Abstract
In the present work, the synthesis of novel azo dye ligand 6-hydroxy-4-methyl-2-oxo-1-propyl-5-[(E)-1,3-thiazol-2-yldiazenyl]-1,2-dihydropyridine-3-carbonitrile (L) and its Cu(II), Co(II) and Ni(II) transition complexes were prepared. The newly formed compounds were characterized by elemental analysis, UV–Vis, FT-IR, 1H NMR, LC–MS, TGA and magnetic susceptibility measurement. The molar conductance indicates that all the metal complexes are non-electrolytic in nature Based on spectral data indicate square planar geometry was deduced for Cu(II) and Ni(II) complexes, Co(II) complex has tridentate chelation of ligand and produce an octahedral geometry around the metal ion. Additionally, the computational study has been performed using density functional theory (DFT) calculation was used to study the electronic structure of synthesized ligand and their complexes. The in vitro antimicrobial activity of the azo dye ligand and its complexes was tested against Gram +ve bacteria (Bacillus subtilis), Gram –ve bacteria (Escherichia coli), yeast (Candida albicans) and fungus (Aspergillus flavus). All the complexes showed enhanced biocidal activity compared to the free ligand. Moreover, azo dye ligand and its metal complexes have been studied for their antioxidant activity. The DNA-binding activity of metal complexes (1a–1c) was studied by electronic absorption spectroscopy and fluorescence spectroscopy. All the complexes bound to CT-DNA through an intercalation mode. Additionally, all the metal complexes act as good cleavage agents against the pBR322 DNA. The computer-aided molecular docking studies of metal complexes with the receptor of GlcN-6-P synthase showed that metal complexes are potent drugs for the target enzymes.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Nitrogen and sulfur bearing heterocyclic compounds have widely studied due to their diverse biological properties and their role as pharmacophores of great historical significance [1,2,3]. Azo dyes are the potential class of organic compounds bearing one or more azo (–N=N–) chromophoric group, which possess unique numerous properties containing molecular aggregation, optical data storage, CD, DVD and Blue-ray discs. In addition to that, they are engaged in various fields such as dying in textile fibers plastics, leather, metal foil food, cosmetics, toys and plastic, biological and clinical investigations and even the organic synthesis [4,5,6]. Thiazolylazo dyes are obtained from 2-aminothiazole are significant because they can form various types of coordination compounds due to several electron rich donor centers with unusual geometrical structural, electronic features and chemical properties, owing to their high sensitivity and selectivity thiazolylazo dyes has been extensively studied [7,8,9,10]. Metal complexes of sulphur–nitrogen chelating ligands are received significant attention due to their fascinating physico-chemical properties and marked bioactivity. In addition to that the metal complexes having thiazole system exhibit a wide range of potential medical applications, including antibacterial, anticancer, antifungal, anticonvulsant and anti-inflammatory activities [11,12,13,14]. The interaction of the azo dye metal complexes with calf thymus DNA was investigated by multi-spectroscopic techniques such as electronic absorption spectroscopy and emission spectroscopy were used to illustrate the binding mode. DNA is the primary pharmacological target in treating a wide array of diseases [15]. Transition metals play an important role in preparing the bioactive compounds, which can interact with DNA by non-covalent interactions [16, 17]. The non-covalent way include three recognized binding modes of the tiny molecule to DNA are electrostatic interactions, groove binding and external electrostatic binding. Among these, the intercalation mode of binding is very significant and coherent to the pharmacological activity of metal complexes [18, 19].
Due to these impending challenges in the last few decades, remarkable step have been taken in designing transition metal based-drugs with lesser toxicity, target specific and improved therapeutic nature [20,21,22,23]. Moreover, azo dye ligand derived from 6-hydroxy-4-methyl-2-oxo-1-propyl-1,2-dihydropyridine-3-carbonitrile have been widely investigated due to their broad range of applications in catalysis and their relevance to bioinorganic systems and ability to form stable complexes with large number of metal ions [24, 25]. In-silico docking study is extensively used to forecast protein–ligand binding and to screen large libraries for compounds that will regulate the activity of a biological receptor [26].
Herein, we present the preparation and characterization of novel azo dye ligand 6-hydroxy-4-methyl-2-oxo-1-propyl-5-[(E)-1,3-thiazol-2-yldiazenyl]-1,2-dihydropyridine-3-carbonitrile (L) and their Cu(II), Co(II) and Ni(II) complexes by different spectroscopic technique. The geometry of the metal complexes was inferred by electronic and thermal techniques. For better understanding, the structural parameter of ligand and their complexes were examined by DFT/B3LYP level studies. The biological screening of azo dye ligand and its metal complexes screened against selected kinds of bacteria, pathogenic fungi and antioxidant activities were also investigated. The calf thymus DNA binding activity of the metal complexes was studied by absorption spectra and emission spectroscopy. As well as their cleavage activity toward pBR 322 DNA has been studied. In addition, in this work, In-silico molecular docking analysis of synthesized complexes was also performed.
2 Experimental Section
2.1 Reagents and Physical Measurements
All the reagent used for the present work are of analytical grade, commercially available, purchased from Aldrich, and they were used as supplied. Carbon, Hydrogen and Nitrogen analyses were performed out on a CHN analyzer Calrlo Erba 1108 analyzer. Melting points of the compounds were identified using Polmon instrument (model No: MP-96). UV–Vis spectra in the region 200‒800 nm were obtained on shimadzu model 1650 UV–Vis double beam spectrometer in DMF solution (10−3 M). The Infrared spectra were recorded from KBr pellet on a Perkin–Elmer 621 FT-IR spectrophotometer in the range 4000–500 cm−1. 1H spectrum of the ligand was performed using Bruker DPX200 FT-NMR spectrometer using tetramethylsilane as the internal reference compound. The mass spectra of the compounds were recorded on LC–MS: water AQUITY-2777C mass spectrometer. The molar conductance measurement was conducted using 10−3 M solutions of the complexes in DMF using an ELICO-CM82 conductivity bridge. Room temperature magnetic susceptibility measurements of the complexes were carried out by Gouy balance model 7550 applying Hg [Co(NCS)4] was employed as calibrant. Thermogravimetric analysis (TGA) of complexes were carried out on SII Exstar TG/DTA 6300 instrument, from room temperature up to 800 °C with a heating rate of 10 °C min.
