Abstract
Purpose
Duplication of chromosome 22q11.2 due to meiotic non-allelic homologous recombination results in a distinct syndrome, chromosome 22q11.2 duplication syndrome that has some overlapping phenotypic features with the corresponding 22q11.2 deletion syndrome. Literature on immunologic aspects of the duplication syndrome is limited. We conducted a retrospective study of 216 patients with this syndrome to better define the key features of the duplication syndrome.
Methods
Single-center retrospective record review was performed. Data regarding demographics, clinical details, and immunological tests were compiled, extracted into a predetermined data collection form, and analyzed.
Results
This cohort comprised 113 (52.3%) males and 103 (47.7%) females. The majority (54.6%) of mapped duplications were between low copy repeat regions A–D (LCR22A to -D). Though T cell subsets were relatively preserved, switched memory B cells, immunoglobulins, and specific antibodies were each found to be decreased in a subset of the cohort. One-fifth (17/79, 21.5%) of patients had at least 2 low immunoglobulin values, and panhypogammaglobulinemia was found in 11.7% (9/79) cases. Four children were on regular immunoglobulin replacement therapy. Asthma and eczema were the predominant atopic symptoms in our cohort.
Conclusion
Significant immunodeficiencies were observed in our cohort, particularly in B cells and antibodies. Our study expands the current clinical understanding and emphasizes the need of immunological studies and multidisciplinary approaches for these patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Genomic band 11.2 of human chromosome 22 harbors well-characterized region-specific low copy repeats (LCRs) which are prone to DNA rearrangements during meiotic crossover through non-allelic homologous recombination [1]. Deletions or duplications occur secondary to misalignment of the LCRs due to high degree of homology between their sequences (> 95%), acting as substrates for non-allelic homologous recombination. The type of misalignment drives the specific copy number variation (CNVs) which can be either deletion, duplication, inversion, or translocation (Fig. 1). While deletion in this region leads to the well-characterized 22q11.2 deletion syndrome (22q11.2del) [2], the less frequently detected duplication results in the infrequently characterized 22q11.2 duplication syndrome (22q11.2dup) [3]. 22q11.2del is estimated to occur in 1:3000 to 1:6000 live births and has variable phenotypes with cardiovascular, immunologic, endocrine, and developmental abnormalities [2]. However, the prevalence of 22q11.2dup has been more difficult to establish, largely due to the technical aspects of diagnosis. A Danish population-based study estimated the prevalence at 1:1606 [4].
22q11.2dup was first described in 1999 [5] and reported subsequently from various centers [6,7,8]. Clinical presentations of 22q11.2dup range from asymptomatic to frank multisystem abnormalities with severe complications. There is phenotypic overlap with the deletion syndrome, particularly cardiac anomalies and developmental delay. Diagnosis of 22q11.2dup is often missed due to the mild phenotype and technical limitations of fluorescent in situ hybridization (FISH) which is relatively insensitive for duplications. The use of microarray and prenatal diagnostic facilities has increased detection recently.
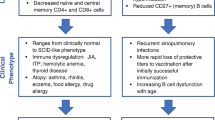
Variable expressivity, incomplete penetrance, and wide clinical phenotypes including neuro-developmental illnesses, learning disorders, cardiac abnormalities, dysmorphic features, gastrointestinal, and endocrine abnormalities have been described [3]. Immunodeficiencies, distinct from those seen in 22q11.2 del, have also been reported in patients with 22q11.2dup, with a recent case series suggesting that T cell counts are unaffected in 22q11.2dup [9,10,11]. However, these studies were limited by sample size. In this study, we performed a retrospective record review of clinical, immunological, and cytogenetic data of patients with 22q11.2dup at our hospital to better characterize the immune phenotype.
