Abstract
Recently, the 5-HT7 receptor has achieved greater attention in research fraternity due to the involvement of neurotransmitter serotonin (5-hydroxytryptamine, 5-HT) in several neurological disorders. Targeting this neuroreceptor, we have synthesized six compounds named as butyl-benzoxazolone substituted piperazinium derivatives (BBOP) derivatives, abbreviated as L1-L6. These compounds have been evaluated for their binding interaction with BSA through photophysical and in-silico approaches. The UV absorption of these compounds with BSA at λmax = 280 nm, showed an optical density (O.D.) in the range of 0.5–0.9, i.e., 21%-53% (L1max = 1.4, L5min = 0.7385) at varied concentrations (17 μM-114 μM). For fluorescence studies, the Ksv value varied inversely with temperature, which confirmed the static mechanism of quenching with L1 showing maximum quenching. The parameters (ΔH, ΔS) obtained from the thermodynamic study for interaction between BSA and L1-L6 were correlated with in-silico (molecular docking) data. The in-silico docking study showed hydrophobic and the Van der Waals forces were the most significant forces. Amino acid residues ARG 217 & TRP 213 (Sudlow Site I) and LYS 116 & GLU 125 (Sudlow Site II) of BSA were primarily involved in H-bonding.Furthermore, the catalytic activity of BSA for hydrolyzingdifferent chemical entities have monitored in the presence of L1-L6 through esterase-like assay with p-NPA as a substrate, to get more insight about the interaction with catalytic residues (LYS 414, LYS 413, and TYR 411) in BSA at site II. These findings showed the potential of these 5-HT7 markers as promising ligands with appropriate drug likeliness characteristics.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Among the receptors for the neurotransmitter serotonin, the most potential target which is involved in the treatment of variety of neuronal disorders is 5-HT7 [1, 2]. Numerous ligands with higher affinity for 5-HT7 receptors were reported but as these receptors share homology with other serotonin receptors so non-selectivity of the ligands remained a problem [3]. We proposed a pharmacophore model for ligands that may be active against 5-HT7 receptors based on the prior literature. It has a one sizable non-aromatic hydrophobic region with an H-bond donor group in addition to the three aromatic hydrophobic regions.
Arylpiperazine (AP) class of compounds are generally known to bind to serotonin receptors efficiently [4], whereas benzoxazolones have better pharmacological profile for targeting on central nervous system (CNS) receptors [5, 6]. In addition, the carbon-chain linker (which connects the two aromatic hydrophobic regions) with more than three carbons was discovered to have a better affinity for 5-HT7 receptors [7].
On the basis of these concepts, we synthesized ligands with an arylpiperazinyl moiety connected to the N-atom of the benzoxazolone moiety via a four-carbon alkyl chain spacer. The aromatic ring attached to the piperazine nucleus was substituted by hydroxy and methoxy groups at all possible three positions ('o'/'m'/'p'). The synthesis and binding efficacy have already been discussed in our previous work [8]. For the synthesis of same, we used methyliodide (CH3I) in the presence of a base for methylation. During this process, it was observed that excess use of CH3I is responsible for the synthesis of a new compound of the quarternary ammonium category. Hence, we obtained six new butyl-benzoxazolone substituted piperazinium derivatives (BBOP) derivatives abbreviated as L1-L6 as shown in Fig. 1.
The selectivity, duration, intensity, effectiveness, toxicity, and rate of drug elimination from the body are crucial factors that must be taken into consideration when using these derivatives for in-vivo studies and investigating their pharmacological profile. One of the most important aspects that determines how a newly developed drug candidate will be distributed throughout the body is how it interacts with a protein found in plasma., therefore we have taken serum protein for our study plan [9].
Serum albumin, which makes up 60% of the plasma protein content, is the primary protein for drug binding. Similar to how enzymes have specific sites for the substrate, it has structurally specific binding sites for drugs [10]. We chose BSA over human serum albumin (HSA) for this study due to the presence of two tryptophan residues, 76% sequence homology with the albumin protein found in humans, cost effectiveness, greater water solubility, and availability in its pure form. The BSA serves as a transporter protein (in-vivo) for most of the biologically active organic heterocyclics. BSA is a 583 amino acid residue cordate molecule with three homologous domains I, II, and III, these domains are further divided into subdomains A and B. Each molecule of BSA consists of six distinct binding sites for different constituents, two of them such as Sudlow site I and II are the most important for drug binding [11, 12]. We used a variety of spectroscopic methods (including absorption, fluorescence, and circular dichroism (CD) spectroscopy) and cheminformatics to visualize the binding mode of the synthesized ligands in order to observe how they interacted with BSA.
Experimental Section
Materials
BSA (fatty-acid free), Benzoxazolone, ‘o’/ ‘m’/ ‘p’- hydroxy/methoxyphehnyl piperazines were purchased from Sigma-Aldrich. Methyliodide, Methanol, Acetonitrile, K2CO3, 1,4-Dibromobutane, and Tetrahydrofuran were purchased from lab line traders, Lucknow. Hi-Media chemicals provided the PBS (phosphate buffer saline).
Preparation of Stock Solution
For photophysical study, BSA (15 μM) was prepared in 1X phosphate buffer solution. Synthesized compounds were dissolved in a small amount of methanol and making the requisite concentration by adding PBS. Each experiment was performed at a fixed concentration of BSA (15 μM) [13]. Compounds L1-L6 were synthesized by our group (Scheme 1).
Synthesis of N-methyl Salts of (arylpiperazinyl-butyl)benzoxazolone Derivatives (L1- L6)
The final compounds 4(a-f) were synthesized as per scheme 1, which consist of three different steps. Two different analogues (hydroxy and methoxy) of the designed compounds were synthesized in which first step involves the N-bromoalkylation of the benzoxazolone nucleus, in the presence of base. TLC (thin layer chromatography) was used to observe the progress of reaction and finally 60% yield of compound 2 was obtained. Further, coupling with hydroxy or methoxy substituted arylpiperazinyl moiety resulted in the formation of the corresponding analogues 3(a-f). The coupling reaction with methoxy substituted arylpiperazine moiety required the presence of base, however the reaction with hydroxyl substituted arylpiperazines occurred without base. Next step involved the methylation, in which for methoxy analogues there was only one site for substitution so reaction proceeded to give compounds 4(d-f) with 71–91% yield. For hydroxy derivatives, there are two possible sites for methylation, one is the hydroxyl group and another one is at the N-atom of piperazinyl moiety but in the absence of base methylation at hydroxyl group is not possible, so methylation at N-atom occurred resulting in the formation of compounds with 4(a-c) 82–94% yield.