2.2 Synthesis of Azo Dye Ligand (L)
6-Hydroxy-4-methyl-2-oxo-1-propyl-5-[(E)-1,3thiazol-2-yldiazenyl] 1,2dihydropyridine-3-carbonitrile was prepared according to the general procedure in the literature [27]. A cold solution of aqueous sodium nitrite in 5 mL distilled water was added drop-wise to the well-cooled solution of 2-aminothiazole (0.01 m) in 20 mL of distilled water and (4 mL) of concentrated hydrochloric acid and stirred for 2 h at 0–5 °C. Further, the cold diazonium solution was added drop wisely to a well stirred solution of (0.01 mol) 6-hydroxy-4-methyl-2-oxo-1-propyl-1,2-dihydropyridine-3-carbonitrile in an aqueous KOH solution (10 mL). The resulting mixture was stirred for another 2 h at the same temperature and the pH was maintained around 5–6. The obtained crude colored azo dye was collected by filtration, washed with distilled water dried and purified by recrystallization in ethanol. The schematic representation for the synthesis of azo dye ligand (L) was displayed in Scheme 1.
2.3 Synthesis of 6-Hydroxy-4-methyl-2-oxo-1-propyl-5-[(E)-1,3-thiazol-2-yldiazenyl] 1,2dihydropyridine-3 carbonitrile (L)
Yield: 68 (%), Color: orange, M.p: 265e273 °C, FTIR data (KBr ncm−1): 3440 ν (O‒H), 1650 ν (C=O), 1480 ν (‒N=N‒). 1H NMR (400 MHz, DMSO-d6, δ ppm):14.30 (s, 1H, OH), 7.65–7.49 (d, 2H, ArH), 2.46 (s, 3H, pyridine –CH3), 3.78 (q, 2H, –CH2–), 0.89–0.87 (t, 3H, –CH3), 1.60–1.51 (m, 2H, –CH2–). LCMS m/z = 306 (M+H). UV–Vis (DMSO) λmax/nm: 445. Anal. Calcd for C13H15N502S (305): C: 69.13; H: 5.12; N: 9.48% Found: C 68.91; H:5.26; N 9.21%.
2.4 Synthesis of Cu(II), Co(II) and Ni(II) Complexes
Ethanolic solution (15 mL) of azo dye ligand (L) (2 mmol) and a solution of hydrated metal chlorides (1 mmol) in a 15 mL of ethanol was added drop wise with the constant stirring. The resulting solutions were then refluxed with stirring for 4 h. The colored solid residue which separated on cooling collected by filtration and washed several time with distilled water and ethanol and dried over anhydrous CaCl2 in the vacuum desiccator.
3 DFT Studies
All computations calculations for azo Schiff base ligand (L) and its complexes were obtained out using the Gaussian 09 software package at density functional theory (DFT) level of theory [25]. The geometrical structure on the ground state were obtained on Backe’s three parameter hybrid exchange functional (B3LYP) and with LANL2DZ basis set [28]. All the geometrical structures were visualized using chem craft 1.7 software. Quantum chemical parameters such as MEP, HOMO–LUMO energy gap (ΔE) and several cycle are calculated for an investigated molecule.
4 Biological Evaluation
4.1 DNA Binding Studies
The DNA binding parameter of metal complexes were carried out using electronic absorption titration spectroscopy. All titration were performed at room temperature in TriseHCl buffer (pH 7.4) gave a ratio of approximately 1.8–1.9. The concentration of the DNA is measured using molar absorption coefficient value of 260 nm (ε = 6600 M−1) [29]. Concentrated stock solutions of the complexes were dissolved in DMSO solvent because of low solubility in buffer solution. Absorption titration experiments were carried out by varying the concentration of the CT DNA (0–60 μM) by keeping the fixed complex concentration (10 μM). While measuring the absorption spectra and to eliminate the absorbance of CT-DNA itself, an equal amount of DNA was added to both the test solution and the reference solutions. From the absorption data, the intrinsic binding constant (Kb) was calculated by following equation.
where εa, εf and εb are the apparent coefficient, free and fully bound forms of the metal complex extinction coefficients, respectively. A plot of [DNA]/(εa– εf) versus [DNA] gave a slope of 1/(εb–εf) and Y-intercept equal to 1/Kb (εb– εf), Kb is the ratio of the slope to intercept.
4.2 Fluorescence Titration
The competitive binding experiments for the metal complexes were carried out using fuorescence spectroscopic studies of ethidium bromide (EB) bound CT-DNA solution in Tris-HCl buffer (pH 7.2) at 25 °C. This experiment was performed out by adding stepwise various concentrations of complexes (0–60 µM) to EB-bound CT-DNA solution. The resulting reaction mixture was incubated at room temperature for 5 min. After successive addition of the metal complexes, the change in fluorescence intensity at 580–620 nm was observed [30]. The relative binding affinity of the complexes with DNA was determined by measuring the quenching constant (KSV) from the slopes of the lines from the plot of fluorescence versus complex concentration using the following Stern–Volmer equation.
where Io and I are the fluorescence intensities in the absence and presence of a quencher, Ksv is the Stern–Volmer quenching constant and r is the concentration of complex to that of DNA.
