Abstract
Five variants of glucokinase (ATP-D-hexose-6-phosphotransferase, EC 2.7.1.1) including wild type and single Trp mutants with the Trp residue at positions 65, 99, 167 and 257 were prepared. The fluorescence of Trp in all locations studied showed intensity changes when glucose bound, indicating that conformational change occurs globally over the entire protein. While the fluorescence quantum yield changes upon glucose binding, the enzyme’s absorption spectra, emission spectra and fluorescence lifetimes change very little. These results are consistent with the existence of a dark complex for excited state Trp. Addition of glycerol, L-glucose, sucrose, or trehalose increases the binding affinity of glucose to the enzyme and increases fluorescence intensity. The effect of these osmolytes is thought to shift the protein conformation to a condensed, high affinity form. Based upon these results, we consider the nature of quenching of the Trp excited state. Amide groups are known to quench indole fluorescence and amides of the polypeptide chain make interact with excited state Trp in the relatively unstructured, glucose-free enzyme. Also, removal of water around the aromatic ring by addition of glucose substrate or osmolyte may reduce the quenching.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glucokinase (ATP-D-hexose-6-phosphotransferase, EC 2.7.1.1) is a member of a family of enzymes that catalyze the phosphorylation of D-glucose by ATP in the first step of the glycolytic pathway. Several lines of evidence point to the pivotal role of glucokinase in regulation of glucose homeostasis in liver and pancreas. The existence of isoenzymes specific to various tissues implies differences in the role of the enzyme [1–5] and point to distinctive regulation of glycolysis in different tissues. The presence of a glucokinase inhibitory regulator protein, specifically active in liver [6] further Indicates that glucokinase has organ specific functions. The most compelling evidence for the importance of this enzyme in health is provided by mutants in human glucokinase that disrupt glucose homeostasis. Inactivating mutants of the enzyme may cause either “maturity onset diabetes of the young”, when one allele is affected (MODY, which is usually a mild form of the disease), or a severe form of “permanent neonatal diabetes mellitus”, when both alleles are affected. Activating mutants cause “hyperinsulinemic hypoglycemia in infancy”, which may be life threatening even if only one allele is altered [7]. Finally, glucokinase activator drugs were developed that have potential for the treatment of patients with type 2 diabetes mellitus [8, 9].
This introductory summary illustrates that understanding the biochemistry of glucokinase (GK) has high translational significance as the enzyme serves as vital glucose sensor in endocrine pancreas and liver. The glucokinase molecule is also of great interest to basic scientists as a protein structure model because of its unique biophysical and biochemical characteristics. Physiologically operating in cells as a monomer, it shows striking sigmoidal glucose dependency of the phosphorylation reaction (nH: 1.7) [5]. It exhibits large but slow conformational transitions when activated by glucose and features an allosteric modifier region that is distinct from the substrate binding site and provides separate allosteric binding sites for inhibitors (GKRP) and activators (GKAs) [9]. These characteristics are currently being investigated by crystallography [10], NMR [11], and tryptophan fluorescence [12, 13] measurements. Here we will apply fluorescence techniques as a means for studying recombinant human glucokinase and several interesting mutants.
Wild-type glucokinase (GK-WT) contains three tryptophans: W257, deeply buried near the glucose binding site, W99 located near the surface in the allosteric modifier region and W167, also close to the surface in the central cleft of the glucose binding site [10]. Zelent, Matschinsky and coworkers have utilized the fluorescence of tryptophan in wild type and in glucokinase mutants in which tryptophan has been changed to isolate conformational changes upon glucose binding to particular regions of the protein and to monitor equilibrium sugar binding [14, 15]. In a recent publication Zelent and coworkers were able to characterize the cooperative protein/protein interaction between glucokinase and its hepatic inhibitor GKRP as influenced by glucose, fructose-phosphates and a GKA using tryptophan fluorescence changes of the native and a disease-causing mutant of GKRP interacting with a tryptophan free, functionally largely intact, mutant glucokinase [13].
In the present paper, we are examining the fluorescence yield, lifetime and spectrum of tryptophan in recombinant gluocokinase. Five types of glucokinase were examined: wild type WT (containing three Trp), three mutants in which two Trp were replaced and only one Trp was expressed at a natural position, and a fourth mutant in which all natural Trp were replaced and one Trp was placed at an unnatural position. In this way, a fluorescent marker is placed at positions 65, 99, 167 and 257.
