Abstract
AML is a highly aggressive malignant clonal disease of hematopoietic origin. Hesperidin as a polyphenol glycoside, Activates the apoptotic pathway and salinomycin as a k + selective ionophore. We examined how hesperidin and salinomycin induce pro-apoptotic effects in KG1a cells. Cells were divided into four groups; 1) control cells (CRTL), 2) cells treated with hesperidin 85 μM, 3) cells treated with 2 μM salinomycin, 4) cells treated with combination of salinomycin and hesperidin. The MTT assay was implemented to determine the IC50 of hesperidin and salinomycin in KG1a cell lines. Propidium iodide staining and flow cytometry were used to analyze the distribution of the cell cycle. The level of ROS was evaluated by fluorescent microscopy and spectrophotometry. Additionally, Akt, XIAP, Bad, and FOXO1 gene expression was analyzed by real-time PCR. Hesperidin/Salinomycin decreased the viability of KG1a leukemic cells more than Hesperidin and Salinomycin separately. Changes in the shape of apoptotic cells and rise in ROS levels were detected after Hesperidin/Salinomycin treatment. Our findings showed that following Hesperidin/Salinomycin treatment, the expression of PI3K/AKT signaling pathway related genes (AKT, PTEN and FOXO1), were in line with the destruction of KG-1a cells. Furthermore, XIAP and BAD mRNA were regulated to trigger apoptosis in cancer cells. The study discovered that hesperidin and salinomycin, could effectively hinder the PI3K/Akt signaling pathway in leukemia cancer cells. Also, the combination of hesperidin and salinomycin has the potential to be a treatment option for acute myeloid leukemia.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Unregulated growth of hematopoietic stem cells in the bone marrow characterizes AMLFootnote 1 as a diverse group of diseases [1]. long-term survival rates of the disease is 30% 1 [2]. Malignant cell growth is recognized by the interference of usual intracellular signaling due to mutations or abnormal external signaling [3]. Intracellular pathways, including the PI3K-AktFootnote 2 pathway, are aberrantly upregulated in cancers such as AML.
One of the factors in this pathway identified as Akt can operate as a proto-oncogene. Avoidance from apoptosis is a necessary step for cancer tumor advancement [4]. One of the exclusive BH3 proteins, BADFootnote 3 plays a role in mitochondrial permeabilization in the apoptotic pathway, which might be over-expressed in cancer cells [5].
FOXFootnote 4 proteins are a superfamily of evolutionarily conserved transcriptional factors that take part in metabolic processes, differentiation, proliferation, cell cycle progression, apoptosis and migration. They inhibit multiple genes involved in cellular processes such as BAD in apoptosis. In this study, we investigate the FOXO1 subfamily. Phospho-FOXO1 Identified in most AML patients due to the phosphorylation of AKT1 signaling [6].
There is another tumor suppressor protein PTEN,Footnote 5 that combats tumor cells and reduces their survival [7]. The important contribution of this tumor suppressor in the progression of myeloid malignancies has been unveiled [8]. Human cancers often display overexpression of XIAP,Footnote 6 which could enable them to evade apoptosis and promote tumorigenesis [9, 10]. One of the drugs that is selected in this study is SALFootnote 7 which impacts the membrane by acting as an ionophore. It exhibits affinity for alkali ions, particularly for potassium ion [11]. SAL possesses anti-neoplastic features in various human cancers, including hematological malignancies [12]. HESPFootnote 8 (7-rutinoside) is a flavonoid in citrus fruits with anti-tumor features. This natural product Stimulates the apoptotic pathway, which has the potential to assist in fighting cancer cells [13]. So far, no research has been done on the effects of HESP/SAL combination on PI3K/Akt signaling pathway and apoptosis in KG-1a cells, so we studied this issue to make our hypothesis come true.
Material and Methods
Materials
We selected a promyeloblast macrophage cell line known as KG-1a was obtained from the bone marrow of a male White patient, age 59, who was suffering from acute myelogenous leukemia. The Pasteur Institute of Iran (Tehran, Iran) provided KG1a cell lines. Gibco (Life Technologies, USA) provided the RPMI-1640 cell culture medium. Fetal bovine serum (FBS) was obtained from PAN Biowest (France). The culture plates were acquired from SPL (City, South Korea). materials to carry out the cell cycle test, PI-propidium iodide and MTT tests (using DMSO-Dimethyl sulfoxide, 3-(4, 5-Dimethyl-2-thiazolyl-2, 5-diphenyl-2-tetrazolium bromide), and also Hoechst 33342, were acquired from Sigma Aldrich (St. Louis, Missouri, USA). Cellular ROS Assay Kit, was provided by Abcam (Japan). The RNA isolation kit was purchased from Sinaclon (Iran). Real-Time PCR requires Real Q Plus 2 × Master Mix Green High ROX™, which was obtained from AmpliQon (Denmark). For cDNA synthesis, the Prime Script™ RT reagent kit (Takara, Japan) was utilized in this study.
