities (No.2014KJJCB32, 2013NT57) and by SRF for ROCS, SEM.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the Topical Collection on Patient Facing Systems
Rights and permissions
About this article
Cite this article
Bie, R., Xu, S., Zhang, G. et al. Efficient Fine Arrhythmia Detection Based on DCG P-T Features. J Med Syst 40, 168 (2016). https://doi.org/10.1007/s10916-016-0519-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10916-016-0519-0