4.3 DNA Cleavage Activity
The DNA cleavage activity of the newly prepared compounds was determined by agarose gel electrophoresis technique using pBR322 plasmid as a target [31]. Here the experiment was carried using supercoiled pBR322 DNA (0.3 μg) in buffer (5 mM Tris-HCl, 50 mM NaCl and pH 7.2) and (40 and 80 μM) metal complexes (1a–1c) were incubated for 3 h at 37 °C. After incubation 5 μL of loading buffer (0.25% Xylene cynol 0.25% bromophenol blue and 30% glycerol) was added. Samples were electrophoresed for 2 h at 50 V in Tris-acetate-EDTA (TAE) buffer using 1% agarose gel containing 1 mg/1 mL ethidium bromide (EB). The gel was photographed under UV transilluminator (280 nm) and documented.
4.4 Antimicrobial Activity
Antimicrobial activity of azo dye ligand (L) and its metal complexes (1a–1c) is studied using agar well diffusion method against the bacterial species of Escherichia coli (gram negative bacteria) and Bacillus subtilis (gram positive bacteria). Ciprofloxine is used as standard antibacterial agent (control). The in vitro antifungal activities were carried out against two fungal strains, Candida albicans and Aspergillius flavus. The test compounds had been dissolved in dimethyl formamide to get a concentration of 0.5 and 1 mg/cm3. Sample poured petri plates were incoculated with microorganism and incubated at 37 °C for 24 h [32, 33]. The minimum inhibitory concentration (MIC) value of the compounds was determined by serial dilution technique.
4.5 Antioxidant Activity
The free radical scavenging activity of the azo dye ligand (L) and its complexes were determined by 1,1-diphenylpicrylhydrazyl (DPPH) as per literature [34, 35]. Different concentrations (5, 10, 15, 20 and 25 μM) of test compounds were dissolved in DMSO. To these solutions, 2 mL of DPPH solution (0.2 mM in methanol) were added. The mixture was shaken vigorously and allowed to stand at room temperature for 20 min. The decrease in the absorbance of DPPH was calculated relative to the measured absorbance of the control. The percentage of radical scavenging activity was calculated by the following formula:
4.6 Molecular Docking
In-silico molecular docking study was performed using HEX 6.1 software. The structures of the synthesized compounds have been drawn by CHEMSKETCH and convert it into PDB format from mol format by online OPENBABEL (https://www.vcclab.org/lab/babel/).The protein targets were acquired for the docking studies from Protein Data Bank (https://www.rcsb.org) [36]. All calculations were carried out on an Intel Pentium 4, 2.4 GHz based machine running MS Windows XP SP2 as operating system. The output results from AutoDock were analysed by ligplot and paymol software. The docking scores proved valuable information regarding the interaction of prepared compounds with active sites of GlcN-6-P synthase [37].
5 Results and Discussion
Azo dye ligand (L) and its Cu(II), Co(II) and Ni(II) complexes were prepared and characterized by using different spectroscopic methods. The synthesized complexes were coloured, solid, and stable towards air and moisture at room temperature and they are readily soluble in DMF and DMSO. The molar conductance measurements of the complexes in DMF (1 × 10−3 M) solution revealed their non-electrolytic in nature [38]. The analytical and physical data of azo dye ligand (L) and their metal complexes (1a–1c) are presented in Table 1. These data of metal complexes indicate that 1:2 stoichiometric ratio of metal to ligand with general a formula [M (L)2] 2H2O for Cu(II), Ni(II) and [M(L2)2H2O] Co(II) complexes. The microanalytical data of the synthesized compound are in good agreement with the proposed stoichiometry of the complexes.
5.1 Infrared Spectra
The binding mode metal chelates were determined by matching FT-IR spectrum of free ligand with its complexes is summarized in Table 2. The free ligand exhibited a characteristic broad band at 3414 cm−1 which can be assigned to stretching vibration of hydroxyl group. Altogether the metal complexes, this band is absent in all the complexes indicating the participation of the hydroxyl group in complex formation [39]. The ligand and its complexes exhibited medium intensity bands in the region 3056–3017 cm−1 is attributed to vibrations of aromatic C–H. A sharp absorption band observed at 1660–1690 cm−1 region is due to carbonyl group ʋ (C=O) group, which appears at the same position in all the complexes, indicates corbonyl not taking part in the coordination [40]. The moderate intensity band observed at 1448 cm−1 is assigned to (‒N=N‒) stretching vibration of the functional azo group and in all the metal complexes this frequency is shifted to lower frequency by 10–25 cm−1 indicating its participation in coordination sphere [41]. Further, the appearance of new medium intensity non-ligand absorption bands in the region 490–460 and 622–555 cm−1 are assigned to (M‒N) and (M‒O) vibration bands respectively, which support the participation of nitrogen and oxygen atom in coordination with metal centre [42]. The existence of broad band within 3455–3450 cm−1 range in all the spectra of complexes are assigned to (–OH) of water molecules connected with the complexes. This was moreover supported the existence of weak band at 715–730 cm−1 is assigned to (H2O) for coordinated water molecules [43]. The obtained FTIR results were in consistent with their proposed molecular structures of the prepared compounds.