Synthesis of Compound 2
To a constantly stirring solution of 1(7.4 mmol) in acetone, 1.2 eq. of K2CO3 was added, followed by the addition of 1,4-dibromobutane (8.88 mmol) after a 5 min interval, refluxed for 4 h, and cooled down gradually to the temperature of the surroundings. Furthermore, acetone was removed by using a rotatory evaporator, and the rest of the solution was extracted using DCM-water. The DCM layer was collected separately, washed with brine solution, and then any moisture present in it was removed by adding a drying agent, sodium sulfate. After the removal of moisture, the collected DCM layer was evaporated to obtain the concentrated residue.Then the purified compound was obtained from the concentrated residue after its purification (column chromatography) using a silica gel of 60–120 mesh. The solvent system used for purification was a mixture of 30% ethyl acetate and hexane.
Synthesis of Compound 3a-c
The compound2 was dissolved in anhydrous acetonitrile with constant stirring. To the stirred reaction mixture, the o/m/p-hydroxy phenylpiperazines (1.782 mmol) were added singly,refluxed for 36 h, and brought back to the normal surrounding temperature. The resulting solution was extracted with ethylacetate-water system. Any moisture present in the extracted solvent was removed by using anhydrous magnesium sulphate. After drying, the solvent was allowed to concentrate at lower pressure using rotavapor. After concentration the impure residue was purified by the same procedure as that of compound 2, but the solvent system used was methanol in chloroform. The purified compounds were characterized by 1H-NMR spectroscopy [8].
Synthesis of Compounds 3d-f
The synthetic procedure for compounds 3d-f followed the same procedure as that of 3a-c, except that the o/m/p-hydroxy phenylpiperazine (1.782 mmol) was substituted by the o/m/p-methoxy phenylpiperazine (1.782 mmol) and 1.2 eq. of K2CO3 was added and the reflux time was reduced for 5 h.
Synthesis of Compound (4a-c)
The compounds 3a-c (0.782 mmol) were dissolved in anhydrous acetonitrile, followed by the addition of methyl iodide with continual stirring for about 48 h. The protocol for purification and characterization (Shown in supplementary file, Fig. S1-S6), of the obtained residues were same as that discussed above.
1H-NMR (DMSO-d6, 500 MHz) (4a)—δ 1.78 (4H, m), δ 3.11(3H, s), δ 3.2(2H, m), δ 3.3(2H, m), δ3.53(7H, m), δ 3.9( 2H, t), δ6.79(2H, m), δ 6.89 (1H, t), δ 6.97 (1H,d), δ 7.15 (1H, t), δ 7.2 (1H, t), δ 7.3 (2H, t), δ 9.3 (1H, s).
IR (thin film) (4a) νmax: 3479, 2943, 2824, 1753, 1514, 1485, 1372, 1239 and 752 cm.−1
13C NMR (DMSO-d6, 100.6 MHz): δ153.91, 149.98, 142.05, 137.68, 130.94, 123.93, 123.85, 122.33, 119.35, 119.21, 115.74, 109.75, 109.26, 59.59, 43.31, 41.05, 24.25 and 18.27.
1H-NMR (DMSO-d6, 500 MHz) (4b)- δ 1.2 (2H, m), δ 1.77 (4H, m), δ 3.09 ( 3H, s), δ 3.50 (8H, m), δ 3.87 ( 2H, t), δ 6.3 (1H, d), δ 6.4 (1H, m), δ 6.44(1H, d), δ 7.03(1H, t), δ 7.14(1H, t), δ 7.23(1H, t), δ 7.4 (2H,d).
IR (KBr) νmax: 3440, 2925,2825, 1778,1663, 1498, 1239, 1023 and 750 cm.−1
13C NMR (DMSO-d6, 100.6 MHz): δ158.10, 153.90, 150.75, 142.03, 130.93, 129.73, 123.94, 122.32, 109.67, 109.27, 107.19, 106.72, 102.78, 61.87, 58.86, 46.18, 41.81, 41.06, 24.23 and 18.31.
1H-NMR (DMSO-d6, 500 MHz) (4c)- δ 1.9 (4H, m), δ 3.2 (3H, s), δ 3.45(2H, m), δ 3.7 (8H, m), δ 4.06 (2H, t), δ 6.8 (2H, t), δ 7.04 (2H, d), δ 7.33 (1H, m), δ 7.4 (1H, m), δ 7.5 (2H, d), δ 9.2 (1H, s).
IR (thin film) νmax: 3132, 2931, 1682, 1591, 1468, 1330, 1126 and 769 cm.−1
13C NMR (DMSO-d6, 100.6 MHz): δ153.91, 151.71, 142.25, 142.03, 130.98, 123.93, 122.31, 118.18, 115.53, 109.75, 109.25, 61.99, 59.22, 46.34, 43.48, 41.13, 24.17 and 18.28.
Synthesis of Compound 4d-f
To the stirred solution of compounds 3d-f(0.782 mmol) in acetonitrile, 5 eq. of methyl iodide was added at 0 ̊C and continued to stirring for 12 h at normal room temperature. The procedure for purification and characterization (Shown in supplementary file, Fig. S4-S6), of the obtained residues were same as that discussed above.
1H-NMR (DMSO-d6, 500 MHz) (4d)- (-δ 1.78 (5H, m), δ 3.11(3H, s), δ 3.28 (2H, m), δ 3.5 (7H, m), δ 3.8 (3H, s), δ 3.89(2H, t), δ 6.92 (2H, d), δ 6.99 (2H, d), δ 7.03 ( 1H, t), δ 7.15 (1H, t), δ 7.2 (1H, t), δ 7.3 (2H, m).
IR (thin film) νmax: 3299, 2922, 2817, 1780, 1661, 1498, 1240, 939 and 749 cm.−1
13C NMR (DMSO-d6, 100.6 MHz): δ153.90, 151.72, 142.16, 142.04, 138.99, 130.94, 123.92, 123.59, 122.31, 120.74, 118.54, 111.89, 109.75, 109.24, 59.53, 55.38, 43.30, 41.03, 24.24 and 18.25.