A major finding of our paper is that fluorescence intensity from Trp of wild type and all variants changed upon addition of glucose. While the intensity of tryptophan fluorescence in GK in WT increases by addition of glucose, the fluorescence lifetimes and absorption and emission spectra are much less dependent upon the binding of glucose. This is in contradiction to the usual “simple” excited state reaction measurement, where fluorescent lifetime and quantum yields are directly proportional to each other. We suggest that this is due to the existence of a dark excited state species.
The existence of dark excited state species [16], or extremely short excited species [17] of tryptophan are well documented. We do not know any literature example in a protein where the lifetime does not significantly change while the intensity changes so much (>200%). We considered how to vary the experimental conditions to gain insights on the nature of the dark complex. We used glycerol and sugars as osmolytes. We found that glycerol and sugar osmolytes have the effect of increasing fluorescence intensity of GK-WT, and increasing the binding affinity for glucose. Substitution of glycerol for water is known to produce more condensed forms of proteins [18] and higher binding affinity by the condensed form accounts for the increased binding affinity. Osmolytes may reduce the local concentration of water around the aromatic ring, as well as stabilize the non-quenching conformation of the amide group relative to the aromatic ring.
Methods
Materials
D-glucose (purity ≥99.5%), glycerol (purity ≥99%), L-glucose, trehalose, sucrose, and N-acetyl-L-tryptophanamide (NATA) and other chemicals were from Sigma Chemical Co. USA.
Generation of Recombinant WT and Mutant Glucokinase Proteins
In all experiments described in this article, recombinant glucokinase was used. Recombinant wild type and mutant human β-cell glucokinases were generated as Glutathione S-Transferase (GST) fusion proteins in E. coli as previously described [14, 19, 20]. Point mutations were introduced into the pGEX-3X vector using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, California, USA). All mutant vectors were transformed into E. coli cells and verified by DNA sequencing. GST-GK fusion proteins were cleaved with factor Xa and submitted to a second round of purification by removing GST with glutathione agarose and Factor Xa with benzamidine Sepharose 6B following protocols provided by the manufacturer.
The enzyme variants made for this project were: GK-W99 (W167F/W257F), GK-W167 (W99R/W257F), GK-W257 (W99R/W167F) and GK-W65 (W99R/W167F/W257F/T65 W). As criterion of purity, all mutant glucokinase proteins showed a single band at 50 kD using gel chromatography.
Sample Preparation
Glucokinase was dissolved in buffer pH 7.2 containing 20 mM K phosphate, 50 mM KCl, 1 mM ethylenediaminetetraacetic acid (EDTA), and 1 mM dithiothreitol (DTT). When glycerol was added, the glycerol percent is expressed as volume/volume.
Absorption and Fluorescence Measurements
UV/Vis absorption spectra were obtained using a Hitachi Perkin-Elmer U-3000 spectrophotometer.
Fluorescence emission spectra were obtained with a Fluorolog-3-21 Jobin-Yvon Spex Instrument SA that uses a 450 W xenon lamp for excitation and a cooled R2658P Hamamatsu photomultiplier tube for detection. For all measurements quartz cuvettes were used that gave paths of 0.4 cm for excitation and 1 cm for emission. Excitation wavelength was at 295 nm and fluorescence emission was measured in the range of 300–500 nm. The instrumental slit width was set to provide a band-path of 4 nm for excitation and 3 nm for emission. Spectra were corrected for instrumental response using Spex instrument software. A thermostatically controlled cell holder maintained sample temperature at 20 °C.
Fluorescence quantum yields of tryptophan, ϕ (the ratio of the number of photons emitted relative to the number of photons absorbed by the fluorophore) were determined for wild type and mutant glucokinase by the following equation [21]:
The subscripts S and R refer to the sample and reference standard, respectively; A R and A S denote the absorbance at the excitation wavelength (A < 0.05 to avoid the inner filter effect); F S and F R denote the integral intensities of the corrected fluorescence spectra measured under identical instrument settings; n S and n R are the refractive indices. (n R (water =1.3334) and n s (glycerol (95%) = 1.465). Fluorescence quantum yields were determined relative to N-acetyl-L-tryptophanamide (NATA) (ϕ = 0.14) in water as reference [22].