5.2 Electronic Spectral Studies
The UV–Visible spectral data of newly synthesized azo dye ligand (L) and their metal complexes were recorded in DMF solution in the range of 200–800 nm and data are presented in Table 3. The free ligand showed intense absorption bands at 22,727 cm−1 is ascribed to \({\text{n}} \to \pi\) * of the azo group. Further, the absorption due to (‒N=N‒) group in all the metal complexes are shifted to lower wavelength indicating coordination of the metal ions with the azo dye ligand. The electronic spectrum of Cu(II) complex showed two bands at 30,303 and 20,161 cm−1 which are attributed to 2B1g → 2A1g transitions, suggesting square planar geometry and this is further emphasized by its magnetic moment value of 1.82 BM [41]. The spectrum of Co(II) complex exhibited three bands at 18,181, 24,937 and 35,714 cm−1. These bands may be attributed to transition 4T1g → 4T2g(F), 4T1g → 4A2g(F) and 4T1g → 4Tg(P), respectively. The observed magnetic susceptibility value 4.38 BM also indicate the octahedral geometry around Co(II) ion [44]. The Ni(II) complex exhibited three d–d bands at 20,080, 22,727 and 30,581 cm−1 that could be assigned to 1A1g → 1B1g, 1A1g → 1A2g, and INCT respectively. The observed magnetic moment value 3.46 BM clearly indicating the square planar geometry for Ni(II) complex [45].
6 1H NMR Spectral Data
The proton NMR spectrum of the azo dye ligand (L) was recorded using DMSO-d6 as a solvent and tetramethylsilane as an internal standard (Supplementary Material S1). The 1H NMR spectrum of the ligand displayed a singlet peak at 14.30 ppm which is assigned to the protons of the phenolic –OH group. The aromatic protons corresponding to 1,3-thiazole group were resonated as two distinguished doublets in the range of 7.65–7.49 ppm. The signal corresponding to CH2 protons attached to the nitrogen atom of the pyridone ring was observed as triplet at 3.78 ppm. Another singlet appeared at 2.46 ppm which corresponds to the methyl group (CH3) attached to the pyridone ring. The methylene and methyl protons of the aliphatic group attached to nitrogen atom appeared as multiplet and triplet within the range of 1.60–1.51 and 0.89–0.87 respectively [46].
6.1 Mass Spectra
The mass spectrum of free ligand displayed well defined molecular ion peak observed at m/z 306 [M+H], which is in good agreement with the expected molecular mass of the azo dye ligand (Supplementary Material S2). In addition, the mass spectra of complexes showed molecular ion peak for Cu(II), Co(II) and Ni(II) complexes of ligand exhibited m/z at 704.35, 699.35 and 670 respectively, Which are coincident with the stoichiometric composition of [M (L)2] type. Further, the observed molecular ion peaks in all the spectra of the synthesized complexes are in good agreement with their proposed empirical formula.
6.2 Thermal Analysis
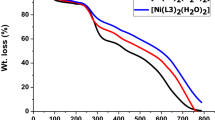
In order to determine the thermal stability and molecular structure of newly synthesized complexes were carried out under nitrogen atmosphere in the temperature range 30–1000 °C. The thermal properties of complexes are given in Table 4 and Fig. 1. The TG graph of Cu(II) complex (1a) showed three successive degradation stages, the first stage of degradation occurred in the temperature range 30–83 °C is corresponding to the loss of lattice water molecules with a weight loss of 2.62% (calcd 2.82%). The second stage of degradation occurred at 83–320 °C with a mass loss of 32.62% (calcd 32.79%) due to the removal of C12H16S2 fragments. The third stage of degradation occurred in the temperature range 320–810 °C corresponds to complete decomposition of organic part C40H36N8O3 with a mass loss of 45.48% (calcd 49.55%). finally leaving one mole of CuO and two mole of carbon as residue. In the TG graph of Co(II) complex, the first step of degradation occurred in the temperature range 28–111 °C is represents the loss of two coordinated water molecule with a mass loss of 5.09% (calcd 5.14%). The second stage of decomposition appeared in the temperature range 111–365 °C with a practical mass loss of 34.91% (calcd 35.76%) corresponds to removal of C12H16N2S2 molecules. The third step of decomposition occurred with a weight loss of 45.34% (calcd 46.14%) due to the complete elimination of organic part C13H8N8O3 with in a temperature 365–685 °C, Finally leaving one moles of CoO and carbon as residue. Similarly, the TG graph of Ni(II) complex, the first stage of decomposition appeared in the temperature range 30–83 °C with a weight loss of 5.62% (calcd 5.96%) is due to elimination of two lattice water molecule. The second stage with weight loss of 31.88% (calcd 32.14%) at 83–350 °C is corresponding elimination of C12H16S2 fragments. The third step with mass loss of 49.23% (calcd 51.40%) at 350–670 °C is assigned to complete dissociation of organic part C13H8N8O3, Finally leaving one mole of NiO as residue.
6.3 Computational Details
6.3.1 Molecular Electrostatic Potential Surface of Ligand and Its Complexes
Molecular electrostatic potential (MEP) is greatly useful method in forecasting to electrophilic and nucleophilic sites in a molecule. This technique is helpful in determining reactivity of molecule and intra or intermolecular interaction in the molecule. Figure 2, displays the MEP of the prepared ligand and its complexes. The negative and positive potential were represented as blue and red color respectively, the blue colours show the positive regions to nucleophilic reactivity and red color indicative the negative region to electrophic reactivity. The most negative site is oxygen close to nitrogen atom and group looks to be red in color with highest electron density [47]. The electrostatic potentials increase in the order as follows red < orange < yellow < green < blue.
6.3.2 Geometrical Optimization
The optimized structures of the azo dye ligand (L) and its complexes (1a–1c) are given in Fig. 3. Selected geometric parameter bond lengths and bond an angle of ligand is calculated at B3LYP/6-31G(d,p) level and depicted in Table 5. The electronic parameter of the compounds was determined from the total energy and Koopman’s theorem. The frontier molecular orbital descriptors such as HOMO–LUMO energy gap, ΔE, absolute electronegativity (χ), chemical potential (Pi), absolute hardness (η), absolute softness (σ), global electrophilicity (ω), global softness (S), and additional electronic charge (ΔNmax). The chemical reactivity parameter for ligand and their complexes are calculated using following equation and tabulated in Table 6.