1H-NMR (DMSO-d6, 500 MHz) (4e)- δ 1.76 (4H, m), δ 3.1 (3H, s), δ 3.5 (10H, m), δ 3.7 (3H, s), δ 3.9 (2H, t), δ 3.9 (2H, t), δ 7.17 (2H, m), δ 7.2 (1H, t), δ 7.4 (2H, t).
IR (thin film) νmax: 3212, 2933, 1774, 1598, 1489, 1202, 968 and 756 cm.−1
13C NMR (DMSO-d6, 100.6 MHz): δ160.24, 153.89, 150.70, 142.03, 130.93, 129.85, 123.92, 122.31, 109.73, 109.25, 108.17, 105.12, 101.93, 61.78, 58.85, 55.04, 46.19, 41.74, 41.04, 24.24 and 18.28.
1H-NMR (DMSO-d6, 500 MHz) (4f)- δ 1.76 ( 4H, m), δ 3.1 (3H, s), δ 3.34 (2H, m), δ 3.50 (8H, m), δ 3.7 (3H, s), δ 3.9 (2H, t), δ 6.87 (2H, d), δ 6.97 (2H, d), δ 7.15 ( 1H, t), δ 7.2 (1H, t), δ 7.36 (2H, d).
IR (thin film) νmax: 3254, 2938, 2697, 1688, 1513, 1258, 836 and 752 cm.−1
13C NMR (DMSO-d6, 100.6 MHz): δ153.89, 153.58, 143.49, 142.04, 130.93, 123.92, 122.31, 117.69, 114.33, 109.74, 109.25, 61.92, 59.10, 55.24, 46.21, 43.02, 41.04, 24.25 and 18.28.
Absorbance Spectroscopy Measurements
The UV–visible spectra were recorded on a uv spectrophotomer (Specord 250 plus, Analytikjena, Germany). The spectra were scanned at 298.15 K, over a wavelength range of 200–450 nm. The reference of the experiment was set using the PBS buffer solution. The spectral measurement was taken by using a constant concentration of BSA (15 μM) against varying concentration of compounds L1-L6 (0-98 μM).Absorbance of each compounds were taken in triplicate [14].
Fluorescence Spectroscopic Study
Fluorescence spectra was taken on a FL6500 (Perkin Elmer) spectrophotometer with 1 cm optical path rectangular quartz cuvette. BSA fluorescence was monitored by changing the concentration of L1-L6 (0 to 114 µM) at a constant BSA concentration (15 μM). To conduct the experiments freshly prepared solutions were used. For eachexperiment, BSA was excited at a wavelength of 295 nm with slit width 2.5 nm, and the emission spectrum was measured between 305–450 nm [15]. The peak for emission spectrum was obtained at 340 nm. The fluorescence intensities of the respective L1-L6 compoundsin buffer solutions were recorded to eliminate the fluorescent background [16, 17]. The following equation was used to adjust all fluorescence data for the inner filter effect:
Here, Iobs and Icorrrepresent the observed and corrected fluorescence respectively. Ab1 represents the absorption fluorescent intensity at emission wavelength and Ab2 represents the absorption fluorescent intensity at excitation wavelength.
After subtracting the L1-L6 spectra from the BSA fluorescence spectra, quantitative analyses of the spectra were carried out. The Stern-volmer (S-V) plot was used to examine the spectral data obtained fluorescence quenching study:
The intensities of fluorescence of BSA, with and without synthesized derivatives were represented by F and Fₒrespectively. The synthesized derivatives act as quencher in this process. Ksv represents the constant known as Stern–Volmer constant, \({\uptau }_{^\circ }\) and Kqdenotes average life-time of protein alone (≈10–9 s) and rate constant of protein in the presence of quencher, and [Q] represents the concentration of synthesized derivatives (quencher).
Synchronous Fluorescence Spectroscopy
Synchronous spectra of protein (BSA) were recorded with band widths of 2.5 nm for varying concentrations of L1-L6 (0 to 114 μM). In this experiment, the spectra were scanned at a constant wavelength interval of Δλ = 15nm, which provides information about the surroundings of TYR residues, and Δλ = 60nm, which provides information about the proximity of TRP residues [18, 19]. The spectroscopic behavior of tyrosine and tryptophan residues of BSA alone was illustrated by the spectrum [20].
Esterase-Like Assay
p-Nitrophenylacetate (p-NPA) has been employed as a substrate to assess the esterase-like activity of BSA in the presence of synthesized derivatives. The absorption of the p-nitrophenol (the reaction product) at 405 nm was measured using a UV-Vis spectrophotometer (Analytik Jena specord-250 plus). The reaction mixture, maintained at 37°C and pH 7.4, included 15 µM BSA, p-NPA at concentrations of 50, 75, and 100 µM, and synthesized derivatives at 17, 51, and 114 µM. The molar extinction coefficient (ε) of p-NPA was considered to be 17,700 M-1 cm-1 [21]. Initial reaction velocities (V0) were determined from the slope of the graph between 0 and 2 minutes. Kinetic parameters were calculated using the Michaelis-Menten equation.
where, the symbols used have their usual meaning as reported in our previous work [14]. The value of Vm obtained from the above equation was used to calculate the Kcat by using the formula:
where, [P] = total concentration of BSA.
CD Spectroscopy
The CD spectrometer (Jasco J-815) was used for scanning the spectra of BSA alone and in the presence of compounds L1-L6. Using milliQ as a solvent, 10X phosphate buffer saline was diluted to 1X with a pH of 7.4. The CD measurement was taken with the successive addition of L1-L6(17, 66, 114μM) to a 15 μM BSA solution. The spectra were scanned over a wavelength ranging from 190 to 340 nm in a cuvette with a 300 μL capacity under continuous nitrogen supply. The scanning speed was set at 100nm/min. The value of mean residual ellipticity at 208 nm (MRE208) obtained from the equation 1 were used to determine the helical content of bound and free proteins [22, 23].
where Ɵ˳ represents observed CD (in milli deg units), the protein concentration was represented by the notion Cp, n is the number of amino acids in BSA, and the path length was represented by l in units of cm.
The α-helical content of protein was determined by using the formula:
where, 33000 and 4000 represents the MRE values of purely α-helical content of protein and β form with random cross at 208 nm [24, 25].
