Abstract
The present study for the first time explores the use of Central composite design (CCD) of RSM to optimize the process parameters of biosynthesis of AgNPs from rhizome extract of Curculigo orchioides based on the absorbance of surface plasmon resonance (SPR) band at 430 nm that corresponds to the synthesis of mono-disperse, spherical AgNPs. A polynomial model was established as a functional relationship between the synthesis of AgNPs and four independent variables such as concentration of AgNO3, % rhizome extract, pH and temperature. The optimum conditions for maximum AgNPs synthesis were 2 mM concentration of AgNO3, 20 % rhizome extract, pH 8, and temperature of 60 °C. A significant correlation (R 2 = 0.8947) was observed between the experimental data and the predicted values indicating the adequacy of the model. Transmission electron microscopy (TEM) revealed spherical particles with size range of 5–28 nm. Selected area electron diffraction pattern and X-ray diffraction analysis confirmed the face-centered cubic structure of metallic silver. The plausible mechanism for the reduction of AgNO3 to AgNPs was proposed following the identification of functional groups by FTIR. The antioxidative activity of AgNPs was demonstrated with scavenging of hydrogen peroxide (H2O2), 1,1-Diphenyl-2-picrylhydrazyl (DPPH) and superoxide radicals.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bio-inspired synthesis of silver nanoparticles (AgNPs) has become the focus of extensive research during the last decade. Owing to its unique size-dependent optical, electronic and biological properties they are now being utilized in various fields and applications in microelectronics [1], photocatalysis [2], photonics [3], lithography [4], biomaterials [5], biosensors [6], drug delivery [7], bio-imaging [8], food preservation [9], and water purification systems [10]. A variety of biological systems including bacteria, fungi, algae and plant extracts have been used for the green synthesis of AgNPs. Compared to chemical and physical methods, the green synthesis of nanoparticles employing the plant extracts has emerged as a simple and viable alternative due to its non-toxic, environmentally benign and renewable nature [11]. Moreover, plant-mediated green synthesis is more preferable than microbial systems since microorganisms used for the synthesis of AgNPs are reported to be pathogenic in nature to either plants and/or humans and the direct use of plant extract in synthesis eliminates the elaborative process of cell culturing [12, 13].
In recent years, medicinal plant mediated synthesis has emerged to be one of the advantageous sources for biosynthesis of AgNPs. Typical medicinal plants utilized for the synthesis of AgNPs are Aloe vera [14], Ocimum sanctum [15], Leucas aspera [16], Cissus quadrangularis [17], Solanum nigrum [18], Azadirachta indica [19]. Secondary metabolites of medicinal importance appeared to act as the main reducing agent responsible for the synthesis of nanoparticles.
A variety of factors such as the concentration of plant extract, AgNO3 solution, temperature, pH, and incubation time regulates the synthesis of AgNPs. The intensity and the position of the surface plasmon resonance (SPR) band are dependent on the nature, size and shape of the particles formed, their—interparticle distances, functional groups adsorbed on the particle surface, and dielectric constant of the medium [20, 21]. A shift in the SPR band due to variations in the reaction conditions gives a measure of the synthesis of nanoparticles in terms of shape, size and aggregation of the particles. The effect of varying concentrations of plant extract and silver salt on the biosynthesis of AgNPs along with the influence of temperature and pH on the process of nucleation have been well demonstrated by several workers [22–24]. However, studies carried out with plant extract mediated synthesis of AgNPs have mostly utilized traditional one-factor-at-a-time (OFAT) approach to optimize the synthesis process. With this univariate approach, the interactions between the process variables have been ignored, and that may lead to ambiguous conclusion with regard to the efficiency of the process. This inefficient approach can be replaced with the multivariate chemometric methods, one of which is response surface methodology (RSM). RSM is a statistically valid method which simplifies the process optimization by analyzing the mutual interactions among the variables over a range of values and predicts the best fit conditions with a minimum number of experiments. It can also explain the effects of individual and interactive test variables on the response. RSM has been applied for optimization of AgNPs biosynthesis in several microbial systems [25, 26], whereas the application of RSM for determining the optimum conditions for the biosynthesis of AgNPs in plants was limited only with the extract of Tridax procumbens and the experimental design used was Box-Behnken matrix [27]. In another instance, Taguchi orthogonal arrays experimental design was used for optimization of biosynthesis of AgNPs by aqueous extract of Eucalyptus oleosa [28]. In RSM category, central composite design (CCD) which is appropriate for fitting second order polynomial with two-level factorial design points, 2k axial points and centre points representing replicate terms has been frequently used for optimizing a variety of biochemical processes. However, RSM with CCD has not yet been adopted for optimizing the biosynthesis of AgNPs using plant extract. Recently, we have successfully applied CCD and optimize the biosynthesis of AuNP from Swertia chirata, an endangered medicinal plant [29].