Materials and Methods
Study Population
We performed this observational study through a retrospective record review of a digital clinical and laboratory repository/database of patients and their parents with 22q11.2dup diagnosed at the Children’s Hospital of Philadelphia from January1997 to March 2022. All the children and adults with positive tests for chromosome 22q11.2 duplication detected by any method were enrolled. Demographics, clinical details, hematologic and immunologic data, cytogenetics, and therapeutics were extracted systematically onto a data collection form and analyzed. Two hundred eighteen patients from 155 families were identified. Two patients were excluded for lack of clinical data. A total of 216 patients were identified with recorded genetic tests and clinical history. Thirty patients did not have any non-genetic laboratory data. A total of 178 patients had undergone at least one type of immune testing. Four (1.9%) children were recorded as deceased. Twenty-six (12%) had no data on current status.
Testing
Diagnosis of 22q11.2dup was made via various genetic testing modalities including microarrays [including array-based genomic hybridization (ACGH)], single nucleotide polymorphism (SNP) microarray, FISH, multiplex ligation-dependent probe amplification (MLPA), genome sequencing, and prenatal genetic screening. Flow cytometry was carried out on Beckman (Cytoflex platform) flow cytometers (Life Sciences, IN, USA) in the hospital’s clinical laboratory. Immunoglobulins were measured by nephelometry, and antibody titers were measured by enzyme-linked immunosorbent assay (ELISA) in the clinical laboratory. Genetic testing methods and flow protocol details are available on request. Standard published normative data were used for the reference ranges of hematological and immunological studies and comparisons [12,13,14]. Absolute counts and percentages were compared with references separately.
Statistical Analysis
Descriptive statistics (frequencies, mean, standard deviation (SD), and proportions) were used for summarizing categorical and continuous variables. All point estimates were supplemented with their corresponding 95% CI (confidence interval). A p value ≤ 0.05 was taken as significant. No correction for multiple comparisons was performed. Association of categorical variables was analyzed using chi-squared test (χ2)/Fisher exact tests whereas comparisons of quantitative variables between two study groups were carried out using independent sample t test (parametric) or Mann-Whitney U test (non-parametric). Comparisons of the subgroup means of variables were performed. One-way analysis of variance (ANOVA) was used to compare means of parametric tests whereas Kruskal-Wallis tests by ranks were applied for comparisons in non-parametric one. Tukey’s HSD (honest significant difference) test was used in conjunction with an ANOVA to find means that are significantly different from each other. Pearson correlation coefficient (Pearson’s r) and Spearman’s rank correlation coefficient (Spearman’s rho ‘ρ’) were also estimated to find out linear association between various continuous variables. Linear correlations were further analyzed graphically using scatter plots. Analysis was performed using ‘SPSS’ version 23.0 software (IBM, USA).
Results
Demographics, Inheritance, and Diagnostics
Our study cohort of 216 patients was comprised of 163 (75.5%) children, 41 (19%) adults, and 12 (5.6%) fetuses (with postnatal follow-up), and the self-reported gender was 113 (52.3%) males and 103 (47.7%) females (M/F ratio = 1.096:1) (Table 1). One of the antenatally diagnosed cases died at the postnatal age of 3 months due to respiratory complications. The majority (70.8%) were non-Hispanic, white. Overall age at diagnosis ranged from prenatal to 59.75 years with a mean of 9.8 years (SD 13.9). The mean age at diagnosis was 4.35 years (SD 5.06) for children (Table 1).
Perinatal details were available in 151 cases. Gestational weeks at birth ranged from 26 to 42 weeks with a mean of 37.9 (SD 3.3) weeks. The majority (76.3%) were born full term with a mean birth weight of 3005 (SD 841.8) grams.
The inheritance pattern was paternal in 39 (18.1%) and maternal in 49 (22.7%) (Table 2). The remainder had either a de novo or an uncertain parental inheritance. Microarray (chromosomal, ACGH, and SNP-based) was the predominant modality used for diagnosis (114 patients (52.8%)). MLPA, FISH, and genome sequencing were used for diagnosis of the remainder. Twelve (5.6%) cases were diagnosed antenatally. More than half (54.6%) of patients had a duplication of LCR22A to -D (Fig. 1). Duplications involving DNA distal to LCR22D site were seen in 46 (21.3%) patients. Among them, 36 (16.7%) had solely distal to LCR22D duplication events whereas 10 (4.6%) patients had a duplication involving regions bridging the A–D and distal regions. One patient each was found to have a duplication before region LCR22A and after LCR22H. A nested duplication (within -A to -D region) was identified in 35 (16.2%) patients. Sixteen (7.4%) patients were diagnosed with FISH only, and therefore, the breakpoints are unknown.