Molecular Docking
The glide module of Schrödinger maestro version 11.2 was used for docking studies. The BSA’s X-ray crystallized structure (PDB id- 4F5S) was imported from the database (http://www.rcsb.org/). The chemdraw software was used to draw the ligand structure. Before proceeding for docking analysis, the structure of both protein (imported from PDB database) and ligand were subjected to the energy minimization. For protein preparation, firstly the protein structure was optimized using the prepwiz module then it was preprocessed after assigning bond order and adding H-atoms. Finally, the entire protein structure was reduced to the lowest feasible energy state using the OPLS3 force field. The Schrodinger ligprep application was used to convert the 2D ligand structure into 3D. The docking study was performed thrice for each ligand and the observed RMSD value was found to be 0.3.Finally docking studies were conducted using the optimized low energy ligand conformations. The values of Glide scoring function was obtained by the equation:
Result and Discussion
For protein-ligand interaction studies, absorption as well as emission (fluorescence) spectroscopic techniques play a very important role. Absorption spectroscopy reveals the information about the groups present within the skeleton of molecules. It also explores the structural change as well as complex-formation during the interaction process. Similarly, spectrofluorimetry is useful in calculating the thermodynamics and equilibrium binding constant between the protein and the interacting drug molecule and also facilitates the calculation of the number of equivalent sets of binding sites present on the protein for drug binding. Through modified Stern-Volmer plots that were correlated with molecular docking and thermodynamic analysis, it also reveals details about the kinds of interaction forces involved in the interaction process.
Absorbance Spectroscopy Measurements
The spectral changes that the protein underwent as a result of the continuously increasing concentration of synthesized derivatives to the 15 μM BSA are depicted in Fig. 2. The maximum peaks for BSA alone, appears at 280 nm, which is due to the π-π* transition in the TRP and TYR residues [26, 27]. The absorption maxima of BSA enhanced in the presence of L1-L6 without any shift in peak position. The maximum increase in O.D. was observed for L1 i.e. between 0.7AU-1.4AU (53%), followed by L2 (0.5-0.9, 47%), L3 (0.6-0.97, 38%), L4 (0.5-0.84, 35%), L6 (0.6-0.9, 26%) and L5 (0.6-0.74, 21%). The increase in optical density demonstrated that compounds L1–L6, though to varying degrees, interacted with BSA. L1 displayed the highest level of interaction, while L5 displayed the lowest level. Furthermore, the addition of compounds L1-L6 increased the optical density of BSA, is confirmed by the inset figures of Fig. 2. This deduced that compounds L1–L6 interacted with BSA and changed the local polarity near the TRP and TYR residues. Fluorescence spectral analysis supported the observations from absorption spectroscopy.
Fluorescence Measurements
The three aromatic fluorophores which impart intrinsic fluorescence to BSA are W (TRP), Y (TYR), and P (PRO). However, W alone can be seen in the fluorescence spectrum for the majority of proteins [28, 29]. This is because P has a low quantum yield, while Y undergoes complete quenching if it is found next to a carbonyl group [30]. Excitation for W occurs at 295 nm, while emission for it is typically positioned around 340 nm [31]. This demonstrated that the interaction of BSA with synthetic derivatives involves W, not Y residue. The fluorescent intensity of BSA decreased gradually without any shift, except for L2 in which slight blue shift was observed by increasing the concentration of compounds L1-L6 (Fig. 3). The maximum decrease (quenching) was observed for L1 (38%), followed by L3 (33%), L2 (18%), L6 (17%), L5 (15%) and negligible quenching was observed in the case of L4 (2%). The quenching in fluorescent intensity suggests that the native structure of protein (BSA) disrupts and compounds L1-L5 may bind to the binding cavity of BSA molecule.
Finally, the study demonstrates that interactions with synthetic derivatives L1-L6 have a significant effect on BSA's intrinsic fluorescence, which is primarily caused by the TRP residue. The observed quenching in fluorescence intensity, particularly with L1, L3, and L2, suggests that these compounds disrupt BSA's native structure and likely bind to its binding cavity. Except for a slight blue shift with L2, there is no fluorescence shift, indicating that non-fluorescent complexes between the BSA and the derivatives have formed. These findings help us better understand the binding interactions between BSA and various synthetic compounds, emphasizing the importance of TRP in fluorescence-based protein interaction studies.
Binding Mechanism Analysis
The fluoroscence quenching for BSA was evaluated using the changes in emission maxima that occurred during interactions with L1-L6. Quenchers and fluorescent probes interacted in one of two ways: statically or dynamically. While the dynamic mechanism of quenching involves the approach of the fluorophore to the quencher in its excited state, the static type of mechanism involves the complex formation of drug with protein in the ground state [32]. The mechanism of quenching can be deduced by fitting the values of fluorescence emission spectra into the stern–volmer equation:
where, the symbols used have their usual meaning as described in Section "Fluorescence Spectroscopic Study".
There are basically two criteria for determining whether the mechanism of quenching followed by the system is dynamic or static in nature. First one is the relationship between temperature and stern–volmer constant (Ksv). If the value of Ksv varies directly with temperature, then the mechanism of quenching followed by the system is dynamic in nature, however the decreased value of Ksv with rise in temperature is indicative of static quenching [28]. Second one is based on the value of bimolecular collisional constant (Kq). The value of Kq is determined by the frequency of collisions between BSA (the fluorophore) and the quencher (L1-L6); the rate of diffusion, concentration, and size of the fluorophore and quencher all affect the frequency of collisions. It is evident that:
‘D’ denotes the overall diffusion coefficients of quencher and fluorophore, NA is the Avogadro’s number, ‘a’ denotes the sum of molecular radii. The maximum value of Kq for diffusion controlled process is 1010 M−1 s−1 [28, 33]. The higher value of Kq suggests that long–ranged interaction exists between L1-L6 and BSA and follows the static mechanism of quenching. The lower value of Kq suggests dynamic quenching.
The calculated value of Ksv and Kq (Fig. 4, Table 1), were found to be decreased with temperature of the system. For L1, on increasing the temperature by 10̊ C, the Ksv value was decreased by 8.26 M−1. Similarly, for rest of the five compounds such as L2, L3, L4, L5, and L6, the decrease in Ksv value was observed to be 8.48, 9.74, 8.57, 9.44 and 2.84 M−1 respectively. The change in Ksv value indicates the sensitivity of compound towards BSA. So, L3 is the most sensitive compound with maximum change in Ksv value and L6 with minimum change was observed to be the least sensitive compound. The Kq value obtained for all the six compounds was of the order of 1011, which was higher than the maximum limit of the diffusion controlled process, thus confirming the nature of quenching as static.