Cell Culture
The culture flasks with a capacity of 25 cm2 contained acute myeloid leukemia (AML) KG1-a cells which were cultured in RPMI 1640 medium supplemented with 10% FBS and penicillin/streptomycin. The standard conditions for culturing cells included a humidified atmosphere, 5% CO2, and a temperature of 37 °C.
Determination of Cytotoxicity through MTT Assay
To determine the IC50 value in cells and study the impact of HESP and SAL on cancer cell viability, the MTTFootnote 9 colorimetric assay was conducted. HESP (concentrations ranging from 10 to 150 μM) and SAL (concentrations ranging from 1 to 10 μM) were used to treat KG1a cells and (2 × 104) cells in each well of 96-well plates for 24, 48 and 72 hours were cultured. Following the IC50 calculation, another MTT experiment was done to study the effects of HESP/SAL combination on KG1a cells. Following the assay, 20 μl of MTT (5 mg/mL) was added to each well and left to incubate for 4 hours at 37 °C. DMSO solution was added after the contents of each well were drained. After shaking the plate slowly for 15 minutes, the absorbance was measured at 570 nM with a multi-well plate reader (Quant BioTek Instruments, USA) [9, 10]. Using GraphPad Prism 9.0.0 software (Graph Pad Software, CA, USA), the IC50 values were calculated from the sigmoidal dose-response graph.
Analysis of the Morphology of Apoptotic Cells Using Hoechst 33258 Staining
First KG1a cells were divided to four treatment groups (Control, 85 μM HESP, 2 μM SAL and SAL/HESP). 12-well plates seeded with 5 × 105 KG1a cells for 48 hours. The next step was to harvest the cells and rinse them with cold phosphate-buffered saline (PBS). Next, cells were treated with Hoechst 33258 solution (1 mg/mL in ddH2O) and the final concentration was adjusted to 100 μg/mL. The cellular morphology was examined using fluorescence microscopy (EUM-5000 FLCD, Labex Instrument).
Cellular ROS Detection Assay
At the beginning of the process cells were placed on a 96 well plate and incubated to attach overnight. In the following stage, cells were treated for 48 hours. Next, the cells were rinsed in 1X Buffer and stained with 25 μM DCFDAFootnote 10 in 1X Buffer for 45 min at 37 °C for triple time. After wards washing with 1X Buffer was executed repeatedly between staining steps. Ultimately, cells were washed in 1X Buffer and changes were determined as a percentage of control after background subtraction. The nucleus of the cells was stained with DAPIFootnote 11 as well. The cells were observed under microscope for DCF fluorescence at the excitation and emission wavelengths of 535/485 nm (green filter) and for DAPI fluorescence at the excitation and emission wavelengths of 457/357 nm (blue filter). the fluorescence emissions were evaluated by the FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA). The provided data were analyzed using FlowJo software (Ashland, OR, USA).
PI Staining and Flow Cytometry-Based Analysis of Cell Cycle Distribution
PI staining method was utilized to estimate the distribution of cells across different cell cycle phases. For each of the four cell groups, 5 × 105 cells were cultured using this method. Following treatment for 48 hours, the cells were exposed to PI in a darkened room for 30 minutes (PI dosage of 50 μg/mL, RNase A dosage of 100 μg/mL, and 0.5% Triton X-100). During the next stage, Flow cytometry (BD Biosciences, San Jose, CA, USA) was used to analyze the stained cells. The sub-G1, G0/G1, S, and G2/M phases cell distributions were displayed as DNA histograms. FlowJo software from Ashland, OR, USA was used to evaluate the cell cycle dispersal.