Eleven percent of patients had multiple genetic variants, possibly modifying their phenotype. An additional 13q12.3 duplication, 15q11.2 duplication, and CYBB mutation were detected in 2 patients each (one male with clinical chronic granulomatous disease and his mother, a carrier). Moreover, isolated deletions noted in each different individual were on chromosome 16p11.2, 16q24.3, 16p13.11, 5p14.1, 13q12.11, 10q21.3, 20p11.21, Xp22.33p22.2, Yp11.2 and an in-frame indel in exon32 of NF1. Duplications in region 5p13.2, 16p13.11, 17q21.31, 20p11.21 and the SMARCB1 3′UTR accompanied 22q11.2 duplication in individual patients. Instances of uniparental disomy of chromosomes 16 (16q22.1q24.3) and 18 (18p11.22q21.1) were also observed in our cohort. One patient had 2 duplications [at 8p22 (paternally inherited) and at 5p13.2 (maternally inherited)] in addition to variants in MYO7A and COL4A3 genes in the hearing loss panel. An inversion in chromosome 9 was also noted in 2 patients.
Clinical Profile
We collected clinical details to seek associations with immunodeficiency. Among 216 subjects, developmental delay in at least 2 sectors of milestones was observed in 48.6% of patients. At least one cardiac issue was present in 33.8% of patients, and 30.5% of patients had endocrine disorders (Table 1). We analyzed the CBC and electrolytes and liver function. These are reported in Supplemental Table S1 and were largely normal.
Immunological Profile
Among 212 patients with immune system-related clinical data, at least one type of immune evaluation was performed in 178 patients. A summary of findings of lymphocyte subsets is depicted in Table 3 and Supplemental Table S2.
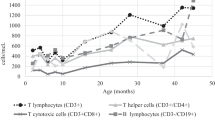
Detailed analyses of lymphocyte subsets were performed on 76 patients. Nine had a low percentage of lymphocytes with eight also having low absolute counts per age-related references. T cell counts were largely preserved with only 2 patients (2.6%) having low CD3 counts for age (clinical features are given in Supplemental Table S3). Different sets of 2 patients each were found to have low CD4 and CD8 T cell counts. T cell counts rose with age largely due to increasing CD8 T cells with age, a pattern not typical in the general population. Two patients had low CD19 B cell counts.
CD4/CD45RA+ naive T cells as a percentage were higher than the normal range for age in 19.6% of patients. Both absolute and percentage counts of CD45RO+ memory T cells were low in 6.2% of patients whereas 56% had low percentages of this subset and no patient had only decreased counts with preserved percentages. T cells in different age groups are illustrated in Fig. 2. Descriptive details are tabulated in Supplemental Table S2, and additional lymphocyte subsets are portrayed in Supplemental Figure 1.
Hypogammaglobulinemia occurred in patients both 22q11.2 deletion and duplication [2, 9]. We therefore examined the B cell subsets in our cohort in the subset of 16 patients who had expanded B cell profiling. Twenty-five percent of patients had low switched memory B cells. None of the patients had low naïve B cells. Higher CD19+CD27+IgM+ B cell counts were seen in 40%. Transitional B cells were found to be low in 13.3% of cases. Conversely, it was found to be raised in 26.6% of patients. Changes in the counts among age groups were not uniform and did not reach statistical significance. Trends of B cells in various age groups are shown in Fig. 3. Decline in naïve B cells and rise in memory B cells, typical for age-related changes, were observed. Details of descriptive analysis of the B cell subset overall and as per age group are illustrated in Table 4.