Tryptophan Fluorescence Lifetime Measurements
Fluorescence intensity decays were measured using time-correlated single-photon-counting with a FluoTime200 fluorimeter (PicoQuant, Inc., Berlin, Germany). Excitation was at 295 nm. Emitted light at 340 nm was selected using a monochromator. To avoid effects of Brownian rotation a polarizer oriented at the magical angle was placed on the emission side. A micro channel plate was used to detect the emitted light. The intensity decay data were fitted with FluoFit software (PicoQuant version 4).
Some fluorescence lifetimes were measured using Fluorolog TCSPC fluorimeter equipped with Nano-LED laser (Horiba Scientific Inc., Edison, NJ). Excitation was at 278 nm. The emitted light at 340 nm was selected using a monochromator. A photomultiplier tube (Hamamatsu 928, Japan) was used to detect emitted light. The intensity decay data were fitted with Data Analysis Software (Horiba Scientific Data Station 2.7).
The data were analyzed in terms of a sum of exponentials:
where αi are the amplitudes of the intensity decay times τi with Σαi = 1.0. The intensity mean lifetime is defined as τmean = Σαiτi 2 / Σαiτi .
Results
Fluorescence Intensity and Spectra of GK’s with Glucose and Glycerol
Tryptophan fluorescence of GK depends on location of the tryptophan residue in the protein and is sensitive to the structural changes upon ligand binding. Figure 1 shows the effect of adding glucose and glycerol to GK in producing a fluorescence increase of tryptophan. Upon binding of glucose, the fluorescence quantum yield of GK-WT goes from 0.094 to 0.187, an increase of almost 2 times. GK-W99 increases 1.6 times, GK-W167 1.8 times, and GK-W257 1.2 times while for GK-W65 glucose binding decreases fluorescence intensity by 15% (0.85 times).
Fluorescence emission spectra and intensity changes for glucokinase and NATA in buffer (black), buffer (b) with 100 mM D-glucose (blue) or 95% glycerol (red). a GK-WT, b GK-W167, c GK-W99, d GK-W257, e W65 and f NATA in water and glycerol. Excitation wavelength: 295 nm. Protein concentration about 1 micromolar
Figure 1 also shows the effect of glycerol at high concentration on tryptophan fluorescence. Structural changes of glucokinase in the presence of glycerol increase tryptophan fluorescence more strongly than glucose binding. The total increase for GK-WT is 2.6 times, and for individual Trp residues are: 4.6 times for GK-W99, 2.3 times for GK-W167, 1.3 times for GK-W257 and 1.1 times for GK-W65. For comparison NATA fluorescence in glycerol increases 2.1 times in relation to its fluorescence intensity in water (Fig. 1F). Glucose at the concentration used has no effect on NATA fluorescence yield.
Absorption and Fluorescence Spectra of GK-WT and NATA in Various Solvents
Absorption spectra for the various proteins are shown in Fig. 2. Lack of a solvent effect is seen for the protein absorption, consistent with the burial of Trp within the protein, and also consistent with the relative insensitivity of absorption spectra to solvent. (The large absorption at <250 nm and below includes contributions from other aromatic groups and amide groups, and was not analyzed).
Glycerol shifts NATA absorption and fluorescence spectra relative to water (Fig. 3c and d). The fluorescence shift of NATA follows decreasing of solvent polarity: water, glycerol, cyclohexane. This sequence of shift with polarity has been long recognized [23]. As seen in Fig. 3a, the peak position of the absorption spectrum of Trp in GK-WT does not shift, but there is a small diminution of the long wavelength emission contribution of GK-WT in glycerol (Fig. 3b).
Solvent effect on the absorption and emission spectra of GK-WT and NATA. a and b GK in buffer and 20% glycerol/water. Dotted black line: spectrum of sample in buffer multiplied by 2.88 to normalize its intensity to the spectrum in glycerol. c and d Absorption and emission of NATA in water (black), glycerol (red) and cyclohexane (gray). Spectra are normalized to the same intensity
The small diminution in long wavelength emission is especially noticeable when the spectra are normalized (red and dotted lines in Fig. 3b). Similar results were obtained for single Trp mutants. The spectra in Fig. 4 are normalized to emphasize the shift with addition of glycerol to single Trp variants. The shift was the largest for GK-W99.