Real-Time PCR Footnote 12 Analysis of Gene Expression
The three treated and control cell groups of KG1a cells were collected after 48 hours of exposure. RNA extraction was performed using RNX-plus reagent (Cinnagen, Iran). The RNA quality and quantity were evaluated using 1% agarose gel electrophoresis and NanoDrop (Thermo Scientific, USA). To synthesize cDNA, one microgram of total RNA was treated with the Prime Script™ RT Reagent Kit (Takara Inc., Kyoto, Japan). The Rotor-Gene Q real-time PCR cycler (Qiagen) was used to perform triplicate reactions, employing Master Mix Green High ROX™ (Ampliqon, Denmark). ACTB (β-actin) was employed as the housekeeping gene to normalize the expression levels of AKT, PTEN, FOXO1, BAD and XIAP. Gene expression was compared between four cell groups. The relative mRNA expression was evaluated using the comparative ΔΔCt method [11]. The real-time PCR primers were synthesized by Oligo (Macrogen, Seoul, South Korea). The Table 1 contains the primer sequences. The following pattern was used to repeat the cDNA synthesis cycles: 95 °C for 30 seconds, 58–64 °C for 30 seconds, 72 °C for 30 seconds, and a final extension of 72 °C for 5 minutes. To test amplification specificity and the existence of primer dimers, a melting curve analysis was conducted on each run. Each primer’s mean amplification efficiencies were also calculated by LinRegPCR 2017.9 version [12].
Statistical Analyses
The experiments were conducted in triplicate and expressed as the mean ± SD. Graph Pad Prism software version 9.0.0 (Graph Pad Software, CA, USA) was utilized to conduct the IC50 calculations. In addition, statistical analysis was performed using an independent sample t-test or one-way ANOVA, with a Tukey’s post hoc test conducted via SPSS 23.0 software (SPSS Inc., Chicago, IL, USA). The results were considered statistically significant at P-values of 0.05 (*), 0.01 (**); 0.001 (***); 0.0001(****).
Results
The Viability of KG1a Leukemic Cells Was Reduced by the SAL/HESP Combination
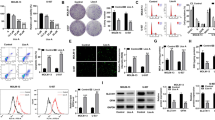
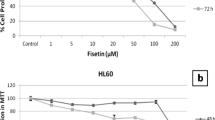
Initially the IC50 of SAL was calculated and the 2 μM for 48 hours was selected (Fig. 1A). In the next stage the selected Ic50 for HESP was 85 μM for 48 hours (Fig. 1B). Eventually the effect of SAL/HESP combination on cancer cell viability was assessed by MTT assay. The doses 85 μM HESP and 2 μM SAL were used in preparation of the combination drug. The results showed that the SAL/HESP combination has far pernicious effects than SAL and HESP separately (Fig. 1C).
The viability of KG1a leukemic cells was reduced by the SAL/HESP combination. The selected KG1a cells were treated with concentrations of SAL ranging from 1 to 10 μM. These cells demonstrated different growth rates after subjection to SAL (Fig. 1A). The data revealed that after HESP treatment, KG1a viability decreased in dosage or time-dependent manner (Fig. 1B). KG1a cell viability was decreased with SAL/HESP combination (Ic50 of each drug was combined) in comparison with SAL and HESP separately (Fig. 1C). The results demonstrated as the mean ± SD of three separate trials. (**P < 0.01; ***P < 0.001; ****P < 0.0001)
Morphology of apoptotic cells Changes after SAL/HESP treatment. These findings confirmed the outcomes of the cell cycle assay. Chromatin compaction and nuclear fragmentation assay were performed with Hoechst 33258 staining as explained in the methods to examine morphological alterations on treated cells. The KG1-a cells that received no treatment displayed round nuclei with normal and spindle-shaped cell membranes (Fig. 2A). The SAL, HESP, SAL/HESP treatment respectively caused greater increase in nuclear fragmentation and chromatin condensation, while avoiding necrosis induction. The changes are shown by arrows in Fig. 2B, C, D
Morphology of Apoptotic Cells Changes after SAL/HESP Treatment
To validate that the sub-G1 fraction is due to apoptosis, a nuclear condensation assay was conducted using Hoechst 33342 staining according to the experimental protocols. Cells are in their steady state without any treatment (Fig. A). Also, SAL, HESP and SAL/HESP was applied to KG1a cell culture at their IC50 concentration and the cell death was increased respectively (Fig. B, C, D).
ROS Level Increased after SAL/HESP Treatment
To explore whether SAL and HESP triggered the production of ROS, the total ROS level was assessed using DCFH-DA through flow cytometry. According to the results, there was a gradual rise in ROS production in HESP, SAL, and HESP/SAL respectively in the KG1a cell line. The statistically significant improvement was observed in the HESP administered group after 48 hours (****P < 0.0001) with a 2.7-fold increase. The KG1a cells treated with SAL showed a 1.7-fold surge after 48 h (*p < 0.05). Furthermore, the SAL/HESP group exhibited a 4.1-fold growth (****P < 0.0001) in comparison to the control group after 48 hours of incubation (Fig. 3).