In evaluating antibody production (N = 80), 19.4% of patients had low IgG, whereas IgA was low in 30% of cases and low IgM was found in 26.3% of cases. At least one immunoglobulin isotype was low in 44% of patients who were tested. Seventeen patients had low levels of any 2 immunoglobulin isotypes (among IgG, IgA, IgM) whereas 11.3% had panhypogammaglobulinemia. However, only 4 (1.8%) patients were on regular subcutaneous (2 patients) or intravenous (2 patients) immunoglobulin replacement therapy. Figure 4 shows the expected rise in IgG and IgA with age with some patients over 5 years of age with an IgG < 500 mg/dl, demonstrating hypogammaglobulinemia (Table 5 provides additional details).
Antibody titers against diphtheria, tetanus and pneumococcus were evaluated in 72 patients. Five [5/65; 7.7%] patients had low anti-tetanus titers whereas anti-diphtheria titers were low in 12% (7/58). Seventeen different anti-pneumococcal antibodies were tested. Low titers in more than 2 subgroups of antibodies panel were found in 8 patients (Table 5 and Supplemental Table S4).
Periodic fevers and recurrent infections were recorded in 5 (2.3%) patients. Infections included sepsis, respiratory (including sinus) infections, gastrointestinal infections, neurological infections, dental, genitourinary tract infection, ear and eye infections. Infections noted among hypogammaglobulinemia patients were recurrent otitis, sinusitis, pneumonia, upper and lower respiratory infections, bronchiolitis, boils, abscesses, thrush, gastrointestinal infections, teeth and nail infections, urinary tract infections and meningitis. Two patients with low antibody titers had bronchiolitis, pneumonia and frequent febrile episodes. Interestingly, 2 patients with 22q11.2dup also had chronic granulomatous disease with documented pathogenic CYBB mutations and were treated as appropriate for chronic granulomatous disease. Asthma and eczema were the prominent atopic symptoms in our cohort. One-fourth of the patients had asthma requiring medication whereas eczema was noted in 20 (9.4%) patients. Allergic rhinitis, reactive airway disease and food allergy were other allergic diatheses noted in our cohort. Autoimmunity was seen in 5 patients (2.3%).One patient each had isolated ITP, autoimmune encephalitis or thyroiditis. Two patients had both ITP and autoimmune encephalitis. The duplication breakpoints and immunologic evaluations were no different in this subgroup with autoimmunity.
Correlations
Logistic regression was performed in two different ways. IgG levels were available in 72 patients of whom 19.4% had low levels for age. IgG values did not correlate significantly with T cells or B cell percentages. Significant positive correlation was observed between IgG level and CD8+ cytotoxic T cells (%) (ρ = 0.3, p = 0.006), but this was eliminated upon the addition of age as a variable. No other lymphocyte subset showed significant correlation with low IgG.
Lymphocyte population percentages as continuous variables were also compared among different sites of duplications. Duplication sites were grouped according to LCR endpoints as well as the regions: standard, nested, distal or standard plus distal. No statistical difference was noted for T cells or B cells among various duplication site categories (p > 0.05, Kruskal-Wallis test). Similarly IgG, IgA or IgM values were not significantly different between various categories of duplication sites (p = 0.32, Kruskal-Wallis test).
Using cross-tabulation of categorical findings, the number of cases with panhypogammaglobulinemia, low IgG, low IgM or two Ig serotypes below the lower limit of normal was not significantly different among various duplication sites. Clinical details of patients with panhypogammaglobulinemia are given in Supplemental Table S3. However, low IgA values were significantly more common in patients with the standard zone of duplication (LCR22A to -D) in comparison to others (p = 0.02, Fisher’s exact test). Differences in numbers of patients with low T cells, B cells and their subsets (counts in %) were not statistically different among various duplication categories.
Discussion
Due to variable expressivity, incomplete penetrance and a wide range of phenotypes, 22q11.2dup may be underdiagnosed and we undertook this study to expressly characterize the immune system in more detail [3]. 22q11.2del, the syndrome due to deletion in the same region, leads to significant immunodeficiency, and several reports have highlighted the surprising concordance of phenotypic features in the patients with the deletion and the patients with the duplication [2,3,4, 8, 10, 15,16,17,18,19]. As in 22q11.2del, duplication in same region was found to result in developmental disorders, immune deficiencies, endocrine disorders and cardiac defects [3, 8, 9, 18].