Analysis of Binding Interactions
The binding parameters such as Kb and n (Table 1) were calculated by using the equation:
where, Kb represents the binding constant and the number of binding sites is represented by n. If the value of Kb is higher than order 105, then it means that the interaction between drug / ligands with BSA is strong enough so that they are less freely available in plasma for in-vivo studies. On the other hand, if the value of Kb is less than order 103, then the interaction is very weak which leads to its poor distribution or bioavailability. The study revealed that with rise in temperature the values of n and Kb decreased which might be due to less stable complexation between L1-L6 with BSA. The binding constant of L1-L6 varied between 102–104 indicating a mild interaction with BSA which might be leads to their poor distribution in the body with low biological half life.
The obtained value of n was close to unity, suggested there might be single binding site on protein for L1-L6 binding, and it also suggested that these derivatives might bind to the hydrophobic pocket of BSA and TRP 212 which is found in the subdomain IIA and responsible for the conformational changes in protein structure. The lower value of n at higher temperatures implied that the system's entropy and vibrational motion increased with temperature, and that it may also have a higher value for the coefficient of diffusion, all of which contributed to the complex's decreased stability (Fig. 5).
Thermodynamic Parameters
Non-covalent molecular interactions are primarily the driving forces for interaction between a protein and a drug. H-bonding, hydrophobic interactions, the Van der Waals force of attraction, and electrostatic interaction are examples of non-covalent forces [34]. The thermodynamic variables were calculated with the help of Van'tHoff equation [35]:
where, R is a constant known as gaseous constant. The values of ΔH and ΔS were obtained from the linear plot of 2.303R Log K vs \(1/T\), following the straight line equation; y = mx + c (supplementary fig. S7). After obtaining these values the Van't Hoff equation was applied to get the change in Gibbs free energy (ΔG).
The magnitude and sign of ΔH and ΔS decides the type of interaction. The negative change in both the enthalpy as well as entropy signifies that among the non-covalent forces, the Vander Waals and H-bonding were mainly involved in the process of interaction. However, the positive contribution of enthalpy and entropy change justifies the hydrophobic interactions. Electrostatic forces are the most important forces when both the conditions are favourable i.e. ΔH is negative and ΔS is positive. The –ive value of ΔG denotes spontaneous process.
This study deduced that both the enthalpy as well as entropy changes were negative, it means the H-bonding along with Van der Waals force of attraction were involved in the binding mechanism. The negative value of ΔG for L1, L5 & L6, signifies that the process is spontaneous and is enthalpically driven. For L2, L3, and L4 the sign of ΔG were positive which suggested that hydrophobic interaction forces were also involved binding interaction analysis and process is entropically driven (TΔS > ΔH) (Table 2).
Synchronous Fluorescence Spectroscopy
The synchronous fluorescence spectra of BBOP derivatives are shown in the Fig. 6a and b. As already discussed in Section 4.2.5, the synchronous spectra provides the information about the environment of TRP and TYR residues, so here firstly we have discussed about the changes around TYR residues and then around the TRP residues. The red shift along with quenching means increase in polarity and decrease in hydrophobicity around the TYR and TRP residues. Blue shift shows the case opposite to that in red shift (decrease in polarity and increase in hydrophobicity). However, when quenching is not accompanied by shift, then there might be interaction between protein and drug which is not responsible for the conformational changes in protein [36, 37].
a Synchronous fluorescence spectra of BSA (15 μM) with BBOP derivatives [L1(A), L2(B), L3(C), L4(D), L5(E) and L6(F)] at Δλ = 15 nm (at varied concentrations 0-114 μM). b Synchronous fluorescence spectra of BSA (15 μM) with BBOP derivatives [L1(A), L2(B), L3(C), L4(D), L5(E) and L6(F)] at Δλ = 60 nm (at varied concentrations 0-114 μM)
Now, for TYR residues (Fig. 6a), the presence of L1 and L2 showed the red-shift in peak position. Compounds L3-L5, on the other hand, showed quenching without any shift, and L6 showed a blue shift. The extent of quenching shown by L1, L2, L3, L4, L5, and L6 were 26% 33%, 27%, 11%, 23% and 29%, respectively. These results suggest that the interaction of BSA with L1 and L2 resulted in the increased polarity around TYR, which might be due to the disruption of the dense hydrophobic binding pocket of BSA. We can also say that the interaction with L1 and L2 resulted in increased hydrophilicity of the protein. In addition, BSA interacted with L3-L5 without undergoing any conformational changes. Similarly, L6 created a more non-polar environment around TYR residues.
Besides that for TRP residues (Fig. 6b), L1 and L4 showed red-shift, L2, L3, and L6 showed blue shift and L5 showed no shift. The quenching for TRP residues were found to be approximately 55%, 12%, 16%, 15%, 58% and 83% for L1, L2, L3, L4, L5 and L6 respectively.
From the above discussion it can be concluded that, BSA interaction with L1 and L2 causes a red shift in TYR residues, indicating increased polarity and protein hydrophilicity. Compounds L3-L5 cause fluorescence quenching with no shift, indicating binding without significant conformational changes, whereas L6 causes a blue shift, indicating a more nonpolar environment around TYR. For TRP residues, L1 and L4 cause a red shift, L2, L3, and L6 cause a blue shift, and L5 causes no shift. The varying quenching effects, particularly with L1 and L6, highlight distinct interaction dynamics and have a significant impact on BSA's native structure.