SAL/HESP Caused Sub-G1 Accumulation in KG1a
The proportion of cell cycle phases is shown in KG1-a control cells (Fig. 4A). Our research revealed that HESP treated cells were distributed 36.39, 36.57, 24.78, and 2.71% in sub-G1, G0/G1, S, and G2/M phases, respectively (Fig. 4B). The proportion of cell cycle phases changed to 47.26, 31.73, 21.01, and 1.54% at sub-G1, G0/G1, S, and G2/M phases, after a 48-hour treatment with 2 μΜ SAL (Fig. 4C). The percentage of the cell cycle phases after SAL/HESP treatment was 48.17, 27.52, 24.97 and 2.45% respectively (Fig. 4D). our results show that the G1 phase arrest has happened after treatment with HESP, SAL and SAL/HESP combination treatment resulted in more cell accumulation in the sub-G1 phase. The information about cell cycle phases (before and after treatment) is comprehensively in the chart (Fig. 4E).
SAL/HESP caused sub-G1 accumulation in KG1a. Flow cytometry was used to examine the cell cycle in untreated and treated KG1a cells. The proportion of cell cycle in KG1-a control cells (4A). Distribution of cell cycle in KG1-a cells following 48 hours of incubation with 85 μM HESP, 2 μM SAL and SAL/HESP (4B, C, D). The statistical quantification of data was illustrated in the diagram after treating with hesperidin (Fig. 4E). The outcome was assessed by FlowJo software. (**P < 0.01; ****P < 0.0001)
Evaluation of Gene Expression Quantity by Real-Time PCR
Following the exposure of KG1a cells to HESP, SAL and SAL/HESP, the gene expression level of BAD, FOXO1, PTEN, AKT and XIAP were measured by real-time PCR. According to our discoveries, after 48 hours of incubation with 85 μΜ of hesperidin, the expression of Bad was remarkably augmented by about 28.13-fold (***P < 0.001) (Fig. 5D). It is apparent that there is a significant upward trend in FOXO1 and PTEN gene expression after treatment with HESP, SAL and SAL/HESP respectively (Fig. 5A, B). Besides, Akt gene displayed the minimum gene expression in HESP group with 0.14-fold smaller amount when compared to the control group (****p < 0.0001) (Fig. 5C). The results also revealed that XIAP showed a significant expression level reduction (0.26-fold) in HESP/SAL group compared to the control (****p < 0.0001) (Fig. 5E). The general trend of the graph is decreasing in Akt and XIAP.
Determination of mRNA expression after exposure to HESP, SAL and SAL/HESP. The q-PCR was utilized to measure fluctuations in transcription levels BAD, FOXO1, PTEN, AKT and XIAP genes. The data is presented as mean ± SD (n = 3); The ANOVA compared the treated and untreated group of the cells (Tukey’s post hoc test) (*p < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001)
Discussion
AML is a malignant disease that progresses rapidly due to the proliferation of immature myeloid leukemia cells [14, 15]. There are several signaling pathways which play role in acute myeloid leukemia. One of the signaling pathways deregulated in AML is the PI3K-Akt pathway which is upregulated in cancerous cells [16]. The PI3K-Akt pathway manages vital functions such as proliferation, differentiation, and survival. [17]. In AML, oncogene activating mutations or inactivation of tumor suppressor genes lead to dysregulation of the PI3K-Akt signaling pathway [17, 18]. There seems to be a connection between the activation of this pathway and decreased overall survival in 60% of AML patients [17, 19]. Akt is a component of the PI3K-Akt pathway, and can function as a proto-oncogene [20]. The findings of our experiment indicated a reduction in akt mRNA expression in KG1a cells following the administration of all three drug categories (SAL, HESP, SAL/HESP), and there was no significant variance among the treatment groups, indicating that the combined therapy of SAL and HESP is not overly effective. Yoon et al. pointed out that deactivating Akt could potentially increase cancer cells sensitivity to SAL [21], which was further explored in later studies [22,23]. The research conducted by Choi et al. suggests that the PI3K/Akt pathway can be targeted by co-treatment with SAL and MK-2206 or LY294002, as potential therapeutic drugs for sensitizing cancer cells [23]. Also Shahbazi et al. showed that Hesperidin suppressed insulin-induced phosphorylation and activation of Akt [24]. Furthermore, Hesperidin treatment reduced the upregulation of Akt and PI3K expression, and effectively preserved the structural integrity of the liver tissues from the development of hepatocellular carcinoma [25]. Additionally, in accordance with the conclusions of another research, hesperidin hinders breast cancer by blocking the NF-κB and Akt signaling pathways [26]. Akt can convey an anti-apoptotic signal by phosphorylating downstream target proteins, including Bad and X-linked inhibitor of apoptosis (XIAP), involved in the control of cell growth [27]. Members of the BH3-Only group consist of several pro-apoptotic proteins such as BAD [28]. We assessed the BAD gene’s expression and