Our analysis illustrates a wide range of clinical phenotypes and multisystem defects in 22q11.2dup. This finding is congruent with prior studies [3, 8, 16, 18, 20,21,22,23]. The frequency of congenital heart disease (CHD) observed in our cohort is consistent with that described in other studies and is clearly lower than that of patients with 22q11.2del. Hypocalcemia was also rare in our cohort with 22q11.2dup compared to published studies with 22q11.2del [2, 24]. The predominant type of duplication in our cohort was an inherited standard LCR22A-D. A study of nested duplications may allow refinement of critical regions dictating specific phenotypic features [17, 25]. One murine study highlighted TXNRD2, COMT and ARVCF as critical for learning differences [26].
Previous studies of patients with 22q11.2dup found conceptually similar immunologic features. Sun et al. found immunodeficiency in 7 patients with 22q11.2dup [9]. In this study, low switched memory B cells and low immunoglobulin levels warranting Ig replacement were reported, consistent with other studies emphasizing the presence of antibody dysfunction [8, 11]. There was relative preservation of T cell numbers in patients with 22q11.2dup [10]. Our study confirms low but detectable population frequencies of abnormal immunologic laboratory studies. These may be interpreted within the context of a population cohort with co-morbidities and reasons for secondary immunodeficiency.
T cell populations in our cohort were relatively preserved with only two patients (2.6%) having low CD3 counts, similar to a previous study [10]. Only 16 patients had extensive B cell evaluations, and one patient had low switched memory B cells. This study was not designed as a comparative study with 22q11.2del, but the T cell populations are clearly more frequently normal in 22q11.2dup compared to published studies of 22q11.2del [2, 10, 27]. A higher number of transitional B cell counts were also observed. These features have also been seen in 22q11.2del where the mechanism is not fully understood [28,29,30].
One notable finding in our study is the trend of increasing CD8 T cell counts with advancing age (Fig. 2). In contrast, our cohort did demonstrate a fall in other lymphocyte subsets with increasing age as observed in children with 22q11.2del and the general population [3, 12].
As was previously reported, hypogammaglobulinemia requiring immunoglobulin replacement occurred in patients with 22q11.2dup [9]. Among 10 such patients, 4 needed regular replacement therapy. Twenty percent of our cases had at least 2 low immunoglobulin isotype levels, and 7–12% had low titers. Antibody dysfunction was seen in all age groups. These data confirm the need for immunologic assessments in patients with 22q11.2dup, targeted toward their specific immunologic features.
The mechanism of immunodeficiency observed in patients with 22q11.2dup is unknown. A possible role of the T-box transcription factor gene (TBX1) is postulated based on patients with the 22q11.2del syndrome [10]. This gene is critical for the organs developing from the pharyngeal arches namely the thymus, parathyroid and cardiac tissues [7, 16, 31]. Defects in the thymus, which in turn leads to defects in T cell development, may contribute to a compromised activation or development of B cells through altered follicular helper T cell function. However, comparatively normal T cell counts in 22q11.2dup suggest that alternative explanations should be entertained. Moreover, duplications in nested, distal or TOP3B(E) regions also led to immune dysfunction suggesting that there must be some other mechanism for the clinical and immunological manifestations. Additionally, further studies are needed to define ethnic and racial differences [2, 21, 32]. Cross-sectional and prospective studies across the globe will help to define modifier effects, and additional studies of epigenetics may be revealing [33].
This is the first study to elaborate on immunological profiles in a large cohort of patients with 22q11.2dup. Though there were two patients with significant T cell lymphopenia, our data confirm that humoral dysfunction is more common. A diagnosis of 22q11.2dup should be considered in patients with humoral dysfunction even if they have limited physical or developmental features suggestive of the syndrome [34] since our data demonstrated a high variability in phenotype.
