Abstract
The histidine-modified EGFP was characterized as a sensing element that preferentially binds nanomolar concentrations of Cu2+ in a reversible manner (Kd = 15 nM). This research aims to determine the causes of nanomolar-affinity of this mutant by investigating significant structural and energetic alterations of the chromophore in the presence of different copper ion concentrations. In order to reveal the unknown parts of the quenching mechanism we have elaborated a specific approach that combines theoretical and experimental techniques. The theoretical experiment included the modeling of potential distortions of the chromophores and the corresponding changes in energy using quantum mechanical calculations. Differences between the modeled energy profiles of planar and distorted conformations represented the energies of activation for the chromophore distortions. We found that some values of the experimental activation energies, which were derived from fluorescence lifetime decay analysis (ex: 470 nm, em: 507 nm), were consistent with the theoretical ones. Thus, it has been revealed similarity between the theoretical activation energy (50 kJmol−1) for 40° phenolate-ring distortion and the experimental activation energy (52.17 kJmol−1) required for histidine-modified EGFP saturation with copper. This chromophore conformation was further investigated and it has been found that the large decrease in fluorescence emission is attributed to the significant charge transfer over the molecule which triggers proton transfer thereby neutralizing the cromophore.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Copper is a catalytic cofactor for many life processes but in elevated or reduced concentrations can cause serious abnormalities in living cells. According to the present investigations the organism inability to metabolize the copper ions affects the brain and neural tissue, i.e., this deficiency leads to dystonia symptoms in ceruloplasmin, gait disturbances, dysarthria, dementia [1–4].
Recognizing the importance of copper, several experiments were carried out for developing competitive measurement system that allows fast, user-friendly and reliable detection. In life sciences it is a challenge the visualization of strictly regulated availability of copper in the tight copper-binding environment of the cell. The selective recognition of these ions is also a great challenge because they have similar magnitude, valence and coordinating properties with some other metals [5–7].
Biochemists have attempted to develop some effective organic molecules that are capable of selective recognition of copper, taking inspiration from processes found in nature. It was found that the genetically encoded fluorescent sensors based on green fluorescent protein can be applied to analyze the availability of copper in intracellular processes. Many of these applications require reporters with specific physicochemical properties which lead to continuous apparition of genetically modified fluorescent protein variants [8–14].
These findings led us to create a histidine mutated version of the EGFP (Enhanced Green Fluorescent Protein) that was shown to be extremely sensitive and selective toward copper(II).
The spectral properties in the absence and presence of copper ions, as well the determination of dissociation constants of this mutant were well characterized in our previous studies [12, 13], but the causes of its extremely high sensitivity remained unclear. In order to unveil some phases of the photoexcited processes that underlie significant changes of the chromophore, ultra-sensitive and complex equipment is needed [14].
The goal of this paper was to reveal with feasible methods some structural features which are responsible for the dramatic emission decrease. The arguments of several studies have driven us to predict that the increased torsional freedom of the chromophore, mainly about the two exocyclic bonds, results in a speeding up of the fast internal conversion pathway which could be the main cause of the nanomolar sensitivity [15–17]. Theoretically, the chromophore may follow different deexitacion pathways. Non-radiative processes may include intersystem crossing from S1 to T1 state or fast internal conversion completed with excited-state intramolecular isomerization. In green fluorescent proteins mainly the latter occurs [17]. Consequently, the evaluation of intramolecular twisting appears to be important for elucidating the alterations caused by the proximity of metal chelated binding site.
Therefore, we have determined the possible distortions of chromophore’s crystal structure and the corresponding energy changes using quantum mechanical calculations. In this manner the theoretical activation energies of the chromophore distortions were evaluated, the heights of which were linked to the experimental activation energies of 2His-EGFP. We have derived the experimental activation energies from fluorescence lifetime decay, in a manner that from these values the radiative and non-radiative decay rate constants were calculated and they were substituted in Arrhenius equation. Thus, we were able to estimate the magnitude of the chromophore distortions induced by each copper concentration.
In summary, by combining the computational and experimental techniques we attempted to fill the unknown parts of the quenching mechanism which are still unclear [18–21]. Hence, this analysis appears to be a new approach in simulating the chromophore behavior during the quenching process and offers a further opportunity to get a powerful insight into the deexcitation mechanism.
Materials and Methods
Construction of Expression Vectors
The construction of vectors, their expression and purification were effectuated with similar techniques that were described in our previous study. However, we have realized some further modifications in each method as it is described below.
Two vectors were designed and constructed containing the genes of wild type and histidine modified EGFPs [12, 13]. These coding genes were inserted in the same vector (pET15b) under the same conditions in order to be able to identify the influence of introduced residues that are part of the copper binding site, as it was described earlier [12, 13].
The mutated gene fragments from pET-EGFP were amplified with NdeI and BamHI primers by PCR. The resulting gene fragments were digested with NdeI and BamHI restriction enzymes and then ligated into the target vectors. The mixture contained 25–45 ng of genes of interest and 55–75 ng digested vectors. The success of the operations was verified by agarose gel electrophoresis.
Expression and Purification of EGFP Variants
Recombinant plasmids were introduced by heat-shock transformation into E. coli BL21STAR host cells (1 μL vector construct was added to 100 μL chemically competent cells). Transformed cells were plated on LB agar containing 100 μg/ml ampicillin, and incubated at 37 °C overnight. A weak expression (leaky expression) was effectuated without induction and well-folded proteins were obtained [13].