The energy gap (ΔE) is examined as a significant stability index, which was used for describing the structure and verification barrier in several molecular systems. While increasing the energies the compound becomes more stable. Moreover, the charge of chemical potential (Pi) was –ve value, whereas electrophilicity index (χ) had +ve value. These data reveal that azo schifff base ligand donated electron to metal ions [48].
6.3.3 Frontier Molecular Orbital Analysis
Chemical reactivity and some physical properties of a typical molecule can be found from its Frontier Molecular Orbitals (FMO) [49]. The energy gap has been calculated from [EHOMO − ELUMO] levels and it is used to clarify the bioactivity of a molecule by comparing the intra-molecular charge transfer. The highest occupied molecular orbital (HOMO) and lowest unoccupied molecular orbital (LUMO) are the main orbital takes part in chemical stability and reactivity. The higher energy electron present in HOMO behaved as an electron donor and while the lower energy electron present LUMO behaved as a electron acceptor. Species with large energy gap indicate a considerable level of intra-molecular charge transfer from the electron-donor groups to the electron‐acceptor groups through the π‐conjugated system. Recently, the lower energy gap HOMO‐LUMO values support the trend of bioactivity. The frontier molecular orbitals (HOMO and LUMO) surfaces of prepared ligand (L) and its complexes showed in Fig. 4. The FMO energies HOMO, LUMO and energy gap (ΔE) are calculated as − 1.771, − 2.362 and − 0.591 eV for (L), − 2.488, − 4.369 and − 1.881 eV for Cu(II) complex. − 4.716, − 9.429 and − 4.668 eV for Co(II) complex and − 4.522, − 7.187 and − 2.635 eV for Ni(II) complex respectively. It may be concluded that the energy gap (ΔE) of ligand is found to be lower than the energy gap of metal complexes that suggest high chemical reactivity of ligand than the ligand than the complexes and hence highest stability for the complexes [50, 51].
6.4 Powder X-ray Diffraction Study
The newly synthesized metal complexes were studied for their crystalline nature by powder X-ray diffraction spectroscopy. The metal chelates are not soluble in common organic solvents, thus single crystal study not achieved and presented in Fig. 5. The powder XRD analysis of the hybrids was obtained in the 2–90 (2θ) range and at wavelength 1.54 Å. The inter-planar spacing was evaluated by well known Bragg’s Equation 2d sinθ = n λ. The calculated inter-planar spacing together with the relative intensities with most intense peak is recorded and depicted in Tables 7, 8 and 9. From all the highly intense peaks, unit cell parameters were calculated for all the complexes and h2 + k2 + l2 were also determined. In all the complexes the calculated values of h2 + k2 + l2, showed the absence of forbidden numbers (7, 15, 23, 71 etc.), which suggests that metal complexes have cubic symmetry. The calculated lattice parameter for the Cu(II), Co(II) and Ni(II) complexes were found to be a = b = c = 6.534, 7.944 and 8.141 respectively.
7 Biological Activities
7.1 DNA Interaction Studies
The DNA binding studies of the metal complexes 1a to 1c were examined by electronic absorption spectroscopy through the variations observed in the absorbance and shift in the wavelength. The changes in absorption spectra of metal complexes (10 μM) were recorded in the absence and presence of CT-DNA and the changes observed in the spectra are shown in Fig. 6. In general, significant hypochromism with red shift/bathochromic shift is due to the strong stacking π–π interaction between the aromatic chromophore of metal complexes and DNA base pairs. The extent of the hypochromism depends upon strength of intercalative binding [52]. In the presence of CT-DNA, upon gradually increasing the concentration the absorption bands of complexes displayed significant hypochromic effect together with moderate bathochromic and hypsochromic shift was observed 1–2 nm and exhibited hypochromism of around be 20‒33%. These results indicate that the metal complex bound with DNA via an intercalation mode of the double helix DNA. When compounds bind into the DNA base pairs, the π* orbital of the intercalators coupled with π orbitals of the DNA base pairs and then leads to decreasing in π–π* transition energies and hence hypochromism observed in all the complexes [52, 53].
To further illustrate the binding strength of metal complexes with DNA, the intrinsic binding constant (Kb) was determined using the Eq. (1). The Kb values are found to be 1.98 × 105M−1, 2.64 × 105 M−1 and 2.86 × 105 M−1 for complexes 1a, 1b and 1c respectively. From the above results reveal that the nickel complex is strongly bind to DNA than the remaining copper and cobalt complexes.
7.2 Fluorescence Quenching Measurements
The fluorescence experiments were performed in order to further support the mode of interaction of the metal complexes with DNA by displacement of ethidium bromide (EB). EB is a sensitive fluorescence probe for DNA structure detection, its fluorescence intensity is increased in the presence of DNA due to strong intercalation between the DNA base pairs. Upon successive addition of metal complex to EB-DNA aduct, the fluorescence intensity is decreases with increase in the concentration of metal complexes. This is owing to their complex competitive binding to EB-bound DNA [54, 55], as displayed in Fig. 7. The EB-bound DNA showed emission band between 310 and 390 nm upon excitation at 270 nm. Quantitatively, the fluorescence quenching constants (KSV) of the complexes have been caluclated using the Stern–Volmer equation and the values are 2.45 × 104 M−1(1a), 1.73 × 104 M−1(1b) and 2.87 × 105 M−1(1c). The results reveal that there are strong interactions between DNA and metal complexes, the fluorescence quenching may be due to the complexes interacting with DNA through an intercalative mode. Moreover, the binding strength of Ni(II) complexes is higher than the Cu(II) and Co(II) complexes and consistent with the results obtained from absorption titration.