Effect of Glycerol, Trehalose, L-Glucose and Sucrose on Binding of D-Glucose to GK
In the above experiments, glycerol was used in high concentration. We examined the effect of glycerol and other substrates on D-glucose binding at lower concentration, the concentration at which the water H-bond network is disrupted by cosolvent [24, 25]. The results are shown in Fig. 5. Glycerol at 20% increases the binding affinity of D-glucose (Table 1) to all mutants. The ratio of equilibrium dissociation constants (KD‘s) is between 4.1 and 4.6. The essentially same effect on all the GK analogues, suggests that the glycerol, as a solvent, is effective in stabilizing the whole protein. Trehalose, L-glucose and sucrose at 1 M concentration have a similar effect (Table 2). An osmotic effect of these compounds will be discussed later.
Fluorescence Lifetimes
In the simplest description of excited state reactions, fluorescence yields and lifetimes are directly proportional to each other. Fluorescence lifetimes were examined by time-correlated decay. Examples of the decay curves for GK are shown in Fig. 6. Time-resolved fluorescence decay of Trp in proteins is usually complex [26]. The fluorescence decay of Trp in GK-WT is also complex, but, as seen by time-correlated single photon counting, it could be fit by two exponential decay functions. The average fluorescent lifetime for GK-WT is 3.68 ns (Fig. 6 top). With glucose the lifetime is 3.96 ns (middle) and in glycerol the value is 4.82 ns (bottom). Therefore, in going from GK-WT in buffer to GK-WT in glycerol, the lifetime increases about 1.31 times. This is considerably less than the increase in fluorescence intensity (about 2.6 times, as documented in Fig. 1a.)
While there is a discrepancy between fluorescence yield and lifetime for Trp in the protein, for NATA in the presence of glycerol the lifetime is increased twice relative to aqueous buffer (Fig. 7), as is the fluorescence yield (Fig. 1f). The lifetime for NATA, therefore, scales proportionately to the increase in fluorescence yield within experimental uncertainty.
Fluorescence lifetimes for the samples studied are given in Table 3. (Some of these values were previously reported [15]. The data presented in the last three columns are especially informative. For NATA, both quantum yield and lifetime are about twice as large in glycerol compared to water. In contrast, the quantum yield of Trp in GK-WT increase is about twice when glucose is bound and about 2.5 times in the presence of glycerol. One might expect a discrepancy between lifetime and quantum yield in cases where there is energy transfer, and since GK-WT contains three Trp, energy transfer is possible. However, energy transfer is not possible in the single Trp mutants, and the pattern is also seen for mutants W99, W167, and W257. Since the absorption coefficient changes slightly under different conditions (Figs. 2 and 3) we can also compare the experimental relative quantum yield with the decay profile, making use of the relationship that the total fluorescence intensity is proportional to the amplitude, α, times the lifetime, τ, for an exponential decay. Again, the observed fluorescence increase by conditions of substrate binding or in the presence of glycerol is higher than that predicted by the lifetime and amplitude changes. Both lifetimes and quantum yields therefore suggest that the resting enzyme is in a conformation in which there is a dark complex. We note that this must be due to a local effect at the Trp site, and is not due to a complex that quenches all Trp, since, in W65, very little change in intensity was observed.
To compare the difference between the model compound NATA and protein, the quantum yield is plotted as a function of lifetime in Fig. 8. The grey trend-line for NATA shows the expected results: lifetime and quantum yield are linearly related. For instance, for W167, the quantum yield increases about 1.8 times when glucose is bound and 2.3 in the presence of glycerol. In contrast, the lifetime for W257 decreases slightly with the binding of glucose and in glycerol increased only about 1.31 times. The largest change was seen in W99, where the fluorescence quantum yield increased over 4.6 times in glycerol relative to the resting enzyme in aqueous buffer.
Data from Table 3. Gray closed circles are GK in buffer, blue closed circles are with the addition of D-glucose, and red closed circles are GK in glycerol (95%). Open circles are NATA in buffer and 95% glycerol. Trend line shows the slope of the data for NATA
Discussion
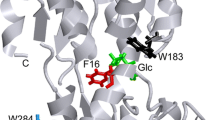
The locations of tryptophan in GK mutants used in this work are shown in Fig. 9.