found that HESP was more potent than SAL, and combining the two did not have a synergistic effect on BAD gene overexpression. Savaee et al. revealed that the simultaneous administration of salinomycin/MK-2206 demonstrated a synergistic effect on prostate cancer cell viability and Akt/Bad expressions [29]. Unusual activation of the PI3K/Akt signaling pathway has been documented to potentially serve as a survival mechanism for breast cancer cells by inhibiting apoptosis through Bad protein phosphorylation [30]. Our results are consistent with the findings of Parajuli et al. [31] regarding salinomycin’s ability to inhibit Akt and Bad expression in ovarian cancer cells [31]. In our research, PTEN was overexpressed by SAL/HESP compared to the other two treatment groups. PTEN stimulates PIP3 dephosphorylation, resulting in the inactivation of the PI3K-AKT signaling. Hesperidin treatment led to the regulation of the PTEN and decrease in activation of the PI3K/Akt pathway, as noted by Saiprasad et al. [32]. Bonneau et al. detected a partial deletion of the PTEN as a clinically relevant genomic modification in 100% of replicate resistance AML cell line [33]. Another study concluded that the mRNA expression level of PTEN had a remarkable reduction in AML patients. The study uncovered that PPARγ upregulation was associated with downregulated PTEN in AML patients compared to the control group. Aggerholm’s initial investigation revealed that PTEN mutations were present in only 1 out of 59 AML patients, indicating that PTEN’s genetic involvement in AML pathogenesis is rare [34]. Similarly, Liu and colleagues, no PTEN mutations were discovered by Liu and colleagues in 62 AML patients, Indicating that irregular splicing machinery may target PTEN [35]. In this study, we have surveyed another gene called FOXO1. Important signaling proteins in growth factor signaling, FOXOs are located in the pathway downstream of AKT. AKT activates transcription factors related to survival by phosphorylating FOXO1, 3, and 4 on three different sites, which causes them to translocate into the cell core [36]. all three treatment groups Demonstrated an increase in foxo1 mRNA expression and the variation between these groups was not significant. The uncontrolled expression of FOXO proteins can alter cell destiny and is a crucial factor in numerous cancers, such as leukemia. There are four constituents in this subfamily: FOXO1, FOXO3, FOXO4 and FOXO6. They manage a broad range of cellular functions including apoptosis, DNA damage repair, cell cycle arrest,and stress response [37]. They trigger or suppress different genes that participate in cellular processes such as BAD in apoptosis. The inactivation of FOXO proteins has been observed in several malignancies, among them being breast cancer, prostate cancer, glioblastoma, and leukemia [6, 38]. The existence of phospho-FOXO1 was discovered to be linked with decreased survival as and a negative unfavorable result in AML patients and it is suggested that phospho-FOXO1 is a significant molecular marker for AML prognosis (63). FOXO1 transcription factor is involved in the regulation of RNOS scavengers, such as superoxide dismutase and catalase, hence its high expression provides protection against oxidative stress [39]. XIAP was the other tumorigenic factor that we opted for mRNA expression analysis. HESP did not affect on XIAP but it’s over expression by treatment with SAL and HESP/ SAL was significant. Its ability to inhibit activated forms of caspase-3, −7, and − 9 is a significant factor in oncogenesis as it prevents cancer cells from undergoing apoptosis [40]. XIAP has been considered an unfavorable gene through a wide range of signal pathways in AML, which includes the caspase pathway and NF-Kb pathway [41, 42]. Suggested by an investigation on 172 primary AML patient samples, there was no relationship between patients and control group [43]. It has been stated that ROS can decrease the expression of XIAP, implying that inhibiting XIAP can enhances the differentiation of AML cells by up regulating ROS [44, 45]. Our finding demonstrated that when the cells are exposed to SAL/HESP the ROS is raised considerably and thus may lead to cancer cell death. The consistent oral treatment of hesperidin in rats with breast cancer showed a noteworthy reduction in lipid peroxidation level in the breast cancer animal model [46]. Also, Yu et al. uncovered that Hesperidin induced apoptosis associated with mitochondria in gastric cancer cells and increased the proportion of intracellular ROS [47]. Elevating the levels of hesperidin results in the cessation of the G0/G1 phase of the cell cycle in cervix cancer cells [48], and this result aligns with ours. Also Xia et al. found that treatment of cancer cells with different doses of hesperidin at different times resulted in a significant increase in the percentage of cells in the G0/G1 phase, suggesting the cell cycle arrest at this stage [49].
