Abstract
The ruthenium(II) complexes having 2,2′-bipyridine and phenanthroline derivatives are synthesized and characterized. The photophysical properties of these complexes at pH 12.5 are studied. The electron transfer reaction of biologically important phenolic acids and tyrosine are studied using absorption, emission and transient absorption spectral techniques. Semiclassical theory is applied to calculate the rate of electron transfer between ruthenium(II) complexes and biologically important phenolic acids.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Antioxidants prevent cells damage by free radicals and molecules like H2O2 that are released during the normal metabolic process of oxidation [1, 2]. Many plant constituents with antioxidant activity have been identified, among these the polyphenols are attracting much attention [3, 4]. The antioxidant activity of polyphenols is mainly due to their redox properties, leading to (i) neutralizing free radicals, (ii) quenching oxygen, (iii) decomposing peroxides [5, 6]. The oxidation potential of a phenol provides an estimate of the energy required to donate an electron; the lower the oxidation potential, lesser the energy required to donate an electron and the higher the expected antioxidant activity [7, 8]. Phenols scavenge reactive oxygen species and free radicals through several proposed mechanisms, including delocalization of electrons, formation of intramolecular hydrogen bonds, and rearrangement of their molecular structure [9]. The number of hydroxyl groups in the aryl moiety leads to substantial changes in the redox potentials of phenols, suggesting that additional hydroxyl groups make polyphenols better electron donors (antioxidants) [10].
As oxidants, the ruthenium(II)-polypyridyl complexes in the excited state have favorable redox potentials and are chemically stable [11–15]. In the past two decades, Rajagopal and coworkers have examined the effect of introducing electron-donating and withdrawing groups in the 4,4′-position of the ligand 2,2′-bipyridine and additional nitrogen atom in the pyridine moiety (2,2′-bipyrazine and pyridyl pyrazine) on the photoinduced electron transfer (ET) reactions of [Ru(NN)3]2+ with phenolate ions [16–23]. Though the excited state electron transfer reactions of most [Ru(NN)3]2+ complexes with neutral phenols are endergonic, the introduction of additional nitrogen atom in the ligand (for example 2,2’-bipyrazine and 4,4’-bipyridine) results in the hydrogen bond formation between phenolic H-atom and N-atom of the ligand of [Ru(NN)3]2+ which facilitates electron transfer from phenol to the excited state [Ru(NN)3]2+ [24, 25]. Further in the presence of a base also the hydrogen bond formation between the phenol and base facilitates electron transfer reaction. Herein we report the observed results on the electron transfer reaction of biologically important phenolic acids with the excited state ruthenium(II)-polypyridine complexes. The oxidation process of tyrosine is of much significance because of its importance in various enzymes [26]. Such redox reactions involving a tyrosine side chain occur in, among others, ribonucleotide reductase, cytochrome c oxidase, galactose oxidase, and photosystem II (PSII) [27, 28]. The side chain of tyrosine participates in several very important electron transfer reactions of this type. Even though, tyrosine is not a phenolic acid, it has phenolic group and carboxylic group altogether and has electron donation capacity and also an important amino acid. Therefore we have also included the electron transfer reaction of tyrosine with [Ru(NN)3]2+ complexes here.
Experimental
The commercial samples of RuCl3.nH2O, 2,2′-bipyridine, 4,4′-dicarboxy-2,2′-bipyridine, 4,7-diphenyl-1,10-phenanthroline disulfonic acid (dpsphen), 4,7-diphenyl-1,10-phenanthroline (dpphen), ammonium hexaflurophosphate and other chemicals were procured from Sigma. The luminophores [Ru(bpy)3]2+ (I), [Ru(bpy)2(dcbpy)]0 (II), [Ru(dcbpy)3]4− (III), [Ru(phen)3]2+ (IV), [Ru(dpphen)3]2+ (V) and [Ru(dpsphen)3]4− (VI) (Chart 1) were prepared using reported literature methods [29–34] and characterized by spectral techniques.
Absorption and Emission Spectral Measurements
Sample solutions of the [Ru(NN)3]2+ complexes and the quenchers were freshly prepared for each measurement. The absorption spectral measurements were carried out using Cyber lab spectrophotometer. Emission intensity measurements were carried out and the emission spectra were recorded using JASCO FP-6300 spectrofluorimeter. Excitation and emission slits with a band-pass of 2.5 or 5 nm are used for all measurements. Care has been taken to minimize solvent and/or water evaporation. The nitrogen gas is purified by passing through Fieser’s solution to remove the oxygen present in the solution. All the sample solutions used for emission measurements were deaerated for about 25 min by dry nitrogen gas purging and keeping the solutions in cold water to ensure that there is no change in volume of the solution.