The transformed bacterial colonies were inoculated into 10 mL LB medium containing 100 μg/ μL ampicillin and incubated for 2 hours, than diluted in 250 mL LB supplemented also with 100 μg/μL ampicillin. The cell suspension was incubated at 37 °C and shaked at 180 rpm for 16/18 h [13]. In order to recover the cells, they were centrifuged at 6000 rpm for 20 min. The supernatant was discarded, and the pellet was resuspended in about 40 mL of MOPS buffer pH 7.5 and incubated at −20 °C overnight. Bacterial suspend was lysed by sonication at 4 °C (4 × 2 min, 70 % amplitude, pulse 10 s, 2 s pause). In order to clarify and separate the soluble proteins from insoluble cell components, the resulting solution was subjected to a new centrifugation at 13,000 rpm for 30 min.
The histidine residues of recombinant proteins, which confer affinity for nickel ions, have enabled the purification by immobilized metal (Ni2+) affinity chromatography (IMAC) which contained Ni-charged polymer matrix named Ni-NTA from Novagen [13]. In purification process non-linear gradient elution was applied using FPLC instrument (Amersham Biosciences). The concentration of target proteins in the effluent was monitored by UV light absorption at 280 nm. The advantage of this method was the components selective elution in the presence of various concentrations of imidazole.
The procedure efficiency was verified using 13,5 % SDS-PAGE. Purified proteins were dialyzed against 2 × 1 L 10 mM MOPS buffer pH 7.5 containing 500 mg/l cation exchange Amberlite CG50 resin (in both cases 48 h)[13]. This method was gentler than the ion exchange chromatography and it is quite effective in removing the residual metal ions from the protein solution.
Sample Preparation
Buffered stock solutions of all used EGFP variants were stored in 10 mM MOPS (pH 7,5) in order to avoid the precipitation of unrequested copper complexes. The final concentrations of EGFP probes were always 2 μM. For the titration experiments Cu(SO4)•5H2O was dissolved in distilled water to 1 M stock solutions which were subsequently diluted and added to the protein solution with different final concentrations in the range from 1 to 5 μM. In order to stabilize the system, all samples were incubated at 20 °C for 40–50 min.
TCSPC
All fluorescence intensity decay kinetics were performed in 1 mL solution that contained 1, 2, 3, 4, 5 μM Cu(SO4)•5H2O and 2 μM protein concentrations at 20 °C. Fluorescence intensity decay kinetics was measured with time-correlated single-photon counting method using LifeSpec from Edinburgh Instruments, U.K. The excitation source was Xe900 450 W Xenon Arc LampXenon which emits light between 230 and 2600 nm. The excitation and the emission wavelengths were adjusted to 470 and 507 nm, respectively. Decay curves were collected with a time resolution of 43 ps/channel for 1024 channels in the multichannel analyzer. We have accumulated more than 5000 photons in the maximum channel. The system was controlled by complex software called F900, developed by Edinburgh Instruments.
Data Processing
Lifetime histograms were obtained by TCSPC (Figs. 4 and 5). As will be explained in more detail in section “Results and Discussions”, the mean lifetime of each component was derived by approximation of multiexponential function, the parameters of which were obtained using delta convolution function method. In order to extract the real decay data we used a program created in Mathlab R2012a software which implies the mathematical algorithms reported by Van den Zegel et al. [22].
Molecular Modeling
In order to simulate ab initio the changes in chromophore with quantum mechanical calculations we utilized Firefly/Gamess package (license program of version 7.1.G was a generous gift from Prof. Alex Granovsky [23]). 3D crystal structure of the chromophore was obtained from the Protein Data Bank, ID: 1EMA (http://www.pdb.org/). The structure of 4-hydroxybenzilidene-2,3-dimethyl-imidazolidinone (Fig. 1) was selected as a reference compound in its anionic form.
The geometry optimization of the anionic chromophore, i.e., the reference energy calculation was performed with closed-shell method and B3LYP 6- 31+G (2d, p) basis set using Cartesian coordinates. In excited state the optimization and single point energy calculation was evaluated at TDDFT/B3LYP 6-31+G (2d, p) level [21, 24]. We chose density functional theory time-dependent (TD -DFT) level because it can be applied to large systems and the results obtained in this manner are in good agreement with those reported previously [24]. In order to investigate the extent of torsions and the corresponding energy profiles, on both the ground and excited states, we have constrained only one of the bridging dihedrals (either γ or β) to a particular angle while allowing the other to relax to the lowest energy configuration. The molecules view, such as the connections distance and the size of the torsion angles, were visualized and designed with Avogadro and MacMolPlt software.
Results and Discussions
Usually, the starting point for discussion about the properties of fluorescent molecules is their light absorption and emission. These properties regarding to wt-EGFP and 2His-EGFP proteins and the eventual alterations occurred as a consequence of the mutagenesis were recorded in our previous study [13]. Accordingly, the overlaid normalized absorbance spectra of the two types of proteins showed identical maxima at 488 and 280 nm that correspond to the chromophores and aromatic amino acids, respectively. The recorded emission spectra of the two proteins having a maximum at 507 nm are completely identical. In regard to sensorial analysis, 2His-EGFP samples presented two-hundredfold increase in affinity toward copper ions in comparison with wt-EGFP [13].
Thus, the valuable properties of our mutant were revealed so far, but one question still remained open: how the chromophore of our mutant loses its emissive properties to such a large extent?
In the literature was already revealed and supported by femtosecond time-resolved spectroscopy that the main non-radiative deexcitation processes may originate from internal conversion associated with torsional motions within the chromophore [17, 25, 26]. Thus, the characterization of the chromophore distortions, mainly about the two exocyclic bonds (β and ɣ) appears to be important for understanding the photo processes of our mutated protein (Fig. 1).