Conclusion
According to the results of this study, HESP and SAL can be considered as cheap and effective drugs in the treatment of leukemia. In some cases, these drugs can be synergistic and strengthen each other’s effects. These natural products make many changes in the expression levels of genes involved in the PI3K/AKT pathway, cell death and oxidative stress in leukemia. More studies in the field of other genes involved in PI3K/AKT pathway as well as their relationship with other signaling pathways such as NF-Kb pathway need further investigations.
Data Availability
No datasets were generated or analysed during the current study.
Notes
Acute myeloid leukemia
Phosphoinositide 3-kinase
BCL-2 antagonist of cell death
Fork head box
Phosphatase and tensin homolog
Chromosome-linked inhibitor of apoptosis protein
Salinomycin
Hesperidin
3-[4,5-dimethylthiazole-2-yl]-2,5-diphenyltetrazolium bromide
Dichlorodihydrofluorescein diacetate
4′,6-diamidino-2-phenylindole
Polymerase chain reaction
References
Hope KJ, Jin L, Dick JE (2003) Human acute myeloid leukemia stem cells. Arch Med Res 34(6):507–514
Zeng AG, Bansal S, Jin L, Mitchell A, Chen WC, Abbas HA et al (2022) A cellular hierarchy framework for understanding heterogeneity and predicting drug response in acute myeloid leukemia. Nat Med 28(6):1212–1223
Anguille S, Van Tendeloo V, Berneman Z (2012) Leukemia-associated antigens and their relevance to the immunotherapy of acute myeloid leukemia. Leukemia 26(10):2186–2196
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144(5):646–674
Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR et al (2005) BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol Cell 17(4):525–535
Lam EW-F, Brosens JJ, Gomes AR, Koo C-Y (2013) Forkhead box proteins: tuning forks for transcriptional harmony. Nat Rev Cancer 13(7):482–495
Patel L, Pass I, Coxon P, Downes CP, Smith SA, Macphee CH (2001) Tumor suppressor and anti-inflammatory actions of PPARγ agonists are mediated via upregulation of PTEN. Curr Biol 11(10):764–768
Tesio M, Oser GM, Baccelli I, Blanco-Bose W, Wu H, Göthert JR et al (2013) Pten loss in the bone marrow leads to G-CSF–mediated HSC mobilization. J Exp Med 210(11):2337–2349
Carter BZ, Mak DH, Wang Z, Ma W, Mak PY, Andreeff M et al (2013) XIAP downregulation promotes caspase-dependent inhibition of proteasome activity in AML cells. Leuk Res 37(8):974–979
Carter BZ, Gronda M, Wang Z, Welsh K, Pinilla C, Andreeff M et al (2005) Small-molecule XIAP inhibitors derepress downstream effector caspases and induce apoptosis of acute myeloid leukemia cells. Blood 105(10):4043–4050
Dewangan J, Srivastava S, Rath SK (2017) Salinomycin: a new paradigm in cancer therapy. Tumour Biol 39(3):1010428317695035
Zhou S, Wang F (2013) T Wong E, Fonkem E, Hsieh T-C, M Wu J, et al. Salinomycin: a novel anti-cancer agent with known anti-coccidial activities. Curr Med Chem 20(33):4095–4101
Novinbahador T, Araj-Khodaei M, Mahdavi M (2023) Evidence for hesperidin as an effective factor in initiating the intrinsic pathway of apoptosis in KG1a leukemia cells. Int J Toxicol 42(2):165–171
Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T et al (2017) Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood J Am Soc Hematol 129(4):424–447
Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM et al (2016) The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood J Am Soc Hematol 127(20):2391–2405
Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR et al (2004) Akt stimulates aerobic glycolysis in cancer cells. Cancer Res 64(11):3892–3899