Transient Absorption Measurement
Transient absorption measurements were made with laser flash photolysis technique using an Applied Photophysics SP-Quanta Ray GCR-2(10) Nd:YAG laser as the excitation source. The time dependence of the luminescence decay is observed using a Czerny-Turner monochromator with a stepper motor control and a Hamamatsu R-928 photomultiplier tube. The production of the excited state on exposure to 355 nm was measured by monitoring (pulsed Xenon lamp of 250 W) the absorbance change. The change in the absorbance of the sample on laser irradiation was used to calculate the rate constant as well as to record the time-resolved absorption transient spectrum. The change in the absorbance on flash photolysis was calculated using the expression
In eqs. (1) and (2) ΔΑ is the change in the absorbance at time t, I0, I and It are the voltage after flash, the pretrigger voltage and the voltage at particular time respectively. A plot of ln(∆At – ∆A∞) vs time gives a straight line. The slope of the straight line gave the rate constant for the decay and the reciprocal of rate constant gave the lifetime of the triplet excited state of [Ru(NN)3]. The time-resolved transient absorption spectrum was recorded by plotting the change in absorbance at a particular time vs wavelength.
Determination of Luminescence Quenching Constants
The sample solutions were purged carefully with dry nitrogen for 3 min. The luminescence measurements were performed at different quencher concentration and the quenching rate constant, kq, values were determined from the Stern-Volmer plot using the equations given below [35].
Here I0 and I are the luminescence intensities and τ0 and τ are the excited state lifetimes of [Ru(NN)3]2+ complexes in the absence and presence of quencher respectively, KSV, the Stern-Volmer constant and kq, the quenching rate constant.
Determination of Binding Constants
UV-Vis Absorption Spectral Titration
The binding constant (Ka) of quencher with the sensitizer in the ground state was evaluated with the aid of Benesi-Hildebrand equation (eq. 5) [36], from the plot of 1/ΔA vs 1/[G].
Here H and G stand for host (sensitizer) and guest (quencher) respectively. ΔA - the change in the absorbance of the [Ru(NN)3]2+ complexes on the addition of [quencher]. Δε - the difference in the molar extinction coefficient between the free [H] and [H] - [G] complex. [H]- the total concentration of host. [G] - the total concentration of guest. For all the guest molecules examined, plots of 1/ΔA values as a function of 1/[G] values give good straight line, supporting the 1:1 complex formation.
Binding Constant Calculated from Luminescence Enhancement
When the phenolate ion is added to [Ru(NN)3]2+ complex we have observed luminescence enhancement in the presence of some quenchers but quenching in the presence of other quenchers. In order to analyze the luminescence enhancement data we have used modified Benesi-Hildebrand equation for the estimation of binding constant in the presence of quenchers [37]. The relevant equation is shown in eq (6)
Here I0 and I are the luminescence intensity of the metal complex in the absence and presence of the quencher, Ka is the binding constant and a and b are constants. From the slope and intercept of the plot of I0/I-I0 against the inverse of the concentration term, [Q]−1, the value of binding constant Kb, is calculated.
Binding Constant Calculated from Luminescence Quenching
We have calculated the binding constant of the quencher with the sensitizer from the luminescence quenching data also by using the following equation which relates the change in emission intensity with the concentration of quencher [35].
ΔI = I0-I, where I0 is luminescence intensity in the absence of the quencher and I is luminescence intensity in the presence of various concentration of the quencher. [Q] – Concentration of the quencher, Ka – binding constant. The plot predicts a linear relationship between 1/ΔI and [Q] with the slope 1/(I0-I) Ka and intercept 1/(I0-I). The ratio intercept/slope is the association constant, Ka.
Results and Discussion
The structures of the metal complexes and the quenchers used in the present study are shown in Chart 1.
Photophysical Properties of [Ru(NN)3]2+ Complexes at pH 12.5
The absorption and emission maxima, luminescence lifetime (τ) and excited state redox potential of all six [Ru(NN)3]2+ complexes at pH 12.5 are presented in Table S1 and the spectra are shown in Figs. S1 and S2. The absorption and emission maxima, excited state lifetime and luminescence quantum yield vary substantially with the change of structure of ligands in [Ru(NN)3]2+. As all the [Ru(NN)3]2+ complexes used in the present study are known metal complexes their photophysical properties have been extensively reported [11, 38, 39]. The value redox potential varies with the change of ligand in [Ru(NN)3]2+ complexes from 0.65 to 1.0 V. It is to be remembered that the electron-withdrawing carboxyl group present in the ligand is in the form of carboxylate ion and the electron–withdrawing power of –CO2 − is less compared to –CO2H which is clearly understood from the Hammett σ values 0.40 and 0.11 respectively [40]. To have a comprehensive study on the dynamics of the electron transfer reaction of electron donor with acceptor there must be a substantial change in the ΔG0 value. The ΔG0 are values collected in Table S2.
Interaction of [Ru(NN)3]2+ Complexes with Phenolic Acids and Tyrosine
Absorption Spectral Changes of [Ru(NN)3]2+ Complexes in the Presence of Phenolic Acids and Tyrosine
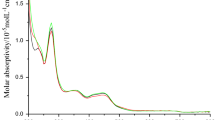
The absorption spectra of [Ru(NN)3]2+ complexes in the presence of phenolic acids and tyrosine are recorded to realize the extent of the ground state complex formation. The absorption spectra of phenolic acids and tyrosine are shown in Fig. S3. The sample absorption spectra of [Ru(NN)3]2+ complexes in the absence and presence of phenolic acids in aqueous medium at pH 12.5 and 298 K are given in Figs. 1 and S4-S6. Since gallic and protocatechuic acids have weak absorption at 454 nm and 424 nm respectively, which are close to the region of MLCT absorption of [Ru(NN)3]2+ complexes, the binding constant of these phenolic acids have been calculated using emission spectral data. The absorption spectra of [Ru(NN)3]2+ complexes in the presence of phenolic acids show 2 to 3 nm blue shift which is due to the ground state complex formation.