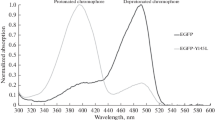
Accordingly, it is reasonable to assume that the two histidine insertion in situs 202 and 204 have altered the torsional barriers for the chromophore distortion and it was evolved as a result the nanomolar sensitivity toward copper ions. This presumption is supported to some extent by the fact that visible absorption spectra of 2His-EGFP in addition of copper has resulted a pronounced drop in the absorption band at 488 nm, which corresponds to the anionic form of the chromophore with a concomitant rise in the energy of neutral state absorption at 395 nm (Fig. 2). The spectral change exhibited a well-defined isobestic point at 430 nm. It also was showed that without disturbing environmental factors, the chromophore of 2His-EGFP is in anionic state, i.e., absorbs at 488 nm and the peak at 395 nm almost disappears.
This illustrates the chromophore conversion during the quenching from anionic to neutral state which, in general, is strongly coupled with the excited-state rotational motion [27, 28]. Theoretically it is possible, whereas femtosecond time-resolved spectroscopic studies have revealed that the two states corresponding to the two major absorption bands can interconvert quickly in the excited state and the presence of an isotope effect implicates proton movement in the interconversion [29].
In an attempt to provide a detailed picture of this phenomenon a crystal structure model (PDB entry: 1EMA) of the chromophore was analyzed with quantum mechanical calculations using Firefly/PC Gamess package. The structure of 4-hydroxybenzilidene-2,3-dimethyl-imidazolidinone (Fig. 1) has been selected as a reference compound which represents the smallest fluorescent moiety of EGFPs.
This structure was optimized at B3LYP/3-21G(d) level followed by frequency calculations to validate that it corresponds to a minimum on the potential energy surface. The excited state energy was calculated by means of time dependent density functional theory at TDB3LYP/6-31+(g,p) level [30]. As a result it was found that in ground state the phenolic and imidazolinone rings both lie in the same plane, i.e., the chromophore optimized structure is planar and the value of minimum energy at which the molecule is stable or is in thermal echilibrum, is −723,5773 kJ/mol (Fig. 1).
Next, we have investigated the changes in energy as a function of geometry distortion by analyzing several variants in which the imidazolinone or phenolate rings were rotated around β and also around γ bonds from 0 to 90° in 10° increments. The constrained optimization showed that in ground state the values of resulting energy due to internal rotation were higher than the energy of planar conformation. In the excited state the planar conformation had the energy maxima, at variance with the ground state. Each constrained optimizations resulted that the bond lengths between atoms remain relatively constant (changes of ≤ 0.1 Angstroms) as did the angles (≤15). A most favorable excited energy state was achieved when the degree of phenolate ring torsion was about 90°, suggesting the existence of conical intersection, i.e., the non-radiative decay channel. This is in good agreement with previous statements according to which in deprotonated state mainly the gamma distortion occurs [31]. From the consideration that the lowest energies of the excited state were obtained with β = 0 and γ = 90°, we have created the energy profile scheme of γ rotated conformations. The structures for intermediate values of angle γ were also characterized in order to illustrate the energy profiles more extensively (Fig. 3).
We already know from lifetime analysis that the fluorescence quenching is not static which means that the proteins with copper ions are not form dark complexes in ground state; they rather interact with each other in excited state. It follows that the loss of fluorescence attributed to the chromophore distortion may occur in S1 state which presumably is not preceded by any structural changes in S0 state, as it is also indicated by the increase of non-radiative decay rate constants. This argument allowed us to relate the changes caused by copper ions to the excited state γ rotation of the chromophore. Moreover, we attempted to define the extent of torsions which may occur during the quenching.
The changes in energies in S1 state caused by excited state intramolecular twisting around the γ-dihedral angle were deduced from the energy profile analyses. In fact, the evaluated energy differences represent the theoretical activation energies for excited state quenching, E a, which seems to be a key parameter for initiating the deexcitation process because it can be associated with fluorescent lifetime decays. The maximum of energy was obtained for planar geometry at γ = β = 0 in excited state, which reflects the highest barrier that must to overcome for quenching process. This barrier is decreasing along the γ-torsion coordinate the magnitude of which represents the theoretical activation energies of any twisted geometries (Table 1).
This experiment was carried out to determine the theoretical activation energies of conformations which can be compared with the experimental ones. The experimental energies required for the excited state quenching can be deduced from fluorescence lifetime analysis. We assume that if any of these values will be in good agreement with the magnitude of theoretical values than the corresponding molecular model will represent the real behavior of the chromophore from 2His-EGFP or wt-EGFP proteins in the presence of certain amount of copper.
Thus, in order to confirm this predict we have measured the average time during which the molecules are in excited state using time correlated single photon counting method. Apart from the environmental factors that could affect the fluorescence, the chromophores of EGFP proteins yield single exponential lifetimes: 2.87 ns for wild-type EGFP and 2.85 ns for histidine modified EGFP. These values correspond to those reported in several studies that were published in this domain. The Fig. 3 shows that there is no significant difference between the lifetime decays of the two proteins. Thus, we have confirmed that the mutation does not have any effect on the photophysical properties of the EGFP.
To correlate the time-resolved fluorescence signal of EGFP with the sensor properties, we performed copper titrations and followed the proteins fluorescence decay. As shown in Figs. 4 and 5, the time-resolved fluorescence intensity was decreased with increasing concentration of copper ions (from 2.8 with blank to 1.17 and 2.25 with 5 μM copper for 2His-EGFP and wt-EGFP, respectively).