In the present study, we have reported the biosynthesis, optimization of the process parameters using CCD of RSM and characterization of AgNPs using rhizome extract of Curculigo orchioides Gaertn., a medicinally important herb belongs to the family Hypoxidaceae. The rhizome extract has been used in many pharmaceutical formulations of herbal industry and reported to possess hypoglycemic, spasmolytic, aphrodisiac, antipyretic, diuretic, antioxidant and anticancer properties [30, 31]. The medicinal property of the plants is mainly due to the presence of cycloartane triterpenes [32, 33]; phenolic glycosides [34] and chlorophenolic glucosides [35].The plant extract was claimed to possess a potent antioxidative activity due to the presence of phenolic glycosides [36]. Thus, the application of AgNPs towards the scavenging potential of reactive oxygen species has also been addressed.
Materials and Methods
Biosynthesis of AgNPs
Healthy plants of Curculigo orchioides Gaertn., grown in the Agricultural Farm of IIT Kharagpur (West Bengal, India) were collected and washed thoroughly with tap water. After drying in laminar air flow cabinet rhizomes were chopped into small pieces. Different amounts of rhizome sample were crushed and boiled in 50 ml of distilled water in a 250 ml Erlenmeyer flask for 30 min to obtain different concentrations of extract solutions (5, 10, 15, 20 and 25 %; w/v). The extract was filtered using Whatman No. 1 filter paper and the volume of the final filtrate was adjusted to 50 ml by adding sterile distilled water. Aqueous silver nitrate (AgNO3) solution (Sigma- Aldrich, India) was prepared using double distilled water. Rhizome extract and AgNO3 solution were mixed in the ratio of 1:9 to form the reaction mixtures and incubated for 8 h. Synthesis of AgNPs was monitored by visual color change and UV–visible absorbance in the wavelength range of 300–700 nm. For the recovery of AgNPs, the reaction mixtures after 8 h of incubation were centrifuged at 12,000 rpm for 30 min, washed three times with double distilled water and lyophilized using a freeze dryer and the dried powder obtained was used for further experiments.
Experimental Design and Optimization of Biosynthesis by Response Surface Methodology (RSM)
The parameters considered for optimization in this study were concentration of AgNO3 (X1), % rhizome extract (X2), temperature (X3) and pH (X4). Design Expert 9.0 (Stat-Ease Inc., USA) was used for RSM following Central composite design (CCD) to evaluate the influence of four independent variables. The central values (zero level) for experimental design were chosen to be 1.5 mM AgNO3, 15 % rhizome extract, temperature 50 °C and pH 7 (Table 1).
The study consisted of 30 trials and the experiments were conducted according to the CCD matrix (Table 2). All the experiments were carried out in duplicate and the absorbance at 430 nm was taken as the dependent variable or response (Y) which corresponds to the surface plasmon resonance band of biogenic AgNPs [25]. A second order polynomial equation was used to fit the experimental results of central composite design using response surface regression method. The equation is as follows:
where predicted response is denoted by Y, regression coefficients by β0, linear coefficient by βi, quadratic coefficients by βii, interaction coefficients by βij and coded level of independent variables by Xi respectively.
The independent variables were coded as X1, X2, X3, and X4, and the second order polynomial equation is as follows:
Characterization of AgNPs
The initial characterization of the biosynthesized AgNPs was carried out by UV–vis spectroscopy. The reaction mixtures were analyzed in the wavelength range of 300–700 nm using Systronics Double Beam UV–vis Spectrophotometer (Ahmedabad, India) at a resolution of 1 nm. The double distilled water was used as a blank. The reaction condition (2 mM AgNO3, 10 % rhizome extract, 50 °C and pH 6.0) that produced the appearance of sharp peak at 430 nm was selected for TEM in order to analyze the size and shape of the synthesized AgNPs. A thin film of the sample was made on the carbon coated copper TEM grids. The copper grid was dried at room temperature and the TEM analysis was done by using an FEI-TECNAI G2 20S-TWIN, FEI, USA equipped with the selected area electron diffraction (SAED) pattern facility and energy dispersive X-ray analysis (EDAX) attachment. TEM was operated at an accelerating voltage of 200 kV. The size of the particles was measured from 100 TEM images at the magnification of 71,000.
X-ray diffraction pattern of dried AgNPs was carried out with Panalytical High—Resolution XRD-I, PW 3040/60 using 2.2 KW Cu Kα radiation (λ = 0.1542 nm). The generator voltage used was 40 kV and current was set at 30 mA. The scanning was recorded in the 2θ range of 10–80 in continuous scan mode with 0.016 step size. The values for interplanar spacing (dh k l) were calculated following the equation:
The lattice constant (A0) of the synthesized AgNPs was also determined from the equation:
The average size of the AgNPs synthesized was estimated from the XRD pattern using the Debye–Scherrer formula [37].
where, “D” is the particle size, “λ” is the wavelength of X-ray (0.1541 nm), “β” is the full width at half maximum (FWHM) of the XRD peak and “θ” is the Bragg angle in radians.