Esterase-Like Assay
Compounds (L1-L6) were evaluated to assess their effects on the esterase-like activity of BSA. The residues LYS 414, LYS 413, and TYR 411, located in site II (subdomain IIIA) of BSA, are responsible for its catalytic activity with p-NPA as the substrate. It is suggested that interactions with synthesized derivatives can alter the local environment around the active center, potentially changing the esterase-like activity of albumin. This activity decreases when ligands bind to the protein in a way that makes the catalytic residues (TYR 411, ARG 410, LYS 413, and LYS 414) less accessible to the substrate. Conversely, the activity increases when ligand binding makes these residues more accessible. These changes were observed through spectral analysis and the data were analyzed using the Michaelis–Menten curve (Fig. 7) and a Lineweaver–Burk plot (Fig. 8), which linearizes the data for increased accuracy. The resulting values are presented in Table 3 [38, 39]. The presence of compound L1 reduced the catalytic efficiency of BSA, but this effect did not vary with increasing concentrations of L1. Similarly, compound L2 caused a decrease in catalytic activity at all tested concentrations (17, 51, 114 μM). In this case, the Km value remained constant at 70 μM, while variations in Vmax were observed, suggesting that compound L2 might inhibit BSA activity non-competitively. Compounds L3 and L5 increased the catalytic activity of BSA at concentrations of 17, 51, and 114 μM, with the maximum increase observed at 17 μM. However, further increases in concentration to 51 μM and 114 μM led to a gradual reduction in catalytic efficiency. Compound L4 slightly reduced catalytic activity at lower concentrations, but showed a significant increase (39.1 μM−1 s−1) at 114 μM. Similarly, compound L6 showed the highest increase in hydrolysis activity at 51 μM, with reduced activity at both higher and lower concentrations. In conclusion, the concentration of these compounds plays a significant role in determining their function in esterase-like activity, acting as either inhibitors or activators depending on the concentration (Scheme 2).
CD Spectroscopy
This technique is used for determining protein secondary structure [40]. CD was used to measure the alterations in arrangement of the BSA polypeptides in the presence of ligands, and the outcomes at varying concentrations of L1-L6 are shown in Fig. 9. On binding of L1-L6, the various inter and intra molecular forces involved in secondary and tertiary structure of proteins get disrupted. The circular dichroism spectra of BSA showed two peaks, one at 208 nm due to π-π*transition and another at 222 nm due to n-π* transition. These two negative absorption peaks represent the α-helical pattern of BSA [41]. Secondary structure constituents of globular proteins (BSA) include the α-helix, turns, β-sheet and random coil. On addition of L1-L6, the α-helical content of BSA increased except for L6 at a concentration of 114 μM where it dropped to 15.4%. The increase in α-helical is indicative of increased ellipticity at peak position. When concentration of L1-L6 increased from 17 μM to 66 μM, then α-helical content decreased for all the derivatives and hence the ellipticity, except for L4 where increase in ellipticity occurred. On further increasing the concentration of L1-L6, from 66 μM to 114 μM the β-sheet content of BSA increased (except for L5 and L6), which signifies that L1-L6 binds effectively with BSA. It is also noticeable, that L1-L6 induces conformational changes in secondary structure of BSA. It might be possible that L1-L6 binds with the hydrophobic pocket of BSA and distort the hellicity of BSA. The maximum change was observed in L5 and L1 showed a minimal change (Table 4).
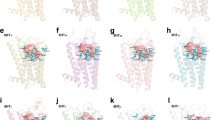
Molecular Docking
A molecular docking is an essential component of cheminformatics, which assists the visualization of orientation of the ligands in drug binding site [42]. The role of important amino acid residues in the binding of BBOP at the different places of BSA is shown in Fig. 10. For Sudlow site I, the common amino acid residues PRO 338, TYR 340, PRO 446, CYS 447 in L1, L4, L5, L6 and residues TYR 149, TYR 156, TRP 213, LEU 218, LEU 237, ILE 289, ALA 290 L2, L3 in L2 and L3 were involved in hydrophobic interactions. The common residues involved in these ligands indicate that it might be possible that the orientation of L1, L4, L5, L6 and L2&L3 were nearly same in hydrophobic pocket of BSA. Also, ARG 194, GLU 293, LYS 294, GLU 443, ASP 450 in L1, L4, L5, L6 and ARG 194, ARG 217, HIP 287, GLU 291 in L2 & L3 were involved in ionic interactions. In addition to hydrophobic and ionic interactions, the residues ASP 450, ARG 217, GLU 291, VAL 342, TRP 213 were involve also stabilizing the complex through H-bond formation. At Sudlow Site II, the common residues involved in hydrophobic interactions were LEU 112, PRO 113, LEU 115, LEU 122, PHE 133, TYR 137, TYR 160 and in ionic interactions were GLU 125, LYS 136, and in H-bonding were GLU 140, LYS 114, GLU 140, LYS 116, PHE 36.. The values of glide score obtained at site I were (-3.845)-(-5.906) KJ/mol and site II were (-4.196)-(-5.412) KJ/mol respectively, as shown in Tables 5 and 6. This suggested that L2 at site I and L3 at site II showed maximum interaction. The above discussion revealed the fact that hydrophobic interactions along with ionic and H-bonding were the main forces involved in binding interaction analysis. These results were consistent with the experimental results which already showed involvement of non-covalent forces in binding process [43, 44].
Conclusion
We have synthesized a total of six Butyl-benzoxazolone substituted piperazinium (BBOP) derivatives on the basis of hypothesized pharmacophore model targeting 5-HT7 receptors. To explore the possible potential to work as drug in future, we performed their pharmacokinetic study via interaction with carrier protein BSA to see its bioavailability. The photophysical data revealed that hydrophobic forces were the major driving force in the interaction process between the BSA and synthesized compounds. These results were confirmed by the thermodynamic and in-silicostudies. Circular dichroism experiments were conducted to show the secondary structural changes in BSA on interaction with synthesized compounds. These results will be helpful in understanding the pharmacokinetics of BBOP derivatives before going to clinical studies.
Data Availability
No datasets were generated or analysed during the current study.