Fluorescence lifetimes of 2 μM wild-type EGFP (2) and 2 μM EGFP/2H mutant (1) in the absence (1. - red line and 2. – purple line) and presence (1. - green line and 2. – red line) of 5 μM CuSO4 (Weighted residuals: (a) EGFP in the absence and (b) EGFP presence of copper ions). Histograms were evaluated with FluoFit software
Wt-EGFP and 2His-EGFP lifetime decays are nearly single exponential at 0 μM copper, as seen from the reasonable values of χ2 for the single exponential fits. However, under conditions of partial Cu2+ saturation the value of χ2 for the single exponential fit is markedly elevated (χ2 ~ 4). At higher χ2 values we have tried to fit the triple exponential equation, but we obtained even higher χ2 values. Therefore, in this experiment it is required the use of two lifetimes for the subsequent analysis.
Biexponential analysis with different lifetimes (short lived and long lived lifetime) was carried out according to Hotzer et al. and Lakowicz et al. [32, 33]:
The mean lifetime is the average time during which the fluorophore remains in the excited state and it is given by:
where a i is the amplitude, τ i is the decay time (Σαi = 1.0) and f i is the fractional intensity of each decay component.
Using these equations, we carried out global multi-exponential intensity decay analysis for all Cu2+ concentrations such that we adjusted the two lifetimes as invariants, while it was allowed the pre-exponential factors variation in response to copper. We assigned the 2.81 and 0.78 ns for 2His-EGFP, 2.79 and 0.64 ns for wt-EGFP to free and bound proteins, respectively. These fits were successful, resulting in a reasonable global χ2. The Cu2+ dependent lifetime components of proteins are summarized in Table 2 along with the pre-exponential factors.
We have calculated in this way the apparent (mean) lifetime in the absence of copper ions and similar results were obtained: 2.81 and 2.79 ns for 2His-EGFP and wt-EGFP, respectively. The decay time of wt-EGFP in the presence of 2 μM Cu2+ was reduced by 16 %. It was observed that the further addition of copper evoked a slow decrease in fluorescence lifetime. This may be caused by the existence of another binding site with very low copper binding affinity or other environmental factors affection. Addition of 2 μM of copper to 2 μM of 2His-EGFP was reduced by half the long lifetime component, from 2.81 to 1.31 ns. At saturated state the lifetime was reduced by 80 %. It was investigated that the saturation occurs when the 2 μM mutant proteins bind 4–5 μM copper. It was found that by further increasing the copper concentration to 25 μM, the values of lifetime decay did not alter significantly. In the case of wild-type protein the results did not show this fact explicitly, because the lifetimes decreased significantly until the addition of 2 μM copper, however, in further addition of copper the decrease was very slow. Thus, it can be assumed that the EGFP protein already had a metal binding site and it was created another one with high binding affinity.
The changes in lifetime indicate that the excited state depopulation process follows different radiative or non-radiative pathways which can be defined by the following equation:
where kobs is the observed decay rate constant that is formed by radiative (kr) and non-radiative (knr) rates.
Thus, the kobs was easily obtained, but for the determination of kr and knr the experimental quantum yields of proteins should be involved. We used the following formula which relates the radiative decay rate in the absence of copper ions to quantum yield:
where, the kr is the radiative rate constant, the τ0 and ϕ0 are the proteins lifetime and quantum yield in the absence of copper, respectively.
The experimental quantum yields for the mutant and wild-type were determined in our previous study, the values of which were: 0.58 ± 0.022 and 0.62 ± 0.12, respectively. Consequently, the radiative rate constants became 0.21 ns−1 for 2His-EGFP and 0.22 ns−1 for wt-EGFP. The radiative rate constants are attributed to the non-bounded proteins and it is assumed that they remain invariables during the copper binding process. It follows that, according to the Eq. 4, the initial values of non-radiative rate constants for both proteins are 0.14 ns−1. In the presence of 5 μM copper the mutant protein’s non-radiative constant increased to 0.64 ns−1. Under the same conditions the wild type protein’s non-radiative constant was only 0.22 ns−1.
The non-radiative pathway may include several deexcitation processes such as excited state intramolecular twisting completed with fast internal conversion or intersystem crossing. Deexciation by intramolecular twisting reflects the alterations caused by copper ions and its course will be represented by quenching rate constant (kq). This value completes the initial value of knr as it is denoted in the following equation [17]:
where the kq is the quenching rate constant, which is part of the non-radiative rate constant and is related to the effect of copper ions on target proteins.
We obtained the kq values for the mutant and wild type protein by substituting the appropriate values into the equation, which were 0.5 and 0.08 ns−1, respectively. The enhancement of quenching rate constant represents more fluorescence loss which may originate from ultrafast internal conversion.
The quenching rate constants can be related to the activation energies of distorted compounds by means of Arrhenius equation:
where a is the preexponential factor, E a is the activation energy for S1 state, k B and T are the Boltzmann constant and the absolute temperature.
In this case the pre-exponential factor is negligible, because the temperature does not change significantly during the experiment. The temperature and the value of Boltzmann constant were 293 K and 1,380 64 × 10−23 J/K, respectively. Thus, the activation energies were evaluated in a function of the quenching rate constants as the Eq. (7) shows. The results are listed in Table 3.
The quenching rate constant regarding to 2His-EGFP mutant was 0.5 ns−1 in the presence of 5 μM copper and, correspondingly, the activation barrier for the radiationless deexcitation process was 52.17 kJ Mol−1. The magnitude of this value in the case of wt-EGFP was 56.61 kJ Mol−1 under the same conditions. Consequently, it has been found that the extent of decrease in activation energy in the presence of 5 μM copper is twice larger for mutant (5.16 kJMol−1) than for wild-type (2.41 kJMol−1).