For Fourier transform infrared spectroscopy (FTIR), lyophilized AgNPs were palletized with kBr and analyzed in the range of 400–4000 cm−1 using a Perkin–Elmer Spectrum-One FTIR spectrophotometer (Perkin–Elmer, USA). The scans of sample were taken at a resolution of 4.0 cm−1.
Antioxidant Assays
Different concentrations of rhizome extract (100, 200, 300, 400 and 500 µg/ml) were prepared by maceration using water and methanol as solvent following the method of Onkar et al. [38], and designated as aqueous (ARE) and methanolic (MRE) extract respectively. Similar concentrations of biosynthesized AgNPs dispersion in deionized water were made by ultrasonic vibration using Sonicator, Rivotek, USA (100 W, 30 kHz).
DPPH Free Radical Scavenging Assay
1,1-Diphenyl-2-picrylhydrazyl (DPPH) free radical scavenging potential of the AgNPs and rhizome extracts was evaluated according to the method of Hanato et al. [39].The reaction mixture containing rhizome extract or AgNPs was mixed with 1 ml of fresh prepared DPPH (0.3 mM) and vortexed thoroughly. Finally, the mixture was incubated in dark for 30 min at 37 °C, and the absorbance was recorded at 517 nm. The DPPH radical scavenging activity was expressed as the percentage of inhibition using the following equation:
where Ac is the absorbance of DPPH solution without nanoparticles and rhizome extract; As is the absorbance of DPPH solution with ARE/MRE/AgNPs/AA. Ascorbic acid (AA) was used as a positive control.
Hydrogen Peroxide Scavenging Assay
The H2O2 scavenging activity was assayed following the modified method of Jayaprakash et al. [40]. A solution of hydrogen peroxide (20 mM) was prepared in phosphate buffer (pH 7.4). Different concentrations of ARE, MRE, AgNPs and AA, were mixed with 2 ml of hydrogen peroxide solution. After an incubation of 10 min at room temperature the absorbance was measured at 230 nm against a blank solution of hydrogen peroxide. The percentage of scavenging activity of H2O2 was calculated using the Eq. 6 by replacing DPPH with H2O2.
Superoxide Radical Scavenging Activity
Superoxide radical scavenging activity was measured by inhibition of nitroblue tetrazolium (NBT) reduction [41]. The reaction mixture consisted of 1 ml of NBT solution (156 μM NBT in 100 mM phosphate buffer, pH 7.4), 1 ml of NADH solution (468 μM in 100 mM phosphate buffer, pH 7.4) and 1 ml of ARE, MRE, AgNPs or AA (100–500 μg/ml). The reaction was initiated by adding 100 μl of phenazine methosulphate (PMS) solution (60 μM PMS in 100 mM phosphate buffer, pH 7.4) to the mixture. After incubation at 25 °C for 5 min, the absorbance of the reaction mixture was measured at 560 nm against the blank sample. The super oxide radical scavenging activity was expressed in percentage of inhibition.
Results and Discussion
Synthesis of AgNPs
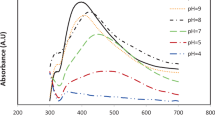
After addition of silver nitrate to the light yellow colored rhizome extract solution, a visual color change was observed. The reaction mixture turned dark brown after 8 h of incubation, indicating the formation of AgNPs. The UV–vis spectra obtained from various reaction mixtures show characteristic absorption bands of AgNPs at around 413–450 nm. The appearance of absorption peaks in this region corresponded to the wavelength of the surface plasmon resonance (SPR) of AgNPs and confirmed the formation of AgNPs [42–44].The position and shape of SPR absorption spectra of nanoparticles are mainly dependent on particle size, stabilizing molecules, state of aggregation and the dielectric constant of the medium [21].
A variation in the reaction condition is known to influence the synthesis of metal nanoparticles [23, 24, 43]. In the present study, the position and shape of the SPR of AgNPs varied with the concentrations of AgNO3, % plant extract, incubation period, temperature and pH. Previous studies using various types of plant extract documented the influence of these parameters on the green synthesis of AgNPs following OFAT approach [23, 24, 45, 46]. The combined effects of the factors and their functional relationships on the biosynthesis of AgNPs were ignored. Based on the SPR band at 430 nm (Fig. 1) which produced spherical shape of AgNPs as confirmed by TEM images, the present study optimized the process parameters using RSM taking into account the intensity of the absorbance representing yield of AgNPs as a dependent variable.
Optimization of AgNPs Biosynthesis Using Response Surface Methodology
On the basis of the experimental design presented in Table 2, a second order polynomial equation in terms of actual factors was developed that illustrates the empirical relationships between the independent variables and the response:
Analysis of variance (ANOVA) was performed to test the significance and adequacy of the model (Table 3). The significance of the model is evident from the model F value of 9.10 with a very low p value (p < 0.0001). There is a 10.75 % chance that the “lack of fit F value” could occur due to the inadequacy of the model. The good predictability of the model is confirmed by a non-significant “lack of fit” value of 3.17.