Strengths and Limitations
To fill the knowledge gap on the immunological features in 22q11.2dup, our study provides critical data on the immune profile of patients. It represents the largest single-center clinical cohort of 22q11.2dup encompassing all clinical, immunological and cytogenetic data. Charts were consistently reviewed by a single reviewer to ensure consistency in recording. However, as with any study design, there were limitations. The sample sizes for the B cell evaluations were smaller than for T cell evaluations. Findings from this hospital may not be true for all regions and types of ethnicity and races. Retrospective data may not represent the true prevalence in society due to possible incomplete documentation, asymptomatic cases or variable rates of hospital visits at different places. Some patients had undergone diagnosis with FISH or MLPA diagnostic evaluations which limited identification of other confounding CNVs contributing to their phenotype. Ascertainment bias with anchoring on patients with features compatible with 22q11.2del may have occurred. Nevertheless, this study has great strength in size and further characterization of clinical features.
Conclusions
Our study showed relative preservation of T cells with a reduction in immunoglobulins and specific antibodies in a subset of patients. Our study provides new details about clinical and laboratory descriptions in a large diverse cohort. It contributes to a better understanding of patients with 22q11.2dup. Though most children with 22q11.2dup had no laboratory evidence of immune deficiency, frequencies observed in this study were significantly higher than in the general population. Additionally, this study leads to important distinctions between the immune phenotype in 22q11.2dup and 22q11.2del.
Data Availability
Data are available upon request.
References
Mikhail FM, Burnside RD, Rush B, Ibrahim J, Godshalk R, Rutledge SL, et al. The recurrent distal 22q11.2 microdeletions are often de novo and do not represent a single clinical entity: a proposed categorization system. Genet Med. 2014;16:92–100.
McDonald-McGinn DM, Sullivan KE, Marino B, Philip N, Swillen A, Vorstman JAS, et al. 22q11.2 deletion syndrome. Nat Rev Dis Primer. 2015;1:15071.
Ou Z, Berg JS, Yonath H, Enciso VB, Miller DT, Picker J, et al. Microduplications of 22q11.2 are frequently inherited and are associated with variable phenotypes. Genet Med. 2008;10:267–77.
Olsen L, Sparsø T, Weinsheimer SM, Dos Santos MBQ, Mazin W, Rosengren A, et al. Rearrangements in the 22q11.2 region: prevalence and population-based risk for neuropsychiatric and developmental disorders. Lancet. Psychiatry. 2018;5:573–80.
Edelmann L, Pandita RK, Spiteri E, Funke B, Goldberg R, Palanisamy N, et al. A common molecular basis for rearrangement disorders on chromosome 22q11. Hum Mol Genet. 1999;8:1157–67.
Ensenauer RE, Adeyinka A, Flynn HC, Michels VV, Lindor NM, Dawson DB, et al. Microduplication 22q11.2, an emerging syndrome: clinical, cytogenetic, and molecular analysis of thirteen patients. Am J Hum Genet. 2003;73:1027–40.
Yu A, Turbiville D, Xu F, Ray JW, Britt AD, Lupo PJ, et al. Genotypic and phenotypic variability of 22q11.2 microduplications: an institutional experience. Am J Med Genet A. 2019;179:2178–89.
Bartik LE, Hughes SS, Tracy M, Feldt MM, Zhang L, Arganbright J, et al. 22q11.2 duplications: expanding the clinical presentation. Am J Med Genet A. 2022;188:779–87.
Sun D, Lee J, Heimall J, Jyonouchi S. Immunodeficiency in 22q11.2 duplication syndrome. J Allergy Clin Immunol Pract. 2021;9:996–998.e3.
Crowley B, Ruffner M, McDonald McGinn DM, Sullivan KE. Variable immune deficiency related to deletion size in chromosome 22q11.2 deletion syndrome. Am J Med Genet A. 2018;176:2082–6.
Traynor R, Butler KM, Cant AJ, Leahy TR. Immunodeficiency in a child with 22q11.2 microduplication syndrome. J Clin Immunol. 2016;36:418–9.