Usually, the decrease in activation energies means that the reaction occurs faster; perhaps it requires low reaction temperatures and/or reaction times, i.e., determines the conditions required to initiate the reaction. In our experiment the conditions were set to be constant therefore the two variables - quenching rate constant and activation energy - are directly dependent on each other. If the non-radiative decay rate constant increases, the structure of fluorescent moiety varies exponentially to lower energy.
In summary, the values calculated with Arrhenius equation were used for defining the real magnitude of gamma torsion barriers during the quenching process with regard to 2His-EGFP and wt-EGFP. As a next step, we looked for those theoretical activation energies which were similar with the values calculated with Arrhenius equation.
Attempts that were made on achieving close quantitative agreement between experimental and calculated activation energies have led to unexpected results. It was found that the energy difference between the planar and twisted geometry, that has a 19° γ-distorted angle, is similar with the activation energy for wt-EGFP in the presence of 5 μM of copper. The experimental Ea for 2His-EGFP under the same conditions is identical with that of theoretical when the γ distortion is equal with 40°. From these results we can infer that the impact of two histidine mutation allowes more flexibility for the chromophore due to which the rate of distorsion is twice as high in the mutant protein.
In this manner we have determined the degrees of γ torsion occurred by addition zof each amount of copper ion (Fig. 6).
It was shown that the activation energies required for the fluorescence quenching for wt-EGFP and 2His-EGFP in the presence of the same concentration of copper differ from each other. This is attributed to the fact that the torsion barrier varies due to 2His mutation. Furthermore, taking into account the extent of quenching, which is up to 60 % for wt-EGFP and about 90 % for 2His-EGFP under the same conditions, it is conceivable that the chromophore might encounter different barriers along the γ-torsion coordinate in S1 state. This can be supported by an alternative explanation, that the excited state relaxation or internal conversion during the complexation with copper ions can take place not only at avoided crossing region (γ = 90°), but also at smaller distortion angles. This can occur if the chromophore surroundings lower the energy of excited-state potential-energy surface during the quenching process. It is also relevant that the internal conversion at a γ-distortion angle smaller than 90° can become more favorable energetically due to the presence of copper. This process was presented in detail and illustrated with a scheme by Huang et al. [34].
The scenario of early energy decrease is also consistent with the expectation that the deexcitation process takes less time during the quenching as it is supported by the experimental non-radiative decay rate constant data. Data listed in Table 3 indicate that the decay of mutant is about six times faster relative to wt-EGFP which means that the γ-torsion barrier has become extremely small.
In order to justify the internal conversion for γ-twisted intermediates (i.e., distorted angle γ by 19 and 40°) a charge transfer analysis is proposed. Significant charge transfer character within the chromophore implies large changes in molecular electron density which triggers the non-radiative decay by transition between anionic and neutral form. This is supported by the increase of λabs for 2His-EGFP-Cu2+ at 395 nm which means that the push−pull electronic character of the appropriate moieties in chromophore is strengthened and thereby the tendency to form a twisted intramolecular charge transfer state is increased [34].
To elucidate this process, we have performed molecular orbital analysis which is suitable to identify the reactive sites within the molecular framework. Calculations of highest occupied MOs (HOMO) and the lowest unoccupied MOs (LUMO) with quantum mechanical method were performed at B3LYP/6-311++G(d,p) level (Fig. 7).
The results regarding to planar geometries are in good agreement with those reported in previous studies [20, 35]. It can be seen, that the HOMO orbital has π-bonding and antibonding character on the γ and β bond, respectively. In LUMO these bonds get π-antibonding character with an increased electronic density around the central carbon atom [21]. The π–π antibonding interaction between γ and β bonds results destabilization of the planar conformation by weakening the double bond character. Consequently, the excitation leads to a slight charge transfer around the central exocyclic bounds allowing the intramolecular torsion and, thus, the activation energy decrease. We mention that the intramolecular torsion will occur if the braking surroundings allow.
In order to find out the charge transfer characteristics of twisted molecules, we have calculated the HOMO and LUMO eigenvalues for the chromophore being distorted by 40° (Fig. 8). In accordance with previous discussions, it is assumed that this structure corresponds to the chromophore of 2His-EGFP being in saturated state by copper ions.
It can be noticed that there are high similarity between the LUMO of twisted and planar geometry, while the HOMO of twisted geometry is mostly localized on phenol ring. Moreover, the probability of finding electrons at imidazolinone ring is negligible in the HOMO, but is significant in the LUMO. The enhanced charge-transfer character entails a reduction in S1 → S0 emission [20] which is supported by the short fluorescence lifetimes for all complexes [34]. From the HOMO delocalization we can infer that the enhanced charge transfer occurs during the deexcitation process and the extra electron density on phenol ring will attracts a proton since it is positively charged. This conversely means that on imidazolinone group due to low electron density the tendency of proton repulsion is enhanced. The higher proton affinity leads to less acidic species. In the present case as the chromophore is twisted, the double bond is broken, the electron density thickens around the oxygen atom and attracts a proton thereby neutralizing the molecule. Consequently, the recovery of the neutral state and thereby the fluorescence quenching becomes more favorable energetically than other deexcitation pathways. This reasoning is consistent with the results obtained from absorption spectra regarding to 2His-EGFP that were recorded in the presence of different copper ion concentrations.
We have tried to collect more pieces of evidence to ascertain that the neutralization by proton transfer due to distortion plays the major role in non-radiative deactivation of chromophore in 2His-EGFP. Therefore, we have detected the reactive sites within the molecular framework by mapping the molecular electrostatic potential distribution of planar and twisted chromophores. Using this method, dipole moments and the sites of hydrogen-bonding interactions can be displayed allowing for us to visualize the variably charged regions of a molecule and predicting the behavior of complex molecules. The total electron density surface mapped with the electrostatic potential of 40° γ-distorted chromophore and the electrostatic potential contour map of planar and twisted geometries was obtained from calculation at B3LYP/6-311+G(2d,p) level which are shown in Fig. 9.