In this model, the linear effects of X 1 (concentration of AgNO3), X 2 (% rhizome extract) and quadratic effects of X 1 X 1 (Concentration of AgNO3, Concentration of AgNO3), X 2 X 2 (% rhizome extract, % rhizome extract) and X 4 X 4 (pH, pH) are significant. On the other hand, the p-value greater than 0.05 suggests that the model parameters are insignificant. Thus, the linear effect of X 3 (temperature), X 4 (pH) and the quadratic effects of X 3 X 3 (temperature, temperature) are insignificant factors.
The goodness of fit of the model was assessed by the determination coefficient (R 2). The R 2 value of the model provides a measure of dispersion between the observed and predicted responses. The closer the R2 value to 1, the better the predictability of the model. The R2 of the model was 0.8947 and it indicates a very good fit between the observed and predicted responses of the model for AgNPs biosynthesis in the present study.
In order to better understand the interactions between variables and to determine their optimum values, response surface contour curves were plotted by fixing one of the variables at the optimum level and varying the other two (Fig. 2). Figure 2a shows the effects of pH and % rhizome extract on AgNPs biosynthesis. An increase in the absorbance was observed with increasing pH of the reaction mixture from 6 to 8. Similar response was also noted with % rhizome extract. A three dimensional plot representing the interactions between temperature and % rhizome extract is shown in Fig. 2b. It is clear that the higher levels of temperature along with % rhizome extract support high yield in terms of absorbance. However, compared to the interaction of pH and % rhizome extract the yield was poor with temperature. There is a gradual increase in AgNPs biosynthesis with increasing pH and AgNO3 concentration (Fig. 2c). The absorption intensity increases with increasing pH and the maximum absorption of 3.07 was obtained with pH 8. The findings indicate that the basic pH enhances the bio-reduction process towards the formation of AgNPs. Moreover, the formation of AgNPs was suppressed at acidic pH i.e. pH < 7. Presumably, the bioavailability of functional groups in the rhizome extract at high pH stimulated the synthesis of AgNPs (24, 27). The interaction between concentration of AgNO3 and % rhizome extract revealed the same trend, although the degree of increase in absorbance was found to be higher than that of cumulative effects of pH and AgNO3 concentration (Fig. 2d). The maximum yield as represented by absorbance of 2.7 was obtained with the interaction of % rhizome extract and concentration of AgNO3. Figure 2e represents the effect of temperature and concentration of AgNO3 on the biosynthesis of AgNPs. Higher levels of AgNO3 influenced the synthesis without any prominent effect of temperature. The interaction between pH and temperature had no remarkable influence on biosynthesis (Fig. 2f). The higher p value of temperature again confirmed the insignificant effect of temperature on the synthesis process. The response surface analysis showed that the pH, AgNO3 concentration and % rhizome extract had significant effects on the biosynthesis of AgNPs. The influence of these factors on the stability of the particles and plasmonic absorption intensity has already been well documented [24, 27, 47]. Previous optimization approach considered the size of the nanoparticles as dependent variable [27, 28]. As it is difficult to obtain uniform size of the particles at particular reaction conditions, the present study optimized the reaction conditions based on the absorbance of SPR band at 430 nm that correspond with the synthesis of spherical AgNPs in the range 5–28 nm as evidenced by TEM analysis. Such optimization approach may lead to the improved synthesis of AgNPs of similar nature.
Response surface plot of biosynthesis of AgNPs (in terms of absorbance at 430 nm) a pH versus rhizome extract, b temperature versus rhizome extract, c pH versus conc. of silver nitrate, d rhizome extract versus conc. of silver nitrate, e temperature versus conc. of silver nitrate and f pH versus temperature
To determine the specific points of optimization numerical optimization was employed. Desirability function was used to find out the optimal conditions for AgNPs synthesis in terms of maximum absorbance. The desirability function converts each response into an individual function that varies over the range (0, 1) and takes the value one when the response is at its target value and less than one if not [48]. To construct desirability indices, five possibilities such as none, maximum, minimum, target and within range were considered as a goal. The criteria for each of the variables like goal, limit, weight and importance are listed in Table 4.
Based on the settings and boundaries mentioned in Table 4, the optimum conditions for maximum AgNPs synthesis obtained were 2 mM concentration of AgNO3, 20 % rhizome extract, 60 °C temperature and pH 8 (Fig. 3). The maximum intensity of the plasmonic resonance of 3.07 at 430 nm obtained from the experimental conditions was found to corresponds well with the absorbance of 3.11 at the same waveband following the optimized model conditions. The good agreement between the experimental value and the predicted value from the polynomial model not only confirms the validity of the model, but it also augments the reproducibility of the experiment for the biosynthesis of AgNPs.