7.3 DNA Cleavage Activity
The Gel electrophoresis is experiment is used to observe the cleaving reaction of plasmid DNA by small molecules. The DNA cleavage capacity of metal complexes with super coiled pBR322 plasmid DNA was performed by using agarose gel electrophoretic technique. When circular plasmid DNA subjected to electrophoresis, the fastest migration will be observed for the intact supercoil form (Form I). If scission occurs on one strand, the supercoils will relax to produce a shorter moving is circular form (Form II). If both strands are cleaved, linear form (Form III) will be generated and migrates in between. The cleavage patterns of synthesised Cu(II), Ni(II) and Co(II) metal complexes are shown in Fig. 8. Lane 1 applies to unrefined pBR322 DNA (control DNA), which was found to be mixture of supercoiled form 1and nicked form II, lane 3(1a) lane 4 (1b) and lane 5 (1c). Metal complex 1a–1c can induce the apperent cleavage of the supercoiled DNA at the concentration of 40 μM and 80 μM. At the concentration of 40 μM metal complex can promote 90% conversion of super coiled DNA (Form I) into nicked form (Form II) DNA. However, on increasing the concentration at 80 μM, the Supercoiled (Form I) of DNA gradually converted into Nicked Circular (Form II) of DNA and then to Linear (Form III) of DNA. It was noticed that, at lower concentration the cleavage capacity of metal complex is more effective when compared to higher concentration. Therefore, all the metal complexes cleave DNA in absence of external agent and offer the proof for intercalation mode of binding between the metal complex and DNA. Thus, we can conclude that the metal complexes inhibit the growth of pathogenic microorganism by cleaving the genome [56, 57].
7.4 Antimicrobial Activity
The antimicrobial activity of the synthesized azo dye ligand (L) and their metal complexes were carried out against Gram-positive bacteria (Bacillus subtilis) and Gram-negative bacteria (Escherichia coli) and two fungal strains (Aspergillus fumigatus and Candida Albicans) by agar well diffusion technique using Ciprofloxacin and Fluconozole as standard drug. The diameter of zone of inhibition (mm) is shown in Table 10 and Fig. 9. From the data, it is observed that all the isolated metal cheated exhibited significantly enhanced antibacterial activity against selected bacterial strains compared to the free ligand. Among all the metal complexes, the complexes 1a and 1b showed grater activity against both the bacteria. Upon chelation, the enhanced bacterial activity of metal complex than the free ligand is due the decrease in polar character of metal ion due to overlapping of the ligand orbitals with the partial sharing of positive charge of the metal ion with the donor atoms and delocalization of electrons over the chelating ring. Thus, enhancing the lipophilic feature of the complex can easily penetrate into the bacterial cell membrane by blocking the metal binding site inside the enzyme of microorganism [58].
Moreover, the efficiency of the prepared compounds against the fungal strains indicates that ligand and its metal complexes had fungal activity and results are tabulated in Table 11. Complex 1b has showed higher antifungal activity compared to free ligand and complexes. The reason for such results can be explained through Tweedy’s chelation theory and is same as discussed in the antibacterial study [59].
7.5 Antioxidant Activity
The antioxidant activity of the azo dye ligand (L) and its complexes (1a–1c) were carried out by using (2,2-diphenyl-1-picrylhydrazyl) DPPH assay, which is a rapid, simple, precise and low-cost technique to spectrophotometrically establish the antioxidant activity [60, 61]. The inhibitory effects of the synthesized compounds on OH radicals are shown in Fig. 10 and Table 12. The inhibitory effect of the prepared compounds against the DPPH radical are observed with an increasing dose-dependent manner, and suppression ratio for OH increases with increasing sample concentration. All the isolated compounds are showed marked antioxidant activity by scavenging DPPH, but lower when compared to ascorbic acid (vitamin C) as standard. Among 1a and 1c complexes exhibited good scavenging efficiency, whereas the complex 1b and ligand (L) showed moderate activity.
7.6 Molecular Docking Studies
Molecular docking is a great method for knowing the interaction between studied complexes and a biological target which is important clinical treatment and nucleic acid molecular recognition [62]. In order to understand the mechanism of drug action. The in silico active pocket prediction of glucose-amine-6-phosphate synthase engaged in binding with the prepared complexes and ciprofloxacin [63]. The docking studies showed a feasible interaction between the investigated complexes and the receptor. All the synthesized metal complexes showed well established hydrophobic interactions to amino acids such as Gly436, Ser438, Ala439, Gly459, Lys460, Asp508, Asp510, Asp512, Ile516, Gly584, Pro1080, His1081, Gly1082 and Arg1122 in the target enzyme active pockets (Fig. 11). The resulting relative binding affinity of the investigated complexes was achieved in the range of − 6.6 kcal/mol to 7.3 kcal/mol, respectively. From the result it is evident that Ni(II) complex showed more –ve relative binding energy in comparison with remaining docked complexes that indicating it may be considered as a good inhibitor glucose-amine-6-phosphate synthase. The obtained data is in good agreement with the results and summarized in Table 13.