The absorption spectra of ruthenium(II) complexes in the absence and presence of different concentrations of tyrosine at pH 12.5 are shown in Fig. S7. Tyrosine has two absorption maxima at 254 and 288 nm and there is no absorption at >320 nm and the spectrum is given in Fig. S3d. The absorption intensity of [Ru(NN)3]2+ complex corresponding to the MLCT transition (~450 nm) increases with an increase in [tyrosinate ion] with 3–7 nm blue shift (Fig. S7). Thus an increase in the absorption intensity at 450 nm with a blue shift on the addition of tyrosine clearly indicates the binding of tyrosine with the photosensitizer [Ru(NN)3]2+. Similar type of adduct formation is reported in the literature for OsIII(bpy)3 3+ with tyrosine [41]. From the increase in the absorption intensity, association constant of tyrosine-[Ru(NN)3]2+ adduct has been estimated using Benesi-Hildebrand (Fig. S8) method and the values are given in Table S3.
Emission Spectral Studies of [Ru(NN)3]2+ Complexes with Phenolic Acids and Tyrosine
The luminescence of all six [Ru(NN)3]2+ complexes has been recorded in the absence and presence of different concentration of the phenolic acids and tyrosine and the change of luminescence intensity with the change of phenolic acid/tyrosine concentration is shown in Figs. 2, 3, and 4 and S9-S14. From the steady state measurements, it is seen that the luminescence of the [Ru(NN)3]2+ complexes is quenched by the added quenchers at pH 12.5. These quenching data are analyzed in terms of Stern-Volmer equation and the experimental bimolecular quenching rate constants, kq for the quenching of [Ru(NN)3]2+ complexes with phenolic acids in aqueous medium at pH 12.5 and the ΔGo values are given in Tables 1 and S2 respectively.
When we analyze the bimolecular quenching rate constants (kq) values obtained for the luminescence quenching of complexes I-VI with polyphenolic acids, the rate increases when one bpy unit in [Ru(bpy)3]2+ is substituted by dcbpy unit [Ru(bpy)2(dcbpy)]0 but the rate decreases when further dcbpy units are introduced in the place of bpy, [Ru(dcbpy)3]4−. These results are interesting because with the introduction of dcbpy and -SO3H units in the metal complex the reaction becomes more exergonic, i.e., ΔGo becomes more negative. Though ΔGo of the reaction is favorable with the introduction of –CO2 − and SO3 − groups in the ligand, it also makes the [Ru(NN)3]2+ complex more bulky and negatively charged species. The rate retardation observed with [Ru(dcbpy)3]4− compared to [Ru(bpy)2(dcbpy)] may be due to the repulsive force operating between negatively charged metal complex and the phenolate anion and also steric effect. In the case of [Ru(dpphen)3]2+ (V), the observed binding constant (Ka) as well as the quenching rate constant (kq), are more than the values observed for [Ru(phen)3]2+ (IV). This is likely due to the π-π stacking and strong hydrophobic interaction when phenyl moiety is introduced in the 4,7-position of the phenanthroline ligand. To the extent that π-π stacking exists between the ligands of [Ru(NN)3]2+ complexes and quenchers the binding becomes stronger and facilitates the photoinduced electron transfer reaction [42]. Interestingly the rate also increases, when the number of hydroxyl groups present in the phenolic acid increases. This is understandable as the introduction of additional hydroxyl groups makes the quencher better electron donor and ΔGo becomes more negative. Therefore the change in the structure of the ligand of the [Ru(NN)3]2+ complex as well as the quencher affects the rate of quenching in this redox system through change of binding constant and ΔG0 values. The quenching reactions carried out in the present study occur by ET mechanism which is substantiated from the transients formed during the flash photolysis study. Contrary to the luminescence quenching observed with five [Ru(NN)3]2+ complexes a substantial enhancement of emission intensity is observed with [Ru(dspphen)3]4− in the presence of gallic and protocatechuic acid at pH 12.5 as shown in the Fig. 3. However for the same [Ru(NN)3]2+ complex the emission intensity is quenched in the presence of p-hydroxybenzoic acid. This enhancement in the emission intensity with the incremental addition of gallic and protocatechuic acid are used to calculate the binding constant of these phenolic acids with [Ru(dspphen)3]4−.
The modified Benesi-Hildebrand [37] equation (eq. 6) is used to calculate the binding constant of [Ru(dspphen)3]4− with gallic and protocatechuic acid at pH 12.5 from the spectral data collected in the Fig. 3. The Benesi-Hildebrand plot is given in Fig. S15. The binding constant values calculated from the slope and intercept of the plot for gallic and protocatechuic acids are 1.6 × 103 M−1 and 1.5 × 103 M−1 respectively. These binding constants show that the binding is fairly strong.