Structure of human glucokinase. Positions of tryptophan residues and amino acids that were mutated are indicated. Co-ordinates were obtained from the Protein Data Bank entries IV4T (no glucose) and IV4S (with glucose). RasMol program (version 2.7.1) was used to create the figure. Data from Kamata et al. [10]
Although tryptophan is the quintessential fluorescent marker for protein conformation, it is long known that indole derivatives in solvents do not exactly mimic the properties of Trp in proteins. The data in this paper clearly illustrate this. NATA shows spectral shifts with increasing glycerol; the presence of glycerol also increases quantum yield (Fig. 3) and lifetime (Table 3). A red shift in the spectrum is attributed to kinetic relaxation of highly dipolar water [27], and increased lifetime and quantum yield can be attributed to the decrease in various non-radiative processes.
In contrast to NATA, upon addition of D-glucose substrate or in the presence of glycerol and non-substrate sugars, Trp in the protein shows very large changes in quantum yield, with little changes in absorption and emission spectra and fluorescence lifetime. We suggest that discrepancy between the model compound and Trp in protein can be explained that by Trp forming an excited state dark complex in the protein. In this model the protein exists as a distribution of conformations, and in one or several conformations the fluorescence of Trp is quenched. Adding substrate shifts the conformation of the closed form, in which the dark complex is either not present or present in a lower concentration. The similar emission spectrum and lifetime could suggest that the substrate-free and substrate are the same species – i.e., a certain fraction of the protein is in the closed form, but this hypothesis is not proven.
The idea that resting glucokinase exists in more than one conformation form is consistent with kinetic data since, in spite of being a monomeric enzyme, liver glucokinase kinetics show cooperativity with regard to glucose concentration [28, 29]. This model is also supported by x-ray scattering [30] and by steady-state fluorescence measurements [31]. Without substrate, x-ray analysis shows an open form for substrate free enzyme and a closed form for glucose-bound enzyme [10]. The protein that may correspond to the open form or may be another conformation, since in solution it is likely that there is a distribution of conformations.
A question arises about the nature of the dark complex. Amino acids can quench by proton transfer and electron transfer. The side chain of His, Lys, Cys, Ser and Tyr are all candidates for quenchers of Trp fluorescence [32, 33]. Carbonyl groups also produce intra-molecular quenching of indole fluorescence [16, 34]. The many interactions that occur for excited state Trp in proteins results in complex decay behavior [34].
In Fig. 10, a blow-up of the area around W99 is shown. In the x-ray determined structure of GK in the glucose-bound form, Trp99 is separated from His 218 by closer to Lys90 than in the glucose-free form. Tyr214 can affect Trp99 and its position is shown on the figure. W167 is in close proximity to Lys169, Lys 172 and Ser174. W257 is in the area of Cys382 and Cys230. W65 is closer to H218 in the glucose-bound form than in the glucose-free form of GK. This perhaps explains the slight decrease in quantum yield from this enzyme when glucose binds.
The data showing that glycerol and non-substrate sugars enhance fluorescence and binding can also be interpreted in the view of the existence of two conformations of GK. Glycerol and sugars act as osmolytes. They remove water from proteins, stabilizing the condensed, more stable conformational forms [18, 35].
Why then does the condensed, more structured forms of GK show higher fluorescence yields than the unstructured forms that are essentially totally quenched? We consider two reasons for this: 1) that in the unstructured form the quencher groups, whether the carbonyl or amino acid side groups, are able to weakly complex with the indole ring and quench the fluorescence. When the protein is structured, the amide groups are involved in H-bonding to form the secondary and tertiary structure and the amino acid side chains that quench are held away from indole ring. 2) Since fluorescence increases when water is excluded, we also need to consider whether water directly plays a role in forming the quenching complex. These two possibilities can be examined with the use of model compounds, as well as computational approaches.
The large fluorescence change upon glucose binding makes GK a potential subject for use as a clinical glucose sensor. Hexokinase from yeast has been considered for this purpose [36]. As noted by Pickup et al., glucose binding to yeast hexokinase results in a relatively small decrease in fluorescence intensity. An increase in fluorescence such as seen in human GK would produce better sensitivity and reliability. Therefore, glucokinase would be a more promising molecule for sensor development. We note also that, with the use of recombinant DNA techniques, human glucokinase can be prepared in large quantities at relatively low cost and that mutants with the desired glucose sensitivity can be tailor-made.