Nepstad I, Hatfield KJ, Tvedt THA, Reikvam H, Bruserud Ø (2018) Clonal heterogeneity reflected by PI3K-AKT-mTOR signaling in human acute myeloid leukemia cells and its association with adverse prognosis. Cancers 10(9):332
Nepstad I, Hatfield KJ, Grønningsæter IS, Aasebø E, Hernandez-Valladares M, Hagen KM et al (2019) Effects of insulin and pathway inhibitors on the PI3K-Akt-mTOR phosphorylation profile in acute myeloid leukemia cells. Signal Transduct Targ Ther 4(1):20
Chen W, Drakos E, Grammatikakis I, Schlette EJ, Li J, Leventaki V et al (2010) mTOR signaling is activated by FLT3 kinase and promotes survival of FLT3-mutated acute myeloid leukemia cells. Mol Cancer 9(1):1–7
Bellacosa A, Testa JR, Moore R, Larue L (2004) A portrait of AKT kinases: human cancer and animal models depict a family with strong individualities. Cancer Biol Ther 3(3):268–275
Kim J-H, Choi A-R, Kim YK, Kim HS, Yoon S (2013) Low amount of salinomycin greatly increases Akt activation, but reduces activated p70S6K levels. Int J Mol Sci 14(9):17304–17318
Choi A-R, Jung M-J, Kim J-H, Yoon S (2015) Co-treatment of salinomycin sensitizes AZD5363-treated cancer cells through increased apoptosis. Anticancer Res 35(9):4741–4747
Choi AR, Kim JH, Yoon S (2014) (2014) Sensitization of Cancer Cells through Reduction of Total Akt and Downregulation of Salinomycin-Induced pAkt, pGSk3β, pTSC2, and p4EBP1 by Cotreatment with MK-2206. BioMed res int 1:295760–295768
Shahbazi R, Cheraghpour M, Homayounfar R, Nazari M, Nasrollahzadeh J, Davoodi SH (2018) Hesperidin inhibits insulin-induced phosphoinositide 3–kinase/Akt activation in human pre-B cell line NALM-6. J Cancer Res Ther 14(3):503–508
Mo’men YS, Hussein RM, Kandeil MA (2019) Involvement of PI3K/Akt pathway in the protective effect of hesperidin against a chemically induced liver cancer in rats. J Biochem Mol Toxicol 33(6):e22305
Kongtawelert P, Wudtiwai B, Shwe TH, Pothacharoen P, Phitak T (2020) Inhibitory effect of hesperidin on the expression of programmed death ligand (PD-L1) in breast cancer. Molecules 25(2):252
Paccosi E, Balzerano A, Proietti-De-Santis L (2023) Interfering with the ubiquitin-mediated regulation of Akt as a strategy for Cancer treatment. Int J Mol Sci 24(3):2809
Zha J, Harada H, Yang E, Jockel J, Korsmeyer SJ (1996) Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-XL. Cell 87(4):619–628
Savaee M, Bakhshi A, Yaghoubi F, Pourrajab F, Chegini KG (2022) Evaluating the effects of separate and concomitant use of MK-2206 and salinomycin on prostate cancer cell line. Rep Biochem Mol Biol 11(1):157
Al-Bazz YO, Underwood JC, Brown BL, Dobson PR (2009) Prognostic significance of Akt, phospho-Akt and BAD expression in primary breast cancer. Eur J Cancer 45(4):694–704
Parajuli B, Lee H-G, Kwon S-H, Cha S-D, Shin S-J, Lee G-H et al (2013) Salinomycin inhibits Akt/NF-κB and induces apoptosis in cisplatin resistant ovarian cancer cells. Cancer Epidemiol 37(4):512–517
Saiprasad G, Chitra P, Manikandan R, Sudhandiran G (2014) Hesperidin induces apoptosis and triggers autophagic markers through inhibition of Aurora-a mediated phosphoinositide-3-kinase/Akt/mammalian target of rapamycin and glycogen synthase kinase-3 beta signalling cascades in experimental colon carcinogenesis. Eur J Cancer 50(14):2489–2507
Bonneau D, Longy M (2000) Mutations of the human PTEN gene. Hum Mutat 16(2):109–122
Aggerholm A, Grønbæk K, Guldberg P, Hokland P (2000) Mutational analysis of the tumour suppressor gene MMAC1/PTEN in malignant myeloid disorders. Eur J Haematol 65(2):109–113
Liu TC, Lin PM, Chang JG, Lee JP, Chen TP, Lin SF (2000) Mutation analysis of PTEN/MMAC1 in acute myeloid leukemia. Am J Hematol 63(4):170–175