The luminescence quenching of [Ru(NN)3]2+ complexes with various [tyrosine] in aqueous medium at pH 12.5 are shown in the Figs. 4 and S14 and the quenching rate constant (kq) values are given in the Table 1. Intramolecular electron transfer from tyrosine to the excited state [Ru(bpy)3]2+ in the presence of external electron acceptor methyl viologen is reported by Sjodin et al. [31]. We have analysed the photoinduced intermolecular (bimolecular) electron transfer reactions of [Ru(NN)3]2+ complexes with tyrosine at pH 12.5. The spectral results in Figs. 4 and S14 show that the luminescence intensity of the [Ru(NN)3]2+ complexes is reduced substantially in the presence of tyrosinate ion. The measured kq values for the quenching of [Ru(NN)3]2+ complexes with tyrosine are given in Table 1. When we analyse the bimolecular quenching rate constants (kq) values obtained for the complexes I-III with tyrosinate ion, we notice that the rate increases slightly from 5.1 × 108 M−1 s−1 to 6.8 × 108 M−1 s−1 with the introduction of one dcbpy unit instead of bpy in the [Ru(NN)3]2+ complex. But the rate decreases to the value of 9.0 × 107 M−1 s−1 when we replace all three bpy units by dcbpy ligand. These results may be explained in terms of ΔGo values, charge and bulk effect. The substantial change in the kq value from 1.5 × 1010 M−1 s−1 to 3.8 × 107 M−1 s−1, with the introduction of sulphanato group in the phenyl ring of 4,7-diphenyl phenanthroline ligand may also be attributed to the bulky and negatively charged nature of the complex [Ru(dspphen)3]4−.
In analogy to the analysis of results observed with the phenolic acid-[Ru(NN)3]2+ adduct we assume that prior to electron transfer, photoexcited [Ru(NN)3]2+ complex forms adduct with tyrosine and ket values of tyrosine collected along with other phenolate ions. The experimental and calculated bimolecular ET rate constant values for the quenching of [Ru(NN)3]2+ complexes with tyrosine are in good agreement.
Adduct Formation and the Determination of Association Constants of Polyphenolate ion with [Ru(NN)3]2+ Complexes
In analogy to the reports by Meyer et al. and Wenger et al. [43, 44], on the reaction of excited state [Ru(NN)3]2+ complexes with 1,4-dihydroxybenzene and phenol we also assume that prior to electron transfer, [Ru(NN)3]2+ complexes and polyphenolic acids form π- stacked adducts. This is proposed earlier by Hoffman et al. [42], based on 1H NMR spectral study. They have substantiated the existence of ground state interaction between [Ru(bpy)3]2+ and phenol due to hydrophobic or face-to-face π- stacking interactions using NMR spectroscopy. Thus following the procedure of Meyer et al. [43], we have calculated the association constant (Ka) for phenolic acid-[Ru(NN)3]2+ adduct and the values are given in the Table 2. Although, similar to the observation of Meyer on the Ru(bpy)2(bpz)2+/1,4-dihydroquinone couple [43], the appreciable association constants observed realize us to conclude that the overall quenching process occurs by a combination of static and dynamic quenching. It appears reasonable to follow Meyer’s assumption that the formation of adducts is responsible for competition between static and dynamic quenching pathway.
In order to obtain first-order rate constant (ket) for reduction in the excited state sensitizer-quencher adducts, one may divide the value of kq by Ka. This procedure yields ket values (Table 2) that are on the order of 105 to 107 s−1 for six [Ru(NN)3]2+ complexes. Binding constant values are on the order of 10 to 104 M−1.
Transient Absorption Spectra
Nanosecond transient absorption difference spectra of [Ru(NN)3]2+ complexes in the aqueous medium at pH 12.5 were recorded after 355 nm laser pulse excitation at various time delay and are shown in Fig. S16. For each solution, a set of kinetic traces were collected for a sequence of monitoring wavelengths at 10 ns time intervals. The spectrum, at each time delay, consists of bleach around 450 nm and a positive absorption with maxima centered around 380 and 510 nm. Figure 5a shows the transient absorption spectra of excited state [Ru(bpy)2(dcbpy)] in the presence of quenchers gallic acid and tyrosine. The absorption band at 380 nm corresponds to the dcbpy anion radical which is in close agreement with our previous report [45]. The bleaching around 450 nm is due to the loss of ground state absorption, dπ-π*(MLCT) transition [46].
The bleaching around 600–700 nm corresponds to the light emission from the excited state to the ground state. The peak formation around 400 nm in the presence of gallic acid is attributed to the phenoxyl radical and another band around 510 nm to the formation of ruthenium(I) species. The peak formation at 400 and 520 nm corresponding to the phenoxyl radical and [Ru(dcbpy)3]+ was also reported by Swarnalatha et al. [16, 18]. Generally, a positive signal originates from the absorption of excited state species or any product generated at the excited state and the positive signal observed around 510 nm is due to the reduction of Ru(II) to Ru(I) [47, 48]. Similar results are observed when the transient absorption spectra are recorded in the presence of tyrosine (Fig. 5b). The transient absorption spectra clearly show that the excited state of Ru(II) complexes undergo rapid ET reactions with polyphenolic acids and tyrosine. After establishing the electron transfer nature of the quenching of *[Ru(NN)3]2+ with polyphenolate and tyrosinate ions using laser flash photolysis technique it is appropriate to propose the mechanism for the process [49]. The luminescence quenching of excited state ruthenium(II)-complexes by phenolate and tyrosinate ions can be explained by Scheme 1.