TEM and EDAX Analysis
The shape and size of the synthesized AgNPs were analysed using TEM. As shown in Fig. 4a. the particles were monodispersed and spherical in shape with different sizes ranging from 5 to 28 nm. The average size obtained was 18.16 ± 5.93 nm with predominant occurrence (inset Fig. 4a).The EDAX analysis of the synthesized AgNPs reveals the presence of elemental silver which also supports the XRD results (Fig. 4b). A strong signal of Ag Peak was obsevered at 3 keV due to surface plasmon resonance and the appearance of the peak indicates the reduction of silver ions to elemental silver [19]. The signal for Cu in the EDAX may arise from Cu TEM grid used in the study. The selected area electron diffraction (SAED) pattern of AgNPs is shown in Fig. 4c. The concentric diffraction rings as bright spots corresponding to the presence of (111), (200), (220), (311) and (222) planes of the face- centered cubic (fcc) AgNPs indicate the crystalline nature of the nanoparticles.
X-Ray Diffraction
The crystal structure of synthesized AgNPs was determined by XRD analysis. The presence of Bragg reflection peaks at 2θ values of 38.1,44.37, 64.59, and 77.48°, respectively were indexed to reflections from (111), (200), (220), (311) planes of face centered cubic (fcc) crystal structure of metallic silver (Fig. 5).The XRD patterns obtained are consistent with the previous reports [19, 24, 47].
The various XRD parameters calculated are shown in Table 5.
The interplanar spacing (d h k l) values and the lattice parameters were matched well with Joint Committee on Powder Diffraction Standards values (JCPDS, silver file No. 04-0783).The average size of the AgNPs synthesized in the bioreduction process is determined using Scherrer`s formula and estimated to be 16.28 ± 0.90 nm which is in accordance with the size observed in TEM analysis. Noteworthy, the derived particle size using the Scherrer’s formula is in the lower limit than that of TEM presumably due to the instrumental broadening.
FTIR Analysis
FTIR spectroscopy was used to characterize the functional groups present in the rhizome extract and as-obtained AgNPs in order to explain the mechanism of bioreduction and particle stabilization. The FTIR spectra of rhizome extract show the presence of strong absorbance bands at 3409, 2926, 1639, 1418, 1320, 1254 and 1049 cm−1 (Fig. 6a). The strong band located at 3409 cm−1 is typical to the O–H stretching vibrations of phenols and carboxylic acids [49]. The peak at 2926 cm−1 corresponds to the symmetric and asymmetric C–H stretching vibrations of aliphatic acids and aldehydes [50]. The band at 1639 cm−1 is characteristic of N–H2 bending, C=N and C=C stretching of amino acids and confirms the presence of proteins in the extract [51, 52]. The peak at 1418 cm−1 may be due to C–H deformation in aliphatics. The presence of peak at 1320 cm−1 indicates the occurrence of C–N stretching band of aromatic functional group of protein [53]. The peak obtained at 1254 cm−1 suggests C–O stretching of esters [54]. The strong peak observed at 1049 cm−1 implied the C–O–C glycosidic symmetric stretching vibration of acetylated polysaccharides [52].
The FTIR spectrum of rhizome extract mediated AgNPs showed distinct peaks at 3397, 2919, 1646, 1544, 1382, 1317, 1244 and 1028 cm−1 (Fig. 6b). Compared to the FTIR profile of rhizome extract, a shift in the bands at 3397, 2919, 1382 and 1028 cm−1 was noted in the as-obtained AgNPs and could be attributed to the binding interactions of functional groups of amines, alcohols, ketones, aldehydes, hydroxyl, and carboxylic acids of rhizome extract with the AgNPs during their synthesis. The peak noticed at 1544 cm−1 in the AgNPs might be due to the NH- stretch vibrations in the amide II linkage of the protein. The presence of this peak along with the band at 1646 cm−1 suggested the stabilization of AgNPs by the surface bound proteins [55].
Previous phytochemical analysis of rhizome extract of C. orchioides revealed the presence of phenolic glycosides [34], cycloartane triterpenes [32, 33], and chlorophenolic glucosides [34]. However, the phenols and phenolic glycosides played a major role in the biological activity of the plant extract [36, 56]. The presence of such compounds may be responsible for the reduction of AgNO3 to AgNPs as they possess hydroxyl group and glycosidal linkages. Based on a previous report [57] that the primary C-6-hydroxy group was selectively oxidized to carboxylic acid in glycosides, Jung et al. [58] synthesized gold nanoparticles from auric acid using glycosides as reducing agents. Among the various sugar containing reductants, phenyl β-d glucoside generated the highest yield of gold nanoparticles and the oxidation site determined was C-6–OH in glycosides. Considering the proposed type of reductant with oxidation site, the major metabolites present in the rhizome extract and the depicted FTIR spectra, the plausible mechanism (s) proposed in the present study for the synthesis of AgNPs are presented in Fig. 7. The oxidation of C-6–OH in phenolic glycosides of C. orchioides (e.g. curculigoside, curculigoside B, C, G) upon reaction with AgNO3 resulted in the synthesis of highly mono-disperse AgNPs (Fig. 7a). The other possibility is the oxidation of hydroxyl group at C 11 present in the curculigosaponins which may contribute to the reduction of Ag+ to Ag0 via hydrogen abstraction [59] (Fig. 7b). The proposed mechanisms of bioreduction are in agreement with the saponin mediated synthesis of AgNPs in Trianthema decandra [60]. Further, coat covering of the AgNPs by proteins present in the extract rendered the stability and prevented agglomeration of the particles.