References
Hedlund PB (2009) The 5-HT7 receptor and disorders of the nervous system: An overview. Psychopharmacology 206:345–354. https://doi.org/10.1007/S00213-009-1626-0
Jaeschke RR, Sowa-Kućma M, Pańczyszyn-Trzewik P, Misztak P, Styczeń K, Datka W (2016) Lurasidone: The 2016 update on the pharmacology, efficacy and safety profile. Pharmacol Reports 68:748–755. https://doi.org/10.1016/J.PHAREP.2016.04.002
Pittalà V, Salerno L, Modica M, Siracusa M, Siracusa MA, Romeo G (2007) 5-HT7 receptor ligands: recent developments and potential therapeutic applications. Ingentaconnect Com 7:945–960. https://doi.org/10.2174/138955707781662663
Bielenica A, Koziol A, Struga M (2013) Binding Modes of Chain Arylpiperazines to 5-HT1a, 5-HT2a and 5-HT7 Receptors, Mini-Reviews. Med Chem 13:1516–1539. https://doi.org/10.2174/1389557511313100012
Salerno L, Pittalà V, Modica MN, Siracusa MA, Intagliata S, Cagnotto A, Salmona M, Kurczab R, Bojarski AJ, Romeo G (2014) Structure-activity relationships and molecular modeling studies of novel arylpiperazinylalkyl 2-benzoxazolones and 2-benzothiazolones as 5-HT7 and 5-HT1A receptor ligands. Eur J Med Chem 85:716–726. https://doi.org/10.1016/j.ejmech.2014.08.023
Poupaert J, Carato P, Colacino E (2005) 2(3H)-Benzoxazolone and Bioisosters as “privileged scaffold” in the Design of Pharmacological Probes. Curr Med Chem 12:877–885. https://doi.org/10.2174/0929867053507388
Rague A, Tidgewell K (2018) Pharmacophore Comparison and Development of Recently Discovered Long Chain Arylpiperazine and Sulfonamide Based 5-HT7 Ligands, Mini-Reviews. Med Chem 18:552–560. https://doi.org/10.2174/1389557517666170913111533
Kumari N, Adhikari A, Singh D, Bhagat S, Ojha H, Tiwari AK (2022) Benzoxazolone-arylpiperazinyl scaffold-based PET ligand for 5-HT7: Synthesis and biological evaluation. Drug Dev Res 83:1024–1033. https://doi.org/10.1002/ddr.21930
Levitt DG, Levitt MD (2016) Human serum albumin homeostasis: A new look at the roles of synthesis, catabolism, renal and gastrointestinal excretion, and the clinical value of serum albumin measurements. Int J Gen Med 9:229–255. https://doi.org/10.2147/IJGM.S102819
Papadopoulou A, Green RJ, Frazier RA (2005) Interaction of flavonoids with bovine serum albumin: a fluorescence quenching study. J Agric Food Chem 53:158–163. https://doi.org/10.1021/JF048693G
Briand C, Sarrazin M, Peyrot V, Gilli R, Bourdeaux M, Sari JC (1982) Study of the interaction between human serum albumin and some cephalosporins. Mol Pharmacol 21:92–99. https://pubmed.ncbi.nlm.nih.gov/7132963/. Accessed 15 Oct 2022
Bhattacharya AA, Curry S, Franks NP (2000) Binding of the General Anesthetics Propofol and Halothane to Human Serum Albumin: high resolution crystal structures. J Biol Chem 275:38731–38738. https://doi.org/10.1074/JBC.M005460200
Rahman AJ, Sharma D, Kumar D, Pathak M, Singh A, Kumar V, Chawla R, Ojha H (2021) Spectroscopic and molecular modelling study of binding mechanism of bovine serum albumin with phosmet. Spectrochim Acta - Part A Mol Biomol Spectrosc. 244:118803. https://doi.org/10.1016/j.saa.2020.118803
Singh D, Kaur L, Rahman AJ, Singh P, Tiwari AK, Ojha H (2022) Binding and mechanistic studies of 5-HT7 specific benzothiazolone derivatives with bovine serum albumin: Spectroscopic and in silico studies. J Mol Liq 366:120143. https://doi.org/10.1016/j.molliq.2022.120143
Ali MS, Al-Lohedan HA (2020) Spectroscopic and Molecular Docking Investigation on the Noncovalent Interaction of Lysozyme with Saffron Constituent “safranal.” ACS Omega 5:9131–9141. https://doi.org/10.1021/acsomega.9b04291
Shi JH, Zhou KL, Lou YY, Pan DQ (2018) Multi-spectroscopic and molecular docking studies on the interaction of darunavir, a HIV protease inhibitor with calf thymus DNA. Spectrochim Acta - Part A Mol Biomol Spectrosc 193:14–22. https://doi.org/10.1016/j.saa.2017.11.061
Wang BL, Pan DQ, Zhou KL, Lou YY, Shi JH (2019) Multi-spectroscopic approaches and molecular simulation research of the intermolecular interaction between the angiotensin-converting enzyme inhibitor (ACE inhibitor) benazepril and bovine serum albumin (BSA). Spectrochim Acta - Part A Mol Biomol Spectrosc 212:15–24. https://doi.org/10.1016/j.saa.2018.12.040
Asadi Z, Mosallaei H, Sedaghat M, Yousefi R (2017) Competitive binding affinity of two lanthanum(III) macrocycle complexes toward DNA and bovine serum albumin in water. J Iran Chem Soc 14:2367–2385. https://doi.org/10.1007/s13738-017-1172-3
Afrin S, Rahman Y, Alhaji Isa M, Ahmed S, Tabish M (2020) Biophysical insights into the binding characteristics of bovine serum albumin with dipyridamole and the influence of molecular interaction with β cyclodextrin. J Biomol Struct Dyn. 38:3046–3058. https://doi.org/10.1080/07391102.2019.1651220
Ghosh N, Mondal R, Mukherjee S (2015) Hydrophobicity is the governing factor in the interaction of human serum albumin with bile salts. Langmuir 31:1095–1104. https://doi.org/10.1021/la504270a
Satish L, Millan S, Sasidharan VV, Sahoo H (2018) Molecular level insight into the effect of triethyloctylammonium bromide on the structure, thermal stability, and activity of Bovine serum albumin. Int J Biol Macromol 107:186–193. https://doi.org/10.1016/j.ijbiomac.2017.08.157
Tabassum S, Al-Asbahy WM, Afzal M, Arjmand F (2012) Synthesis, characterization and interaction studies of copper based drug with Human Serum Albumin (HSA): Spectroscopic and molecular docking investigations. J Photochem Photobiol B Biol 114:132–139. https://doi.org/10.1016/j.jphotobiol.2012.05.021
Sun Q, He J, Yang H, Li S, Zhao L, Li H (2017) Analysis of binding properties and interaction of thiabendazole and its metabolite with human serum albumin via multiple spectroscopic methods. Food Chem 233:190–196. https://doi.org/10.1016/j.foodchem.2017.04.119