Application of Theory of Electron Transfer
After establishing electron transfer nature of the reaction of polyphenolate and tyrosinate ions with [Ru(NN)3]2+ experimentally now we would like to compare the experimental rate constant values with the values that can be calculated from the theories of electron transfer. To meet this end we have calculated the rate constants for the ET reaction of phenolate and tyrosinate ions to the excited state [Ru(NN)3]2+ complexes using semiclassical theory of ET [50, 51]. The semiclassical expression of ET (eq. 8) popularized by Closs and Miller [52] has been applied to this photoinduced ET reaction.
In eq. (8) HDA is the electronic coupling matrix element, λo is the solvational contribution to the reorganization energy, λi is the vibrational contribution to λ, s = λi/hυ, υ is the high energy vibrational frequency associated with the acceptor, and m is the density of product vibrational levels. As all the ET reactions, the reaction of phenolic acids and tyrosine with six [Ru(NN)3]2+ complexes, in the present study fall in the normal region (−ΔGo < λ) (vide infra) the above equation is simplified to eq., (9)
The total driving force, ΔG, includes electrostatic corrections to ΔGo for the work required to bring the products and reactants together (ΔG = ΔGo– wp – wr). The solvational reorganization energy, λo, can be estimated from the classical dielectric continuum model (eq. (10).
where Dop and Ds are the optical and static dielectric constants of the solvent respectively and rA and rB are the reactants radii and r is the sum of rA and rB. The values of solvent parameters have been collected from the literature, [53, 54] and radii of the [Ru(NN)3]2+ complexes were calculated from MM3 model. The value of λo calculated using eq. (10) is 0.9 eV. The value of λi is taken as 0.2 eV from our previous studies [16, 18] and is employed in the calculation of the rate constant for ET reaction [54]. Thus the value of the reorganization energy (λ) for this redox system is 1.10 eV.
The free energy change (∆G0) of ET reaction can be calculated by using the expression (11).
Where wp and wr are electrostatic correction terms for product and reactant. The ∆G0 values calculated from this equation are collected in Table S2 and the values are in the range of 0.21 to −0.75 eV. By using eqn. (9) the rate constants for electron transfer, ket, have been calculated and compared with experimental values collected in the Table 2. Comparison of ∆G0 and λ values shows that the title ET reaction falls in the normal region (i.e. -ΔGo < λ).
The calculated rate constants for ET from phenolate ions to *[Ru(NN)3]2+ by applying semiclassical expression of ET along with experimentally observed values for the above ET reaction are on the order of 105 to 107 s−1. The ket data in the Table 2 show that the values calculated from semiclassical theory are in fair agreement with experimental values (i.e., the difference between them is in the range 2–10 times). Binding constant of phenolate ions with [Ru(NN)3]2+ varies in the range of 10 to 104 M−1. The magnitude of Ka indicates the strength of the binding of quencher with [Ru(NN)3]2+ in the following order, phenol < pHBA < tyrosine < DHBA < GA at pH 12.5. Thus the overall quenching process is influenced by both binding constant (Ka) and the rate of electron transfer within the excited state [Ru(NN)3]2+- quencher adduct (ket) but more predominantly by Ka values. This result is slightly different from the observation made by Wenger et al. [44] for simple phenols where the overall rate of quenching is decided by Ka values. Because of the presence of more number of –OH groups in the aryl moiety the rate of electron transfer as well as binding constant vary substantially in the present study.
Effect of pH on the Rate of Quenching Reaction
We also report here the results related to the influence of the pH (pH 8, 8.5, 9, 9.5, 10, 10.5, 11.5 and 12.5) on the luminescence quenching of the [Ru(NN)3]2+complexes with gallic acid as quencher. The changes of emission intensity with increasing concentration of gallic acid at pH in the range 8.0 to 12.5 have been studied and the sample spectral changes at different pH are given in the Figs. 2a, 6, S9a, S10a, S11a, S12a and S17-20.
Interestingly the quenching rate constant (kq) data given in Table S4 show that the quenching of the complexes I – V with gallic acid is moderately sensitive to pH and the rate increases substantially, when the pH of the medium is varied from pH 8.5 to pH 11.5 and almost remains constant when pH >11.5. These results on the effect of pH on the rate of the electron transfer quenching reaction of the excited state [Ru(NN)3]2+ complex with gallic acid can be explained by the EPR spectra of gallic acid at different pH (Fig. 7). The spectrum at high pH is explained by a radical that is totally deprotonated, thereby providing a symmetrical structure with two identical hydrogens. At pH values lower than 10, a doublet of triplet was observed. At pH values greater than 10 a spectrum with a 1:2:1 triplet splitting pattern was observed, consistent with all phenolic OH groups being ionized. Figure 7 shows the EPR spectrum of gallic acid consistent with hyperfine spliting due to three protons (aH = 1.00G, aH = 0.23 G and aH = 0.28 G) and at pH 11, 1:2:1 spectrum is observed with two identical hyperfine splitting constant aH = 1.07 G. The pH- dependence of these spectral shapes suggests that these spectra are due to species of different protonation. The larger hyperfine splittings also suggest a greater spin density in the aromatic ring upon loss of a second phenolic proton, as would be expected [55]. However, the spectrum observed at lower pH having three different hyperfine splitting suggests an unsymmetrical structure with three protons; spectra with similar shapes from gallate-derivatives have been observed. The intensity of the EPR signal of the fully ionized species increases significantly in the more alkaline environment. Gallic acid has four potential acidic protons having pKa values of 4.0 (carboxylic acid), 8.7, 11.4, and >13 (phenolic OHs) [56]. The known pKa values of the gallate radical are ≈4 for the carboxyl group and 5.0 for phenolic hydrogen. Thus, under the two pH conditions (9 and 11), the carboxylic acid as well as a phenolic-OH of the semiquinone will be ionized; the proposed structures of these radicals are as presented in the Fig. 7.