Antioxidant Assay
DPPH Free Radical Scavenging Activity
The DPPH scavenging ability of ARE, MRE and AgNPs are shown in Fig. 8a and compared to that of AA. The percent of DPPH radical inhibition of ARE varied from 15 to 42 %. A similar range (17–47 %) was also obtained with MRE. However, the percent inhibition was found to be higher in MRE than that of ARE. The increased activity might be due to more availability of functional groups for antioxidant activity in MRE than ARE. Compared to ARE and MRE, inhibition was found to be high in AgNPs and ranged from 22 to 61 %. The scavenging potential of AgNPs was evident from the UV–vis spectra of DPPH in the absence and presence of AgNPs (Fig. S1). A significant increase in percent inhibition was evident up to 300 µg/ml concentration in all the tested samples. At 300 µg/ml concentration, AgNPs resulted in 53.5 % inhibition as against 36.7 and 41.2 % respectively for ARE and MRE. The DPPH scavenging potential followed the order of AA > AgNP > MRE > ARE. The higher scavenging ability of AgNPs could be attributed to more reactivity of the functional groups adhered to the large surface area of the AgNPs which could entrap a large number of free radicals than the rhizome extract. DPPH free radical scavenging activity of AgNPs synthesized from medicinal plants of Solanum torvum [61], Helicteres isora [62], Cassia tora [63], and Abutilon indicum [64] has also been reported with similar efficiency.
Hydrogen Peroxide Scavenging Assay
In living cells, uncontrolled accumulation of hydrogen peroxide leads to the formation of peroxides and hydroxyl radicals which damage the cell membrane components. The H2O2 scavenging activity of AgNPs and rhizome extracts along with AA as standard is shown in Fig. 8b. The maximum scavenging activity of 60, 63 and 68 % was obtained with ARE, MRE and AgNPs respectively at 500 µg/ml concentration. The percent inhibition of AgNPs was comparable to that of standard AA. The findings are well in accordance with the earlier reports of hydrogen peroxide scavenging activity [62, 64].
Superoxide Radical Scavenging Activity
The superoxide radical scavenging activity of ARE, MRE, and AgNPs was assessed by their ability of NBT reduction using AA as a positive control. Superoxide anion reduced NBT into formazon and formazon generation was measured spectrophotometrically at 560 nm. The degree of NBT reduction was found to be directly proportional to superoxide radical scavenging activity. The scavenging ability of ARE, MRE, AgNP and AA is shown in Fig. 8c. A gradual increase in percent inhibition was noted with increasing concentrations of the tested samples. There was no significant difference in superoxide radical quenching ability of ARE, MRE and AgNPs and the percent inhibition estimated was 71.4, 72.4 and 75.2 respectively at 500 µg/ml concentration. The findings indicate the availability of common functional groups in the reaction mixture prepared from ARE, MRE and AgNPs and their involvement in scavenging superoxide radical in a similar manner. The AgNPs mediated superoxide radical scavenging have also been reported in Solanum torvum [61] and Abutilon indicum [64].
Conclusions
In this study, we demonstrated the biosynthesis of AgNPs from Curculigo orchioides, a pharmaceutically important medicinal plant. Response surface methodology was applied to optimize the process parameters for the biosynthesis of AgNPs. This report for the first time explores the use of experimental design methodology to model and optimize the parameters of biosynthesis of AgNPs in higher plants utilizing absorbance of SPR band as a dependent variable. The derived polynomial model illustrates the functional relationship between the synthesis of nanoparticles in terms of plasmonic resonance intensity at 430 nm and four independent variables: AgNO3 concentration, % rhizome extract, pH and temperature. The optimum conditions for maximum AgNPs synthesis obtained were 2 mM AgNO3, 20 % rhizome extract, 60 °C temperature and pH 8. There was a good agreement between the predicted value and the experimental value which validates the model adequacy. AgNPs thus synthesized were characterized by UV–vis spectroscopy and TEM-EDAX. The TEM-EDAX studies reveal monodispersed particles, spherical in shape and the presence of elemental silver. The SAED pattern and the X-ray diffraction study confirmed the crystalline nature of the AgNPs. XRD parameters such as 2θ peak position, d-spacing values and lattice constant of biosynthesized AgNPs are well matching with the theoretical values of silver. FTIR analysis suggests the presence of biomolecules which play a role in bioreduction of AgNO3 and capping the synthesized AgNPs. Biogenic AgNPs with their plant derived phenolic glycosides and curculigosaponins exhibited potent in vitro antioxidant activities. This optimization approach for biosynthesis of AgNPs along with their ability to scavenge free radicals facilitates the possibility of large-scale production and utilization in agricultural and medicinal applications.