8 Conclusion
In this study, synthesis of novel azo dye ligand (L) and its metal complexes with Cu(II), Co(II) and Ni(II) ions were prepared and characterized by conventional spectroscopic and analytical techniques. The synthetic procedure in this work revealed the formation of complex in 1:2 metal to ligand molar ratio. The molar conductance data suggested that these complexes were non-electrolytes. Based on the results of UV–Vis and magnetic susceptibility data indicate square planar geometry was deduced for Cu(II) and Ni(II) complexes and octahedral geometry of Co(II) complex. The covalent nature of the copper complex was confirmed by EPR study. The thermal properties of metal complexes are investigated by TGA, result showed high thermal stability and suggested molecular formulae of the complex. The molecular structure of azo dye ligand and its complexes were determined by theoretical method (DFT at B3LYP level) and calculated some important parameter viz. geometrical optimization, frontier molecular orbitals and molecular electrostatic potential by same basis set. The binding strength of synthesized metal complexes ascertained with calf thymus DNA, using electronic absorption titrations and fluorescence spectroscopy. The results reveal that complexes bound with CT DNA through intercalation. The prepared complexes have efficiently cleaved supercoiled DNA in the absence of external reagent. All the complexes displayed significant antioxidant activity compared to free ligand. Amid the investigated complexes, Ni(II) complex showed promising antioxidant activity. The free ligand and their complexes were tested against Gram-negative bacteria, Gram-positive bacteria and two strains of fungi, in comparison with the ligand all the complexes showed enhanced activity and can be used as a potent antimicrobial agent against selective pathogens. Furthermore, in silico docking studies have also indicated that the Ni(II) complex showed more –ve relative binding energy and have strong binding affinity with target receptor GlcN-6-P synthase and act as good inhibitor.
References
M. Gaber, S.K. Fathalla, H.A. El-Ghamry, Appl. Organomet. Chem. 33, e4707 (2019)
F.A. Saad, H.A. El-Ghamry, M.A. Kassem, Appl. Organomet. Chem. 33, 4965 (2019)
A.Z. El-Sonbati, M.A. Diab, A.A. El-Bindary, A.F. Shoair, N.M. Beshry, J. Mol. Liq. 218, 400–420 (2016)
A. Tupys, J. Kalembkiewicz, Y. Ostapiuk, V.M.O. Tymoshuk, E. Woz nicka, L. Byczyn ski, J. Therm. Anal. Calorim. 127, 2233–2242 (2017)
A. Penchev, D. Simov, N. Gadjev, Dyes Pigm. 16, 77 (1991)
A.Z. El-Sonbati, G.G. Mohamed, A.A. El-Bindary, W.M.I. Hassan, A.K. Elkholy, J. Mol. Liq. 209, 625–634 (2015)
S.H. Alotaibi, A.S. Radwan, Y.K. Abdel-Monem, M.M. Makhlouf, Spectrochim. Acta A. 205, 364–375 (2018)
Ö.F. Öztürk, Trans. Met. Chem. 32, 224–227 (2007)
K.J. AL-Adilee, A.K. Abass, A.M. Taher, J. Mol. Struct. (2016). https://doi.org/10.1016/j.molstruc.2015.11.038
J. Joseph, B.H. Mehta, Russ. J. Coord. Chem. 33, 124–129 (2007)
Fawaz A. Saad, Hoda A. El-Ghamry, M.A. Kassem, Appl. Organomet. Chem. 33, 4965 (2019)
K. Singh, M.S. Barwa, P. Tyagi, Eur. J. Med. Chem. 42, 394–402 (2007)
S.S. Chavan, V.A. Sawant, J. Mol. Struct. 965, 1–6 (2010)
K.J. Al-Adilee, B.A. Hatem, J. Adv. Chem. 11(3), 3412–3425 (2015)
S. Dhar, D. Senapati, P.K. Das, P. Chatopadhyay, M. Nethaji, A.R. Chakravarty, J. Am. Chem. Soc. 125, 12118–12124 (2003)
N. Chitrapriya, V. Mahalingam, M. Zeller, K. Natarajan, Inorg. Chim. Acta 363, 3685 (2010)
M. Sirajuddin, N. Uddin, S. Ali, M.N. Tahir, Mol. Biomol. Spectrosc. 111, 116 (2013)
S. Kathiresan, S. Mugesh, J. Annaraj, M. Murugan, New J. Chem. 41, 1267 (2017)
H. Xu, K.C. Zheng, Y. Chen, Y.Z. Li, L.J. Lin, H. Li, P.X. Zhang, L.N. Ji, Dalton Trans. 3, 2260–2268 (2003)
R. Kramer, Coord. Chem. Rev. 182, 243–261 (1999)
R.F. Tabor, T.M. McCoy, H. Yingxue, B.L. Wilkinson, Bull. Chem. Soc. Jpn 91, 932–939 (2018)
P. Weis, S. Wu, Macromol. Rapid Commun. 39, 1700220 (2018)
N. Venugopal, G. Krishnamurthy, H.S. Bhojyanaik, P. Murali Krishna, J. Mol. Struct. 183, 37–51 (2019)
W. You, H.-Y. Zhu, W. Huang, B. Hu, Y. Fan, X.-Z. You, Dalton Trans. 39, 7876–7880 (2010)