On the upper part of the figure are shown the electrostatic potential contour maps of planar (left side) and 40° γ-twisted chromophores (right side). In the lower part the total electron density surface mapped with molecular electrostatic potential of 40° γ-twisted geometry is presented (optimized at B3LYP/6-311++G(d,p) level). Contour value: 0.007. The color scheme is as follows: red: electron rich, partially negative charge; blue: electron deficient, partially positive charge; green: neutral
It can be seen that the planar geometry is largely dominated by red color which symbolizes the presence of negative potential over the anionic structure. The map of 40° γ-twisted structure revealed that this degree of torsion largely changes the electrostatic potential distribution over the molecular frameworks. The total electron density surface analysis of this geometry clearly shows the electron density delocalization, i.e., a significant increase of electron density around the O14, O5 and N2 atoms. The map also implies that the green region predominates which corresponds to a transitional state between the electronegative and positive potential (Fig. 9, lower part). These alterations may weaken the aromatic nature of the benzene nucleus and promotes the electrophilic reactivity around the O14 atom which may trigger the intramolecular proton transfer thereby neutralizing the molecule. Consequently, as the chromophore is twisted, a high electron density is collected on the O14 that will want to grab onto the proton since it is positively charged, a result of which the molecule becomes less acidic.
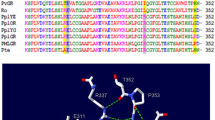
On the assumption that it can be reproduced the alterations in molecular framework we have compared the crystal structures and their surroundings of the neutral and anionic state. This is supported by the fact that the shapes of 2His-EGFP protein’s absorption spectra detected in the presence of copper ions correspond to the absorption properties of neutral chromophore. The chosen models were presented by Laino et al., which were confirmed by more recent studies to be the realistic version of anionic and neutral chromophore and its environment from EGFP [31].
According to this scheme, in anionic state four hydrogen bond donors, i.e., His148, Tyr145, Thr203 and a water molecule are connected to phenol ring. In neutral state these bonds break with exception of water molecule to which the His148 residue binds back. In this conformation the glutamine also changes its position, it connects to the Ser205 forming a closed hydrogen-bond network around the chromophore enabling the protonation of phenolate ring. Other residues retain their position or their structure.
Theoretically, the hydrogen-bond donors being adjacent to the chromophore are included in the transition from anionic to neutral state by shifting the absorption energy. During the process, the chromophore locked into the anionic state is released by the subsequent rotation of Thr203 and by hydrogen bond breaking between the Thr203-OH and phenolate oxygen. This change is supported by an additional rearrangement of His148 and Glu222 OH group [36].
Interactions in absorption spectrum of those residues were determined by realizing mutations as it was presented in several previous studies [18, 37, 38]. Some common empirical rules can be extracted from these results, for instance, mutations of T203V, T203I or T203Y eliminate a hydrogen bond on the chromophore phenolate ring in anionic state and shift to redthe absorption energy [18]. Moreover, it was shown that the absorption peak for wild-type GFP in anionic state is eliminated when Thr203 is replaced by other amino acid residue. Therefore, Thr203 should play an important role in stabilizing the deprotonated phenolic ring of the chromophore. On the other hand, the anionic/neutral state change also involves the conformational changes of residue Glu222, its mutation can cause the loss of the absorption peak at 397 nm [38, 39]. As it was presented by the crystal structure, the Arg96 and Gln94 within the anionic and neutral state remain in the same position [38]. The most significant alteration in the spectrum is a blue-shift by more than 100 nm and a remarkable reduction of the absorption intensity at 488 nm which are induced by the protonation of the phenolic oxygen.
Finally, we have concluded, as it is indicated by the modified map for the gamma torsion, that this magnitude of torsion induces a recovery to the neutral state which is more favorable than other deexcitation pathways.
Conclusions
The 2His-EGFP mutant that exhibited unexpected fluorescence response, specifically, nanomolar sensitivity toward copper ions (200 fold increase) was subjected to an extensive analysis. The starting point of experiment was the absorption spectra monitoring about our mutated protein in addition of copper which leaded to a pronounced increase in the neutral state absorption and a proportional decrease in the energy of anionic state absorption. It demonstrates that the proximity of metal chelated binding site induces the transition from anionic to neutral state, including the protonation of phenolic oxygen during the complexation process with copper ions. The protonation of phenolic oxygen can take place when the chromophore, locked into the deprotonated state, is released by losing its torsional barriers as a consequence of hydrogen bond breaking and rearrangement of several residues.
In this light, energy profile calculations were carried out about the chromophore possible distortions which may occur during the relaxation process from excited state, using quantum mechanical calculations. The maximum of energy was obtained for planar geometries at γ = β = 0 in excited state which is the highest barrier that must to overcome during the reaction. This barrier is decreasing due to intramolecular rotation, i.e., the decay dynamics follow a mechanism of photoisomerization which is responsible for internal conversion. The evaluation of rotation profiles has resulted that the significant changes in energy occurred during the γ rotation and the minimum value of energies was attained at γ = 90° while the β bond was kept at 0°. During the distortion, the structure of fluorescent moiety varies to lower energy and the barrier is changing, which means, that in the presence of different copper ion concentrations the chromophore distortion might encounter different energy barriers along the γ-torsion coordinate in the excited state. Thus, we have determined the energy differences between planar and twisted geometries which resulted, in fact, theoretical activation energies for each distorted cromophore.