References
J. Li, L. Liu, D. Zhang, D. Yang, J. Guo, and J. Wei (2014). Synth. Met. 192, 15.
M. Sofianou, B. Nikos, V. Tiverios, and T. Christos (2014). Appl. Catal. B. Environ. 158, 91.
C. P. Tsangarides, A. K. Yetisen, F. da Cruz Vasconcellos, Y. Montelongo, M. M. Qasim, T. D. Wilkinson, C. R. Lowe, and H. Butt (2014). RSC Adv. 4, 10454.
R. Kanai and I. Eiji (2014). Thin Solid Films 554, 127.
A. Taglietti, C. R. Arciola, A. D’Agostino, G. Dacarro, L. Montanaro, D. Campoccia, L. Cucca, M. Vercellino, A. Poggi, P. Pallavicini, and L. Visai (2014). Biomaterials 35, 1779.
J. Fu, D. Li, G. Li, F. Huang, and Q. Wei (2014). J. Electroanal. Chem. 738, 92.
C. Kumar, Y. Ganesh, and Y. Poornachandra (2014). Colloid Surf. B 125, 110.
A. M. Fayaz, K. Balaji, M. Girilal, R. Yadav, P. T. Kalaichelvan, and R. Venketesan (2014). Nanomed. Nanotechnol. Biol. Med. 6, 103.
A. F. Mohammed, K. Balaji, M. Girilal, P. T. Kalaichelvan, and R. Venkatesan (2009). J. Agric. Food Chem. 57, 6246.
M. Rai, A. Yadav, and A. Gade (2009). Biotechnol. Adv. 27, 76.
S. Iravani (2011). Green Chem. 13, 2638.
S. Silver (2003). FEMS Microbial Rev. 27, 341.
X. Li, H. Xu, Z. S.Chen, and G. Chen (2011). J. Nanomat. 8.
S. P. Chandran, M. Chaudhary, R. Pasricha, A. Ahmad, and M. Sastry (2006). Biotechnol. Progr. 22, 577.
K. Mallikarjuna, G. Narasimha, G. R. Dillip, B. Praveen, B. Shreedhar, C. S. Lakshmi, and B. D. P. Raju (2011). Digest J. Nanomat. Biostruct. 6, 181.
J. J. Antony, M. Nivedheetha, D. Siva, G. Pradeepha, P. Kokilavani, S. Kalaiselvi, and S. Achiraman (2013). Colloid Surf. B 109, 20.
J. S. Valli and B. Vaseeharan (2012). Mater. Lett. 82, 171.
A. Rawani, A. Ghosh, and G. Chandra (2013). Acta Trop. 128, 613.
P. Velusamy, J. Das, R. Pachaiappan, B. Vaseeharan, and K. Pandian (2015). Ind. Crop Prod. 66, 103.
C. J. Murphy, A. M. Gole, S. E. Hunyadi, J. W. Stone, P. N. Sisco, A. Alkilany, B. E. Kinard, and P. Hankins (2008). Chem. Commun. 544.
C. Krishnaraj, E. G. Jagan, S. Rajasekar, P. Selvakumar, P. T. Kalaichelvan, and N. Mohan (2010). Colloids Surf. B 76, 50.
M. Sathishkumar, K. Sneha, and Y. S. Yun (2010). Bioresource Technol. 101, 7958.
S. Kaviya, J. Santhanalakshmi, and B. Viswanathan (2011). J. Nanotechnol. 1.
N. Yang and W. H. Li (2013). Ind. Crop. Prod. 48, 81.
N. E. A. El-Naggar and N. A. Abdelwahed (2014). J. Microbiol. 52, 53.
S. Roy and T. K. Das (2014). Nanosci. Nanotechnol. Lett. 6, 181.
E. O. Nyakundi and M. N. Padmanabhan (2015). Spectrochim.Acta A 149, 978.
S. M. Pourmortazavi, M. Taghdiri, V. Makari, and M. Rahimi-Nasrabadi (2015). Spectrochim. Acta A 136, 1249.
N. Saha and S. Dutta Gupta (2016). J. Clust. Sci. 27, 1419.
S. R. Kurma and S. H. Mishra (1995). Indian Drugs 33, 20.
K. P. S. Chandel in S. Ganesh and N. Sharma (eds.), Biodiversity in medicinal and aromatic plants in India (ICAR publication, New Delhi, 1996), p. 60.