B.G. Devika, B.H. Doreswamy, N.M. Mallikarjuna, H.C. Tandon, J. Mol. Struct. (2019). https://doi.org/10.1016/j.molstruc.2019.02.029
S. Yousef-Ebrahimipour, I. Sheikhshoaie, J. Castro, M. Dusek, Z. Tohidian, V. Eigner, M. Khaleghi, RSC Adv. (2015). https://doi.org/10.1039/c5ra17524k
A. Vogel, Textbook of Practical Organic Chemistry, 5th edn. (Longman Group, London, 1989)
M.J. Frisch, Gaussian 09, Revision A02 (Gaussian Inc., Wallingford, 2009)
A.D. Becke, J. Chem. Phys. 98, 5648 (1993)
M.F. Reichmann, S.A. Rice, C.A. Thomas, P. Doty, J. Am. Chem. Soc. 76, 3047 (1954)
A. Blask, T.C. Bruice, Acc. Chem. Res. 32, 475–484 (1999)
A.A.A. Aziz, S.H. Seda, J Fluoresc 27, 1051–1066 (2017)
A. Bauer, W. Kirby, J. Sherris, M. Turck, Am. J. Clin. Pathol. 45, 493 (1966)
M. Pfaller, L. Burmeister, M. Bartlett, M. Rinaldi, J. Clin. Microbiol. 26, 1437 (1988)
H. Wu, J. Kong, Z. Yang, X. Wang, F. Shi, Y. Zhang, Trans. Met. Chem. 39, 951–960 (2014)
X. Qiu, S. Abdel-Meguid, C. Janson, R. Court, M. Smyth, D. Payne, Protein Sci. 8, 2529 (1999)
S. Mouilleron, M.A. Badet-Denisot, B. Golinelli-Pimpaneau, J. Mol. Biol. 377(4), 1174–1185 (2008)
W. Geary, Coord. Chem. Rev. 81, 7 (1971)
F.A. Saad, J.H. Al-Fahemi, H. El-Ghamry, A.M. Khedr, M.G. Elghalban, N.M. El-Metwaly, J. Therm. Anal. Calorim. 131, 1249–1267 (2018)
A.Z. Sonbati, G.G. Mohamed, A.A. Bindary, W.M.I. Hassan, M.A. Diab, S.M. Morgan, A.K. Elkholy, J. Mol. Liq. 212, 487–502 (2015)
M.S. Masoud, S.S. Hagagg, Alaa E. Ali, N.M. Nsar, J. Mol. Struct. 1014, 17–25 (2012)
X. He, J.J. Liu, H.M. Guo, M. Shao, M.X. Li, Polyhedron 29, 1062 (2010)
K.Y. El-Baredie, Monatsh. Chem. 136, 1139 (2005)
H. Liu, H. Wang, F. Gao, D. Niu, Z. Lu, J. Coord. Chem. 60, 2671–2678 (2007)
N. Venugopal, G. Krishnamurthy, H.S. Bhojyanaik, M. Giridhar, J. Mol. Struct. 1191, 85–94 (2019)
D.N. Sathyanarayana, Electronic Absorption Spectroscopy and Related Technique (University Press (India), Hyderabad, 2001)
Song H, Chen K, Tian, H, Dyes Pigm. 53, 257–262 (2002)
N. Lotfi, I. Sheikhshoaei, S.Y. Ebrahimipour, H. Krautscheid, J. Mol. Struct. 432, 1149 (2017)
W.H. Mahmoud, N.F. Mahmoud, G.G. Mohamed, Appl. Organomet. Chem. 1, 31 (2017)
I. Fleming, Frontier Orbitals and Organic Chemical Reactions (Wiley, New York, 1976)
W.H. Mahmoud, R.G. Deghadi, G.G. Mohamed, J. Therm. Anal. Calorim. 127(3), 2149 (2016)
P. Thanikaivelan, V. Subramanian, J.R. Rao, B.U. Nair, Chem. Phys. Lett. 59, 323 (2000)
K. Dhahagani, M.P. Kesavan, G.G.V. Kumar, L. Ravi, G. Rajagopal, J. Rajesh, Mater. Sci. Eng. (2018). https://doi.org/10.1016/j.msec.2018.04.032
N. Deepika, Y.P. Kumar, C.S. Devi, P.V. Reddy, A. Srishailam, S. Satyanarayana, J. Biol. Inorg. Chem. (2013). https://doi.org/10.1007/s00775-013-1018-0
J. Chen, X. Wang, Y. Shao, J. Zhu, Y. Zhu, Y. Li, Q. Xu, Z.J. Guo, Inorg. Chem. 46, 3306–3312 (2007)
R. Ramesh, S. Maheswaran, J. Inorg. Biochem. 9, 457–462 (2003)
K.R. Sangeetha Gowda, H.S. Bhojya Naik, B. Vinay-Kumar, C.N. Sudhamani, H.V. Sudeep, T.R. Ravikumar Naik, G. Krishnamurthy, Spectrochim. Acta A. 105, 229–237 (2013)
N. Dharmaraj, P. Viswanathamurthi, K. Nataraj, Trans. Met. Chem. 26, 105–109 (2001)
A.G. Bharathi Dileepan, T. Daniel Prakash, A. Ganesh Kumar, P. Shameela Rajam, V. Violet Dhayabaran, R. Rajaram, J. Photochem. Photobiol. Sci. (2018). https://doi.org/10.1016/j.jphotobiol.2018.04.029
M.A. Gyamfi, M. Yonamine, Y. Aniya, Gen. Pharmacol. 32, 661–667 (1999)
G. Marinova, V. Batchvarov, Bulg. J. Agric. Sci. 17, 11–24 (2011)
T. Manjuraj, G. Krishnamurthy, Y.D. Bodke, H.S. Bhojya-Naik, J. Mol. Struct. 1148, 231–237 (2017)
B. Koohshekan, A. Divsalar, M. Saiedifar, A.A. Saboury, B. Ghalandari, A. Gholamian, A. Seyedarabi, J. Mol. Liq. 216, 8–15 (2016)
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Venugopal, N., Krishnamurthy, G., Bhojya Naik, H.S. et al. DNA Binding, Molecular Docking and Antimicrobial Evaluation of Novel Azo Dye Ligand and Their Metal Complexes. J Inorg Organomet Polym 30, 2608–2625 (2020). https://doi.org/10.1007/s10904-019-01394-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10904-019-01394-8