Conclusion
Six [Ru(NN)3]2+ complexes of varying charges and hydrophobicites with ligands bipyridine, phenanthroline and their derivatives have been synthesized and their phtophysical properties and photoinduced electron transfer reactions with polyphenolate and tyrosinate ions studied. The absorption and luminescent spectral data indicate a ground state complex formation between [Ru(NN)3]2+ and polyphenolate/tyrosinate ions. The transient absorption spectra and luminescence quenching data clearly show that the excited state of [Ru(NN)3]2+ complexes undergo ET reactions with phenolic acids and tyrosine at pH 12.5. The quenching rate constant is higher for phenolic acids containing more number of hydroxyl groups and an enhancement in the luminescence of the complex VI with gallic and protocatechuic acids. These interesting results are analyzed in terms of ∆G0 values, charge and bulk effect.
References
Ozyürek M, Güngör N, Baki S, Güclü K, Apak R (2012) Development of a silver nanoparticle-based method for the antioxidant capacity measurement of polyphenols. Anal Chem 84:8052–8059
Bani D, Bencini A (2012) Developing ROS scavenging agents for pharmacological purposes: recent advances in design of manganese-based complexes with anti-inflammatory and anti- nociceptive activity. Curr Med Chem 19:4431–4444
Mubarak A, Swinny EE, Ching SYL, Jacob SR, Lacey K, Hodgson JM, Croft KD, Considine MJ (2012) Polyphenol composition of plum selections in relation to total antioxidant capacity. J Agric Food Chem 60:10256–10262
Vassallo N (2008) Polyphenols and health: new and recent advances. Nova Science Publishers, Incorporated
Quideau S, Deffieux D, Douat-Casassus C, Pouységu L (2011) Plant polyphenols: chemical properties, biological activities, and synthesis. Angew Chem Int Ed 50:586–621
Charles DJ (2012) Antioxidant properties of spices. Springer, Herbs and Other Sources
Costentin C, Louault C, Robert M, Savéant J-M (2009) The electrochemical approach to concerted proton–electron transfers in the oxidation of phenols in water. Proc Natl Acad Sci 106:18143–18148
Dickinson BC, Chang CJ (2011) Chemistry and biology of reactive oxygen species in signaling or stress responses. Nat Chem Biol 7:504–511
Beloborodova N, Bairamov I, Olenin A, Shubina V, Teplova V, Fedotcheva N (2012) Effect of phenolic acids of microbial origin on production of reactive oxygen species in mitochondria and neutrophils. J Biomed Sci 19:89–97
Song N, Stanbury DM (2012) Oxidation of phenol by tris(1,10-phenanthroline)osmium(III). Inorg Chem 51:4909–4911
Kalyanasundaram K (1982) Photophysics, photochemistry and solar energy conversion with tris(bipyridyl)ruthenium(II) and its analogues. Coord Chem Rev 46:159–244
Campagna S, Puntoriero F, Nastasi F, Bergamini G, Balzani V (2007) Photochemistry and photophysics of coordination compounds: ruthenium. In: Balzani V, Campagna S (eds) Photochemistry and photophysics of coordination compounds I. Springer, Berlin Heidelberg, pp. 117–214
Balzani V, Juris A (2001) Photochemistry and photophysics of Ru(II)-polypyridine complexes in the bologna group. From early studies to recent developments. Coord Chem Rev 211:97–115
Baudin HB, Davidsson J, Serroni S, Juris A, Balzani V, Campagna S, Hammarström L (2002) Ultrafast energy transfer in binuclear ruthenium–osmium complexes as models for light-harvesting antennas. J Phys Chem A 106:4312–4319
Badjić JD, Balzani V, Credi A, Silvi S, Stoddart JF (2004) A molecular elevator. Science 303:1845–1849
Swarnalatha K, Rajkumar E, Rajagopal S, Ramaraj R, Banu IS, Ramamurthy P (2011) Proton coupled electron transfer reaction of phenols with excited state ruthenium(II) – polypyridyl complexes. J Phys Org Chem 24:14–21
Rajkumar E, Rajagopal S, Ramamurthy P, Vairamani M (2009) Photophysics of ruthenium(II) complexes carrying amino acids in the ligand 2,2′-bipyridine and intramolecular electron transfer from methionine to photogenerated Ru(III). Inorg Chim Acta 362:1629–1636