J. P. Xu and R. S. Xu (1992). Phytochemistry 31, 2455.
J. P. Xu, R. S. Xu, and X. Y. Li (1992). Planta. Med. 58, 208.
Z. H. Wang, J. Huang, X. C. Ma, G. Y. Li, Y. P. Ma, N. Li, and J. H. Wang (2013). Fitoterapia 86, 64.
J. P. Xu and R. S. Xu (1992). Acta. Pharmacol. Sin. 27, 353.
Q. Wu, D. X. Fu, A. J. Hou, G. Q. Lei, J. Z. Liu, J. K. Chen, and T. S. Zhou (2005). Chem. Pharm. Bull. 53, 1065.
R. W. Cheary and A. Coelho (1992). J. Appl. Crystallogr. 25, 109.
P. Onkar, J. Bangar, and R. Karodi (2012). J. Appl. Pharm. Sci. 6, 209.
T. Hanato, H. Kagawa, T. Yasuhara, and T. Okuda (1998). Chem. Pharm. Bull. 36, 2090.
G. K. Jayaprakasha, R. L. Jaganmohan, and K. K. Sakariah (2004). Bioorg. Med. Chem. 12, 5141.
M. Nishimiki, N. Appaji, and K. Yagi (1972). Biochem. Biophys. Res. Commun. 46, 849.
I. Pastoriza-Santos and L. M. Liz-Marzán (2002). Nano Letters 2, 903.
A. Rai, A. Singh, A. Ahmad, and M. Sastry (2006). Langmuir 22, 736.
P. S. Pimprikar, S. S. Joshi, A. R. Kumar, S. S. Zinjarde, and S. K. Kulkarni (2009). Colloid Surf. B 74, 309.
H. Bar, K. D. Bhui, P. G. Sahoo, P. D. P. S. Sarkar, and A. Misra (2009). Colloid Surf. A 339, 134.
U. B. Jagtap and V. A. Bapat (2013). Ind. Crop. Prod. 46, 132.
S. A. Aromal and D. Philip (2012). Spectrochim. Acta A 97, 1.
D. C. Montgomery, R. H. Myers, W. H. Carter, and G. G. Vining (2005). Qual. Reliab. Eng. Int. 21, 197.
S. Das, S. Kar, and S. Chaudhuri (2006). J. Appl. Phys. 99, 1.
S. Li, Y. Shen, A. Xie, X. Yu, L. Qiu, L. Zhang, and Q. Zhang (2007). Green Chem. 9, 852.
S. Raja, V. Ramesh, and V. Thivaharan (2015). Ara. J. Chem. doi:10.1016/j.arabjc.
A. Synytsya, J. Copikova, P. Matejka, and V. Machovic (2003). Carbohyd. Polym. 54, 97.
V. Gopinath, D. Mubarak Ali, S. Priyadarshini, N. Meera Priyadharshini, N. Thajuddi, and P. Velusamy (2012). Colloid Surf. B 96, 69.
A. Ricci, K. J. Olejar, G. P. Parpinello, P. A. Kilmartin, and A. Versari (2015). Appl. Spectrosc. Rev. 50, 407.
P. Velusamy, J. Das, R. Pachaiappan, B. Vaseeharan, and K. Pandian (2015). Ind. Crop. Prod. 66, 103.
V. Lakshmi, K. Pandey, A. Puri, R. P. Saxena, and K. C. Saxena (2003). J. Ethnopharmacol. 89, 181.
A. E. Boelrijk, J. H. T. Dorst, and J. Reedijk (1996). Recl. Trav. Chim. Pays-Bas 115, 536.
J. Jung, S. Park, S. Hong, M. W. Ha, H. G. Park, Y. Park, and Y. Park (2014). Carbohyd. Res. 386, 57.
M. Villanueva-Ibáñez, M. G. Yañez-Cruz, R. Álvarez-García, M. A. Hernández-Pérez, and M. A. Flores-González (2013). Rev. Mater. Brazil 18, 1501.
R. Geethalakshmi and D. V. L. Sarada (2013). Ind. Crop. Prod. 51, 107.
C. H. Ramamurthy, M. Padma, I. Daisy, R. Mareeswaran, A. Suyavaran, M. Suresh Kumar, K. Premkumar, and C. Thirunavukkarasu (2013). Colloid Surf. B 102, 808.
S. Bhakya, S. Muthukrishnan, M. Sukumaran, and M. Muthukumar (2015). Appl. Nanosci. 1.
A. Saravanakumar, M. Ganesh, J. Jayaprakash, and H. T. Jang (2015). J. Ind. Eng. Chem. 28, 277.
R. Mata, J. R. Nakkala, and S. R. Sadras (2015). Colloid Surf. B 128, 276.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.

10876_2016_1050_MOESM1_ESM.jpg
Supplementary Fig. S1 The UV–vis spectra of DPPH in the absence and presence of AgNPs (0, 100, 200, 300, 400 and 500 µg/ml). Supplementary material 1 (JPEG 18 kb)
Rights and permissions
About this article
Cite this article
Saha, N., Trivedi, P. & Dutta Gupta, S. Surface Plasmon Resonance (SPR) Based Optimization of Biosynthesis of Silver Nanoparticles from Rhizome Extract of Curculigo orchioides Gaertn. and Its Antioxidant Potential. J Clust Sci 27, 1893–1912 (2016). https://doi.org/10.1007/s10876-016-1050-7
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10876-016-1050-7