In conclusion, GK fluorescence increases upon glucose binding or in the presence of osmolytes without a concomitant change in fluorescence lifetime. We attribute this to indicate that in the unstructured form of the enzyme, a dark complex exists between excited state Trp and a quencher molecule.
References
Grossbard L, Schimke RT (1968) Multiple hexokinases of rat tissues. Purification and comparison of soluble forms. J Biol Chem 241:3546–3560
Matschinsky FM, Ellerman JE (1968) Metabolism of glucose in the islets of Langerhans. J Biol Chem 243:2730–2736
Schimke RT, Grossbard L (1968) Studies isozymes of hexokinase in animal tissues. Ann N Y Acad Sci 151:332–350
Matschinsky FM (1990) Glucokinase as glucose sensor and metabolic signal generator in pancreatic beta-cells and hepatocytes. Diabetes 39:647–652
Matschinsky FM (2005) Glucokinase, glucose homeostasis, and diabetes mellitus. Curr Diab Rep 5:171–176
Van Schaftingen E, Detheux M, Veiga de Cunha M (1994) Short-term control of glucokinase activity: role of a regulatory protein. FASEB J 8:414–419
Osbak KK, Colclough K, Saint-Martin, C, Beer NL, Bellanne-Chantelot C, Ellard S, Gloyn AL (2009) Update on mutations in glucokinase (GCK), which cause maturity-onset diabetes of the young, permanent neonatal diabetes and hyperinsulinemic hypoglycemis. Hum Mutat 30:1512–1526
Grimsby J, Sarabu R, Corbett WL, Haynes NE, Bizarro FT, Coffey JW, Guertin KR, Hilliard DW, Kester RF, Mahaney PE, Marcus L, Qi L, Spence CL, Tengi J, Magnuson MA, Chu CA, Dvorozniak MT, Matschinsky FM, Grippo JF (2003) Allosteric activators of glucokinase: potential role in diabetes. Science 301:370–373
Matschinsky FM (2013) GKAs for diabetes therapy: why no clinically used drugs after two decades of trying? Trends Pharmacol Sci 34:90–99
Kamata K, Mitsuya M, Nishimura T, Eiki Jun-ichi, Nagata Y (2004) Structural basis for allosteric regulation of the monomeric allosteric enzyme human glucokinase. Structure 12:429–438
Larion M, Salinas RK, Bruschweiler-Li L, Bruschweiler R, Miller BG (2010) Direct evidence of conformational heterogeneity in human pancreatic glucokinase from high-resolution nuclear magnetic resonance. Biochemistry 49:7969–7971
Zelent B, Buettger C, Grimsby J, Sarabu R, Vanderkooi JM, Wand AJ, Matschinsky FM (2012) Thermal stability of glucokinase (GK) as influenced by the substrate glucose, and allosteric glucokinase activator drug (GKA) and the osmolytes glycerol and urea. Biochim Biophys Acta 1824:769–784
Zelent B, Raimondo A, Barrett A, Buettger CW, Chen P, Gloyn AL, Matschinsky FM (2014) Analysis of the co-operative interaction between the allosterically regulated proteins GK and GKRP using tryptophan fluorescence. Biochem J 459:551–564
Zelent B, Odili S, Buettger C, Shiota C, Grimsby J, Taub R, Magnuson MA, Vanderkooi JM, Matschinsky FM (2008) Sugar binding to recombinant wild-type and mutant glucokinase monitored by kinetic measurement and tryptophan fluorescence. Biochem J 413:269–280
Zelent B, Odili S, Buettger C, Zelent DK, Chen P, Fenner D, Bass J, Stanley C, Laberge M, Vanderkooi JM, Sarabu R, Grimsby J, Matschinsky FM (2011) Mutational analysis of allosteric activation and inhibition of glucokinase. Biochem J 440:203–215
Chen RF, Knutson JR, Ziffer H, Porter D (1991) Fluorescence of tryptophan dipeptides: correlations with the rotamer model. Biochemistry 30:5184–5195
Xu J, Knutson JR (2009) Quasi-static self-quenching of Trp-X and X-Trp dipeptides in water: ultrafast fluorescence decay. J Phys Chem B 113:12084–12089
Vagenende V, Yap MG, Trout BL (2009) Mechanisms of protein stabilization and prevention of protein aggregation by glycerol. Biochemistry 48:11084–11096