Swarnalatha K, Rajkumar E, Rajagopal S, Ramaraj R, Lu Y-L, Lu K-L, Ramamurthy P (2005) Photoinduced electron transfer reactions of ruthenium(II) complexes containing 2,2′-bipyridine-4,4′-dicarboxylic acid with phenols: steric and charge effects. J Photochem Photobiol A Chem 171:83–90
Rajendran T, Thanasekaran P, Rajagopal S, Gnanaraj GA, Srinivasan C, Ramamurthy P, Venkatachalapathy B, Manimaran B, Lu K-L (2001) Steric effects in the photoinduced electron transfer reactions of ruthenium(II)-polypyridine complexes with 2,6-disubstituted phenolate ions. Phys Chem Chem Phys 3:2063–2069
Thanasekaran P, Rajagopal S, Srinivasan C (1998) Photoredox reactions of tris(2,2′-bipyrazine)-, tris(2,2′-bipyrimidine)- and tris(2,3-bis[2-pyridyl]pyrazine)ruthenium(II) cations with phenolate ions in aqueous acetonitrile. J Chem Soc Faraday Trans 94:3339–3344
Thanasekaran P, Rajendran T, Rajagopal S, Srinivasan C, Ramaraj R, Ramamurthy P, Venkatachalapathy B (1997) Marcus inverted region in the photoinduced electron transfer reactions of ruthenium(II) − polypyridine complexes with phenolate ions. J Phys Chem A 101:8195–8199
Rajendran T, Rajagopal S, Srinivasan C, Ramamurthy P (1997) Micellar effect on the photoinduced electron-transfer reactions of ruthenium(II)-polypyridyl complexes with phenolate ions. Effect of Cetyltrimethylammonium cHloride. J Chem Soc Faraday Trans 93:3155–3160
Rajagopal S, Gnanaraj GA, Mathew A, Srinivasan C (1992) Excited state electron transfer reactions of tris(4,4′-dialkyl-2,2′-bipyridine)ruthenium(II) complexes with phenolate ions: structural and solvent effects. J Photochem Photobiol A Chem 69:83–89
Bronner C, Wenger OS (2011) Kinetic isotope effects in reductive excited-state quenching of Ru(2,2′-bipyrazine)3 2+ by phenols. J Phys Chem Lett 3:70–74
Nomrowski J, Wenger OS (2015) Photoinduced PCET in ruthenium–phenol systems: thermodynamic equivalence of Uni- and bidirectional reactions. Inorg Chem 54:3680–3687
Styring S, Sjöholm J, Mamedov F (2012) Two tyrosines that changed the world: interfacing the oxidizing power of photochemistry to water splitting in photosystem II. Biochim Biophys Acta 1817:76–87
Umena Y, Kawakami K, Shen J-R, Kamiya N (2011) Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 a. Nature 473:55–60
Yu Y, Lv X, Li J, Zhou Q, Cui C, Hosseinzadeh P, Mukherjee A, Nilges MJ, Wang J, Lu Y (2015) Defining the role of tyrosine and rational tuning of oxidase activity by genetic incorporation of unnatural tyrosine analogs. J Am Chem Soc 137:4594–4597
Muthu Mareeswaran P, Rajkumar E, Sathish V, Rajagopal S (2014) Electron transfer reactions of ruthenium(II)–bipyridine complexes carrying tyrosine moiety with quinones. Luminescence 29:754–761
Ramanathan S, Ramdass A, Rajkumar E, Rajagopal S (2015) Photoinduced electron transfer reactions of ruthenium(II) phenanthroline complexes with dimethylaniline in aqueous and micellar media. J Fluoresc 25:147–157
Sjodin M, Ghanem R, Polivka T, Pan J, Styring S, Sun L, Sundstrom V, Hammarstrom L (2004) Tuning proton coupled electron transfer from tyrosine: a competition between concerted and step-wise mechanisms. Phys Chem Chem Phys 6:4851–4858
Ramanathan S, Ramdass A, Rajkumar E, Rajagopal S (2015) Micellar effect on the photophysics of heteroleptic ruthenium(II)–phenanthrolinedisulfonato complexes. Luminescence. doi:10.1002/bio.2917
Muthu Mareeswaran P, Babu E, Rajagopal S (2013) Optical recognition of anions by ruthenium(II)-bipyridine-calix[4]arene system. J Fluoresc 23:997–1006
Muthu Mareeswaran P, Babu E, Sathish V, Kim B, Woo SI, Rajagopal S (2014) p-sulfonatocalix[4]arene as a carrier for curcumin. New J Chem 38:1336–1345
Lakowicz JR (2006) Principles of fluorescence spectroscopy, 3rd edition, Springer
Connors KA (1987) Binding constants: the measurement of molecular complex stability. John Wiley & Sons, Ltd, Chichester, U.K.