The experimental activation energies in the presence of certain amount of copper were calculated using Arrhenius equation. This evaluation required fluorescence lifetime analysis. Non-radiative decay rate constants were derived from fluorescence lifetime decay data, and furthermore, the quenching rate constants were also determined by deducting from these data the effect of other environmental factors. By substituting the quenching rate constants in Arrhenius equation we obtained the experimental activation energies for the copper induced fluorescence quenching. Defining the theoretical and experimental activation energies was crucial because by comparing their magnitude to each other it became possible to determine the real chromophore distortions caused by certain amount of copper. The activation energies required for the reaction of 2His-EGFP and wt-EGFP proteins with 5 μM copper were 52.17 and 56.61 kJ Mol−1, respectively. These values are in good agreement with the theoretical activation energies of 40 and 19° γ-distorted chromophores. In order to justify that this is conceivable, we have performed charge transfer analysis on 40° γ-distorted cromophore. It was revealed that this degree of torsion entails a significant charge transfer due to which the probability of finding electrons at imidazolinone ring is negligible in the HOMO, but is significant in the LUMO. Large changes in molecular electron density trigger the proton transfer in excited state and thereby the tendency of transition is enhanced between anionic and neutral form.
By mapping the molecular electrostatic potential distribution of planar and twisted chromophores, the sites of hydrogen-bonding interactions and dipole moments became visible. It was concluded that this degree of torsion largely changes the electrostatic potential distribution over the molecular frameworks, i.e., the significant increase of electron density around the O14, O5 and N2 atoms may weaken the aromatic nature of the benzene nucleus and promotes the electrophilic reactivity around the O14 atom which may trigger the molecule neutralization. In this way, it was demonstrated that the 40° γ-distorted chromophore that corresponds to the 2His-EGFP protein being under saturation with 5 μM copper may induce the recovery to the neutral state, moreover, it can be more favorable energetically than other deexcitation pathways.
In summary, we proposed a new method for the examination of EGFP mutants which serves to define the chromophore behavior during the complexation with quenching agents. This method provides an opportunity to associate the experimental results to the quantum mechanical evaluations by which we can learn other possible actions that alter during the process.
References
Hsiu-Chuan LV, Chien MT, Tseng YY, Ou KL (2006) Assessment of heavy metal bioavailability in contaminated sediments and soils using green fluorescent protein-based bacterial biosensors. Environ Pollut 142(1):17–23
Hartter DE, Barnea A (1988) Brain tissue accumulates 67copper by two ligand-dependent saturable processes. A high affinity, low capacity and a low affinity, high capacity process. J Biol Chem 263:799–805
Lovell MA, Robertson JD, Teesdale WJ, Campbell JL, Markesbery WR (1998) Copper, iron and zinc in Alzheimer's disease senile plaques. J Neurol Sci 158(1):47–52
Desai V, Kaler S (2008) Role of copper in human neurological disorders. Am J Clin Nutr 88(3):855S–858S
Huang GG, Yang J (2003) Selective detection of copper ions in aqueous solution based on an evanescent wave infrared absorption spectroscopic method. Anal Chem 75(10):2262–2269
Lin M, Cho M, Choe WS, Son Y, Lee Y (2009) Electrochemical detection of copper ion using a modified copolythiophene electrode. Electrochim Acta 54(27):7012–7017
Yang M, Yu Y, Shen F, Dierks K, Fang W, Li Q (2010) Detection of copper ion with laser-induced fluorescence in a capillary electrophoresis microchip. Anal Lett 43(18):2883–2891
Isarankura-Na-Ayudhya C, Tantimongcolwat T, Galla HJ, Prachayasittikul V (2010) Fluorescent protein-based optical biosensor for copper Ion quantitation. Biol Trace Elem Res 134(3):352–363
Kneen M, Farinas J, Li Y, Verkman AS (1998) Green fluorescent protein as a noninvasive intracellular pH indicator. Biophys J 74(3):1591–1599
Szabó (P) M, Petres J, Szilágyi L, Miklóssy I, Ábrahám B, Lányi S (2012) Possible application of metal sensitive red fluorescent proteins in environmental monitoring. Environ Eng Manag J 11(1):193–198
Richmond TA, Takahashi TT, Shimkhada R, Bernsdorf J (2000) Engineered metal binding sites on green fluorescence protein. Biochem Biophys Res Commun 268(2):462–465
Palfi M, Kovács E, Miklóssy I, Szilágyi L, Ábrahám B (2009) Engineered green fluorescent protein as a potential metal sensor. Studia Universitatis Babes-Bolyai Chemia, Special Issue: 35–45
Bálint EÉ, Petres J, Szabó M, Orbán CK, Szilágyi L, Ábrahám B (2013) Fluorescence of a histidine-modified enhanced Green Fluorescent Protein (EGFP) effectively quenched by copper(II) ions. J Fluoresc 23(2):273–281
Baker M (2011) Microscopy: bright light, better labels. Nature 478:137–142
Chen MC, Lambert CR, Urgitis JD, Zimmer M (2001) Photoisomerization of green fluorescent protein and the dimensions of the chromophore cavity. Chem Phys 270:157–164