Liang Y, Kesavan P, Wang LQ, Niswender K, Tanizawa, Y, Permutt, MA, Magnuson MA, Matschinsky FM (1995) Variable effects of maturity-onset-diabetes-of-youth (MODY)-associated glucokinase mutations on substrate interactions and stability of the enzyme. Biochem J 309:167–173
Davis EA, Cuesta-Munoz A, Raoul M, Buettger C, Sweet I, Moates M, Magnuson MA, Matschinsky FM (1999) Mutants of glucokinase cause hypoglycaemia and hyperglycaemia syndromes and their analysis illuminates fundamental quantitative concepts of glucose homeostasis. Diabetologia 42:1176–1186
Lakowicz JR (2006) Principles of fluorescence spectroscopy. Springer, New York
Szabo AG, Rayner DM (1980) Fluorescence decay of tryptophan conformers in aqueous solutions. J Am Chem Soc 102:554–563
Konev SV (1967) Fluorescence and phosphorescence of proteins and nucleic acids. Penum Press, New York
Dashnau JL, Sharp KA, Vanderkooi JM (2005) Stereochemical aspects of aldohexopyranose hydration as studied by water-water hydrogen bond angle analysis. J Phys Chem 109:24152–24159
Zelent B, Vanderkooi JM (2009) Infrared spectroscopy used to study ice formation: the effect of trehalose, maltose and glucose on melting. Anal Biochem 390:215–217
Alcala JR, Gratton E, Prendergast FG (1987) Fluorescence lifetime distributions in proteins. Biophys J 51:597–604
Toptygin D, Brand L (2000) Spectrally- and time-resolved fluorescence emission of indole during solvent relaxation: a quantitative model. Chem Phys Lett 322:496–502
Niemyer H, de la Luz Cardenase M, Rabajille E, Ureta T, Clark-Turri L, Penaranda J (1975) Sigmoidal kinetics of glucokinase. Enzyme 20:321–333
Storer AC, Cornish-Bowden A (1976) Kinetics of rat liver glucokinase. Co-operative interactions with glucose at physiologically significant concentrations. Biochem J 159:7–14
Liu S, Ammirati MJ, Song X, Knafeis JD, Zhang J, Greasley SE, Pfefferkorn JA, Qui X (2012) Insights into mechanism of glucokinase activation: observation of multiple distinct protein conformations. J Biol Chem 287:13598–13610
Kim YB, Kalinowski SS, Marcinkeviciene J (2007) A pre-steady state analysis of ligand binding to human glucokinase: evidence for a preexisting equilibrium. Biochemistry 46:1423–1431
Chen Y, Barkley MD (1998) Toward understanding tryptophan fluorescence in proteins. Biochemistry 37:9976–9982
Callis PR, Vivian JT (2003) Understanding the variable fluorescence quantum yield of tryptophan in proteins using QM-MM simulations. Quenching by charge transfer to the peptide backbone. Chem Phys Lett 369:409–414
Albani JR (2014) Origin of tryptophan fluorescence lifetimes. Part 2: fluorescence lifetimes origin of trytophan in proteins. J Fluoresc 24:105–117
Gekko K, Timasheff SN (1981) Mechanism of protein stabilization by glycerol: preferential hydration in glycerol-water mixtures. Biochemistry 20:4667–4676
Pickup JC, Hussain R, Evans BD, Rolinski OJ, Birch DJS (2005) Fluorescence-based glucose sensors. Biosens Bioelectron 20:2555–2565
Acknowledgements
The work was supported by grants from NIDDK no. 22122 and 19,525 (F.M.M.), NSF grant CBET-1264608 (I.G.) and National Research Initiative or Agriculture and Food Research Initiative Grant no. 2014–67,017-2149 (R.D.L., B.Z.& M.G.C.) from USDA National Institute of Food and Agriculture, Improving Food Quality.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Zelent, B., Bialas, C., Gryczynski, I. et al. Tryptophan Fluorescence Yields and Lifetimes as a Probe of Conformational Changes in Human Glucokinase. J Fluoresc 27, 1621–1631 (2017). https://doi.org/10.1007/s10895-017-2099-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10895-017-2099-x