Upadhyay SP, Pissurlenkar RRS, Coutinho EC, Karnik AV (2007) Furo-fused BINOL based crown as a fluorescent chiral sensor for enantioselective recognition of phenylethylamine and ethyl ester of valine. J Organomet Chem 72:5709–5714
Kalyanasundaram K (1992) Photochemistry of polypyridine and porphyrin complexes. Academic, London
Pal AK, Hanan GS (2014) Design, synthesis and excited-state properties of mononuclear Ru(II) complexes of tridentate heterocyclic ligands. Chem Soc Rev 43:6184–6197
Shimidzu T, Iyoda T, Izaki K (1985) Photoelectrochemical properties of bis(2,2'-bipyridine)(4,4'-dicarboxy-2,2'-bipyridine)ruthenium(II) chloride. J Phys Chem 89:642–645
Fecenko CJ, Meyer TJ, Thorp HH (2006) Electrocatalytic oxidation of tyrosine by parallel rate-limiting proton transfer and multisite electron - proton transfer. J Am Chem Soc 128:11020–11021
Li C, Hoffman MZ, Pizzocaro C, Maihot G, Bolte M (1998) Ground-state interactions between ruthenium(II) − diimine complexes and phenol and monochlorophenols in aqueous solution. Inorg Chem 37:3078–3082
Concepcion JJ, Brennaman MK, Deyton JR, Lebedeva NV, Forbes MDE, Papanikolas JM, Meyer TJ (2007) Excited-state quenching by proton-coupled electron transfer. J Am Chem Soc 129:6968–6969
Kuss-Petermann M, Wolf H, Stalke D, Wenger OS (2012) Influence of donor–acceptor distance variation on photoinduced electron and proton transfer in rhenium(I)–phenol dyads. J Am Chem Soc 34:12844–12854
Rajkumar E, Rajagopal S (2008) Photoinduced electron transfer reaction of tris(4,4′-dicarboxyl-2,2′-bipyridine)ruthenium(II) ion with organic sulfides. Photochem Photobiol Sci 7:1407–1414
Liu Y, Hammitt R, Lutterman DA, Joyce LE, Thummel RP, Turro C (2008) Ru(II) complexes of new tridentate ligands: unexpected high yield of sensitized 1O2. Inorg Chem 48:375–385
Zanarini S, Ciana LD, Marcaccio M, Marzocchi E, Paolucci F, Prodi L (2008) Electrochemistry and electrochemiluminescence of [Ru(II)-tris(bathophenanthroline-disulfonate)]4− in aprotic conditions and aqueous buffers. J Phys Chem B 112:10188–10193
Ciana LD, Zanarini S, Perciaccante R, Marzocchi E, Valenti G (2010) Neutral and dianionic Ru(II) bathophenanthrolinedisulfonate complexes: a route to enhance electrochemiluminescence performance in aqueous media. J Phys Chem C 114:3653–3658
Ito A, Kang Y, Saito S, Sakuda E, Kitamura N (2012) Photophysical and photoredox characteristics of a novel tricarbonyl rhenium(I) complex having an arylborane-appended aromatic diimine ligand. Inorg Chem 51:7722–7732
Barbara PF, Meyer TJ, Ratner MA (1996) Contemporary issues in electron transfer research. J Phys Chem 100:13148–13168
Balzani V (2001) Electron transfer in chemistry: catalysis of electron transfer, heterogenous systems, gas-phase systems, volume 4, Wiley-VCH
Closs GL, Miller JR (1988) Intramolecular long-distance electron transfer in organic molecules. Science 240:440–447
Yoshimura A, Uddin MJ, Amasaki N, Ohno T (2001) Low quantum yields of electron-transfer reaction of photoexcited Ru(bpydc)3 4− with Co(tpy)2 3+ and methyl Viologen2+ (bpydc:2,2′-bipyridine-4,4′-dicarboxylate and tpy:2,2′:6′,2″-terpyridine). J Phys Chem A 105:10846–10853
Schrauben JN, Cattaneo M, Day TC, Tenderholt AL, Mayer JM (2012) Multiple-site concerted proton–electron transfer reactions of hydrogen-bonded phenols are nonadiabatic and well described by semiclassical marcus theory. J Am Chem Soc 134:16635–16645
Eslami AC, Pasanphan W, Wagner BA, Buettner GR (2010) Free radicals produced by the oxidation of gallic acid: an electron paramagnetic resonance study. Chem Central J 4:15–18
Severino JF, Goodman BA, Kay CWM, Stolze K, Tunega D, Reichenauer TG, Pirker KF (2009) Free radicals generated during oxidation of green tea polyphenols: electron paramagnetic resonance spectroscopy combined with density functional theory calculations. Free Radic Biol Med 46:1076–1088
Acknowledgments
Prof. S. Rajagopal thanks UGC, New Delhi for sanctioning UGC-BSR Faculty and UGC Emeritus Fellowships. A. Rajeswari thanks UGC, New Delhi and the Management of Fatima College, Madurai for sanctioning permission to avail the benefits of Faculty Development Programme (FDP). A. Ramdass is the recipient of UGC Meritorious fellowship under the Basic Scientific Resear;ch (BSR) Scheme.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(DOC 2780 kb)
Rights and permissions
About this article
Cite this article
Rajeswari, A., Ramdass, A., Muthu Mareeswaran, P. et al. Electron Transfer Studies of Ruthenium(II) Complexes with Biologically Important Phenolic Acids and Tyrosine. J Fluoresc 26, 531–543 (2016). https://doi.org/10.1007/s10895-015-1738-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10895-015-1738-3