Zhang X, Tang N, Hadden TJ, Rishi AK (2011) Akt, FoxO and regulation of apoptosis. Biochimica et Biophysica Acta (BBA)-molecular. Cell Res 1813(11):1978–1986
Zhu H (2014) Targeting forkhead box transcription factors FOXM1 and FOXO in leukemia. Oncol Rep 32(4):1327–1334
Yang J-Y, Hung M-C (2009) A new fork for clinical application: targeting forkhead transcription factors in cancer. Clin Cancer Res 15(3):752–757
García Rubiño ME, Carrillo E, Ruiz Alcalá G, Domínguez-Martín AA, Marchal J, Boulaiz H (2019) Phenformin as an anticancer agent: challenges and prospects. Int J Mol Sci 20(13):3316
Inoue H, Shiraki K, Murata K, Sugimoto K, Kawakita T, Yamaguchi Y et al (2004) Adenoviral-mediated transfer of p53 gene enhances TRAIL-induced apoptosis in human hepatocellular carcinoma cells. Int J Mol Med 14(2):271–275
Surapally S, Jayaprakasam M, Verma RS (2020) Curcumin augments therapeutic efficacy of TRAIL-based immunotoxins in leukemia. Pharmacol Rep 72:1032–1046
Zhou J, Lu X, Tan TZ, Chng WJ (2018) X-linked inhibitor of apoptosis inhibition sensitizes acute myeloid leukemia cell response to TRAIL and chemotherapy through potentiated induction of proapoptotic machinery. Mol Oncol 12(1):33–47
Fulda S (2012) Exploiting inhibitor of apoptosis proteins as therapeutic targets in hematological malignancies. Leukemia 26(6):1155–1165
Paul T, Roy R, Sarkar RD, Sinha S, Biswas N (2021) H2O2 mediated FLIP and XIAP down-regulation involves increased ITCH expression and ERK-Akt crosstalk in imatinib resistant chronic myeloid leukemia cell line K562. Free Radic Biol Med 166:265–276
Zhao R, Yu Q, Hou L, Dong X, Zhang H, Chen X et al (2020) Cadmium induces mitochondrial ROS inactivation of XIAP pathway leading to apoptosis in neuronal cells. Int J Biochem Cell Biol 121:105715
Nandakumar N, Jayaprakash R, Rengarajan T, Ramesh V, Balasubramanian MP (2011) Hesperidin, a natural citrus flavonoglycoside, normalizes lipid peroxidation and membrane bound marker enzymes in 7, 12-Dimethylbenz (a) anthracene induced experimental breast cancer rats. Biomed Prevent Nutr 1(4):255–262
Yu W, Xie X, Yu Z, Jin Q, Wu H (2019) Mechanism of hesperidin-induced apoptosis in human gastric cancer AGS cells. Trop J Pharm Res 18(11):2363–2369
Wang Y, Yu H, Zhang J, Gao J, Ge X, Lou G (2015) Hesperidin inhibits HeLa cell proliferation through apoptosis mediated by endoplasmic reticulum stress pathways and cell cycle arrest. BMC Cancer 15:1–11
Xia R, Sheng X, Xu X, Yu C, Lu H (2018) Hesperidin induces apoptosis and G0/G1 arrest in human non-small cell lung cancer A549 cells. Int J Mol Med 41(1):464–472
Acknowledgments
The authors appreciate the support by the research council of the University of Tabriz, Tabriz, Iran.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
Conceptualization: MM
Data curation: MM
Formal analysis: SA
Funding acquisition: MM
Investigation: SA
Methodology: MM, SA
Project administration: MM, MN
Resources: SA
Supervision: MM, MN
Validation: SA
Writing – original draft: SA
Writing – review & editing: MM, SA.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
All authors consent to the publication of the article in this journal.
Competing Interests
The authors declare no competing interests.
Conflict of Interest
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Abroon, S., Nouri, M. & Mahdavi, M. Hesperidin/Salinomycin Combination; a Natural Product for Deactivation of the PI3K/Akt Signaling Pathway and Anti-Apoptotic Factors in KG1a Cells. J Fluoresc (2024). https://doi.org/10.1007/s10895-024-03808-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10895-024-03808-4