West CW, Hudson AS, Cobb SL, Verlet JRR (2013) Communication: autodetachment versus internal conversion from the S1 state of the isolated GFP chromophore anion. J Chem Phys 139:071104–1–071104–4
Follenius-Wund A, Bourotte M, Schmitt M, Iyice F, Lami H, Bourguignon JJ, Haiech J, Pigault C (2003) Fluorescent derivatives of the GFP chromophore give a new insight into the GFP fluorescence process. Biophys J 85(3):1839–1850
Kummer AD, Wiehler J, Rehaber H, Kompa C, Steipe B, Michel-Beyerle ME (2000) Effects of threonine 203 replacements on excited-state dynamics and fluorescence properties of the Green Fluorescent Protein (GFP). J Phys Chem B 104(19):4791–4798
Jung G, Wiehler J, Zumbusch A (2005) The photophysics of green fluorescent protein: influence of the key amino acids at positions 65, 203, and 222. Biophys J 88(3):1932–1947
Cheng CW, Huang GJ, Hsu HY, Prabhakar C, Lee YP, Diau EWG, Yang JS (2013) Effects of hydrogen bonding on internal conversion of GFP-like chromophores. II. The meta-amino systems. J Phys Chem B 117:2705–2716
Paul BK, Guchhait N (2013) Looking at the Green Fluorescent Protein (GFP) chromophore from a different perspective: a computational insight. Spectrochim Acta A Mol Biomol Spectrosc 103:295–303
Van Den Zegel M, Boens N, Daems D, DeSchryver FC (1986) Possibilities and limitations of the timecorrelated single photon counting technique: a comparative study of correction methods for the wavelength dependence of the instrument response function. Chem Phys 101:311–335
Granovsky AA, Firefly version 8.0.0, http://classic.chem.msu.su/gran/firefly/index.html
Marques MA, López X, Varsano D, Castro A, Rubio A (2003) Time-dependent density-functional approach for biological chromophores: the case of the green fluorescent protein. Phys Rev Lett 90(25):258101-1–258101-4
Fang C, Frontiera RR, Tran R, Mathies RA (2009) Mapping GFP structure evolution during proton transfer with femtosecond Raman spectroscopy. Nature 462:200–204
Lossau H, Kummer A, Heinecke R, Pöllinger-Dammer F, Kompa C, Bieser G, Jonsson T, Silva CM, Yang MM, Youvan DC, Michel-Beyerle ME (1996) Time-resolved spectroscopy of wild-type and mutant green fluorescent proteins reveals excited state deprotonation consistent with fluorophore–protein interactions. Chem Phys 213:1–16
Voityuk AA, Kummer AD, Michel-Beyerle ME, Rösch N (2001) Absorption spectra of the GFP chromophore in solution: comparision of theoretical and experimental results. Chem Phys 269:83–91
Polyakov IV, Grigorenko BL, Epifanovsky EM, Krylov AI, Nemukhin AV (2010) Potential energy landscape of the electronic states of the GFP chromophore in different protonation forms: electronic transition energies and conical intersections. J Chem Theory Comput 6:2377–2387
Philips GN Jr (1996) Structure and dynamics of green fluorescent proteins. Curr Opin Struct Biol 7(6):821–827
Epifanovsky E, Polyakov I, Grigorenko B, Nemukhin A, Krylov AI (2009) Quantum chemical benchmark studies of the electronic properties of the green fluorescent protein chromophore. 1. Electronically excited and ionized states of the anionic chromophore in the gas phase. J Chem Theory Comput 5:1895–1906
Laino T, Nifosí R, Tozzini V (2004) Relationship between structure and optical properties in green fluorescent proteins: a quantum mechanical study of the chromophore environment. Chem Phys 298:17–28
Lakowicz JR (2006) Principles of fluorescence spectroscopy, 3rd edn. Springer Science+Business Media, New York
Hötzer B, Ivanov R, Altmeier S, Kappl R, Jung G (2011) Determination of copper(II) Ion concentration by lifetime measurements of green fluorescent protein. J Fluoresc 21:2143–2153
Huang GJ, Cheng CW, Hsu HY, Prabhakar C, Lee YP, Diau EWG, Yang JS (2013) Effects of hydrogen bonding on internal conversion of GFP-like chromophores. I. The para-amino systems. J Phys Chem B 117:2695–2704
Hasegawa JY, Fujimoto K, Swerts B, Miyahara T, Nakatsuji H (2007) Excited states of GFP chromophore and active site studied by the SAC-CI method: effect of protein-environment and mutations. J Comput Chem 28:2443–2452
Lill MA, Helms V (2001) Proton shuttle in green fluorescent protein studied by dynamic simulations. PNAS 99(5):2778–2781
Arpino JAJ, Rizkallah PJ, Jones DD (2012) Crystal structure of enhanced green fluorescent protein to 1.35 a° resolution reveals alternative conformations for Glu222. PLos One 7(10):e4132/1-8
Sniegowski JA, Phail ME, Wachter RM (2005) Maturation efficiency, trypsin sensitivity, and optical properties of Arg96, Glu222, and Gly67 variants of green fluorescent protein. Biochem Biophys Res Commun 332:657–663
Voityuk AA, Michel-Beyerle ME, Rösch N (1998) Quantum chemical modeling of structure and absorption spectra of the chromophore in green fluorescent proteins. Chem Phys 231:13–25
Acknowledgments
The work has been funded by the Sectorial Operational Programme Human Resources Development 2007–2013 of the Romanian Ministry of Labour, Family and Social Protection through the Financial Agreement POSDRU/88/1.5/S/60203 and by the University of Sapientia, Department of Bioengineering.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Péterffy, J.(., Szabó, M., Szilágyi, L. et al. Fluorescence of a Histidine-Modified Enhanced Green Fluorescent Protein (EGFP) Effectively Quenched by Copper(II) Ions. Part II. Molecular Determinants. J Fluoresc 25, 871–883 (2015). https://doi.org/10.1007/s10895-015-1567-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10895-015-1567-4