Ariga GG, Naik PN, Chimatadar SA, Nandibewoor ST (2017) Interactions between epinastine and human serum albumin: Investigation by fluorescence, UV–vis, FT–IR, CD, lifetime measurement and molecular docking. J Mol Struct 1137:485–494. https://doi.org/10.1016/j.molstruc.2016.12.066
Bi S, Song D, Tian Y, Zhou X, Liu Z, Zhang H (2005) Molecular spectroscopic study on the interaction of tetracyclines with serum albumins. Spectrochim Acta - Part A Mol Biomol Spectrosc. 61:629–636. https://doi.org/10.1016/j.saa.2004.05.028
Duan ST, Liu BS, Li TT, Cui MM (2017) Study of the interaction of cefonicid sodium with bovine serum albumin by fluorescence spectroscopy. J Appl Spectrosc 84:431–438. https://doi.org/10.1007/s10812-017-0488-1
Khatun S, Riyazuddeen S, Yasmeen A, Kumar N (2018) Subbarao, Calorimetric, spectroscopic and molecular modelling insight into the interaction of gallic acid with bovine serum albumin. J Chem Thermodyn 122:85–94. https://doi.org/10.1016/j.jct.2018.03.004
Lakowicz R (2006) Principles of fluorescence spectroscopy.https://doi.org/10.1007/978-0-387-46312-4
Shi JH, Pan DQ, Jiang M, Liu TT, Wang Q (2017) In vitro study on binding interaction of quinapril with bovine serum albumin (BSA) using multi-spectroscopic and molecular docking methods. J Biomol Struct Dyn 35:2211–2223. https://doi.org/10.1080/07391102.2016.1213663
Sułkowska A (2002) Interaction of drugs with bovine and human serum albumin. J Mol Struct 227–232. https://doi.org/10.1016/S0022-2860(02)00256-9
Štefl M, James NG, Ross JA, Jameson DM (2011) Applications of phasors to in vitro time-resolved fluorescence measurements. Anal Biochem 410:62–69. https://doi.org/10.1016/j.ab.2010.11.010
Siddiqui S, Ameen F, Kausar T, Nayeem SM, Ur Rehman S, Tabish M (2021) Biophysical insight into the binding mechanism of doxofylline to bovine serum albumin: An in vitro and in silico approach. Spectrochim Acta - Part A Mol Biomol Spectrosc 249. https://doi.org/10.1016/j.saa.2020.119296
Chu Q, Bao B, Wu W (2018) Minireview mechanism of interaction between phenolic compounds and proteins based on non-covalent and covalent interactions minireview. Med Res 2:1–6. https://doi.org/10.21127/yaoyimr20180014
Chen WY, Huang HM, Lin CC, Lin FY, Chan YC (2003) Effect of temperature on hydrophobie interaction between proteins and hydrophobic adsorbents: studies by isothermal titration calorimetry and the van’t hoff equation. Langmuir 19:9395–9403. https://doi.org/10.1021/la034783o
Chen YC, Wang HM, Niu QX, Ye DY, Liang GW (2016) Binding between saikosaponin C and human serum albumin by fluorescence spectroscopy and molecular docking. Molecules 21. https://doi.org/10.3390/molecules21020153
Gandhi S, Roy I (2019) Synthesis and characterization of manganese ferrite nanoparticles, and its interaction with bovine serum albumin: A spectroscopic and molecular docking approach. J Mol Liq 296:111871. https://doi.org/10.1016/j.molliq.2019.111871
Bhattacharya AA, Grüne T, Curry S (2000) Crystallographic analysis reveals common modes of binding of medium and long-chain fatty acids to human serum albumin. J Mol Biol 303:721–732. https://doi.org/10.1006/jmbi.2000.4158
Wang Y, Zhang H, Kang Y, Cao J (2016) Effects of perfluorooctane sulfonate on the conformation and activity of bovine serum albumin. J Photochem Photobiol B Biol 159:66–73. https://doi.org/10.1016/j.jphotobiol.2016.03.024
Kragh-Hansen U (1981) Molecular aspects of ligand binding to serum albumin. Pharmacol. Rev 33:17–53. https://pharmrev.aspetjournals.org/content/33/1/17.short. Accessed 8 Aug 2022
Trynda-Lemiesz L, Karaczyn A, Keppler BK, Kozlowski H (2000) Studies on the interactions between human serum albumin and trans-indazolium (bisindazole) tetrachlororuthenate(III). J Inorg Biochem 78:341–346. https://doi.org/10.1016/S0162-0134(00)00062-3
Lu Y, Cui F, Fan J, Yang Y, Yao X, Li J (2009) Interaction of human serum albumin with N-(4-ethoxyphenyl)-N′-(4-antipyrinyl) thiourea using spectroscopies and molecular modeling method. J Lumin 129:734–740. https://doi.org/10.1016/j.jlumin.2009.02.005
Chadha N, Tiwari AK, Kumar V, Milton MD, Mishra AK (2015) In silico thermodynamics stability change analysis involved in BH4 responsive mutations in phenylalanine hydroxylase: QM/MM and MD simulations analysis. J Biomol Struct Dyn 33:573–583. https://doi.org/10.1080/07391102.2014.896240. 2015.s, Pak. J. Pharm. Sci. 31 (2018) 2645–2654
Miller JN (1984) Recent developments in fluorescence and chemiluminescence analysis: Plenary lecture. Analyst 109:191–198. https://doi.org/10.1039/AN9840900191
Bujacz A (2012) Structures of bovine, equine and leporine serum albumin. Acta Crystallogr Sect D Biol Crystallogr 68:1278–1289. https://doi.org/10.1107/S0907444912027047
Acknowledgements
We thank Department of Chemistry, B B A University for providing computational studies.
Funding
CO/P/FP/G85/2020-NCSTC DST (Department of Science and Technology, Gov. of India).
Author information
Authors and Affiliations
Contributions
DS excuted the experiments with VKS.NK was involved in synthesis and characterization. HO wrote the manuscript and AKT supervised and analysed all the work
Corresponding authors
Ethics declarations
Consent for Publication
All the authors of the manuscript have approved this manuscript for submission.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Singh, D., Singh, V.K., Kumari, N. et al. Exploring the Binding Mechanism of 5-HT7 Specific Benzoxazolone alkyl Piperazinium Derivatives: A Comprehensive Analysis Using Spectroscopic and Computational Approaches. J Fluoresc (2024). https://doi.org/10.1007/s10895-024-03846-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10895-024-03846-y