Garcia-Prat M, Álvarez-Sierra D, Aguiló-Cucurull A, Salgado-Perandrés S, Briongos-Sebastian S, Franco-Jarava C, et al. Extended immunophenotyping reference values in a healthy pediatric population. Cytometry B Clin Cytom. 2019;96:223–33.
Luning Prak ET, Ross J, Sutter J, Sullivan KE. Age-related trends in pediatric B-cell subsets. Pediatr Dev Pathol. 2011;14:45–52.
Ritchie RF, Palomaki GE, Neveux LM, Navolotskaia O, Ledue TB, Craig WY. Reference distributions for immunoglobulins A, G, and M: a practical, simple, and clinically relevant approach in a large cohort. J Clin Lab Anal. 1998;12:363–70.
Portnoï M-F. Microduplication 22q11.2: a new chromosomal syndrome. Eur J Med Genet. 2009;52:88–93.
Hasten E, McDonald-McGinn DM, Crowley TB, Zackai E, Emanuel BS, Morrow BE, et al. Dysregulation of TBX1 dosage in the anterior heart field results in congenital heart disease resembling the 22q11.2 duplication syndrome. Hum Mol Genet. 2018;27:1847–57.
Wincent J, Bruno DL, van Bon BWM, Bremer A, Stewart H, Bongers EMHF, et al. Sixteen new cases contributing to the characterization of patients with distal 22q11.2 microduplications. Mol Syndromol. 2010;1:246–54.
Butensky A, Rinaldis CP, Patel S, Edman S, Bailey A, McGinn DE, et al. Cardiac evaluation of patients with 22q11.2 duplication syndrome. Am J Med Genet A. 2021;185:753–8.
Pebrel-Richard C, Kemeny S, Gouas L, Eymard-Pierre E, Blanc N, Francannet C, et al. An atypical 0.8 Mb inherited duplication of 22q11.2 associated with psychomotor impairment. Eur J Med Genet. 2012;55:650–5.
Hoeffding LK, Trabjerg BB, Olsen L, Mazin W, Sparsø T, Vangkilde A, et al. Risk of psychiatric disorders among individuals with the 22q11.2 deletion or duplication: a Danish nationwide, register-based study. JAMA. Psychiatry. 2017;74:282.
Wentzel C, Fernström M, Öhrner Y, Annerén G, Thuresson A-C. Clinical variability of the 22q11.2 duplication syndrome. Eur J Med Genet. 2008;51:501–10.
Vaz SO, Pires R, Pires LM, Carreira IM, Anjos R, Maciel P, et al. A unique phenotype in a patient with a rare triplication of the 22q11.2 region and new clinical insights of the 22q11.2 microduplication syndrome: a report of two cases. BMC Pediatr. 2015;15:95.
Verbesselt J, Zink I, Breckpot J, Swillen A. Cross-sectional and longitudinal findings in patients with proximal 22q11.2 duplication: a retrospective chart study. Am J Med Genet A. 2022;188:46–57.
Campbell IM, Sheppard SE, Crowley TB, McGinn DE, Bailey A, McGinn MJ, et al. What is new with 22q? An update from the 22q and You Center at the Children’s Hospital of Philadelphia. Am J Med Genet A. 2018;176:2058–69.
Woodward KJ, Stampalia J, Vanyai H, Rijhumal H, Potts K, Taylor F, et al. Atypical nested 22q11.2 duplications between LCR22B and LCR22D are associated with neurodevelopmental phenotypes including autism spectrum disorder with incomplete penetrance. Mol Genet. Genomic Med. 2019;7:e00507.
Suzuki G, Harper KM, Hiramoto T, Funke B, Lee M, Kang G, et al. Over-expression of a human chromosome 22q11.2 segment including TXNRD2, COMT and ARVCF developmentally affects incentive learning and working memory in mice. Hum Mol Genet. 2009;18:3914–25.
Jawad AF, McDonald-Mcginn DM, Zackai E, Sullivan KE. Immunologic features of chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/velocardiofacial syndrome). J Pediatr. 2001;139:715–23.
Zemble R, Prak EL, McDonald K, McDonald-McGinn D, Zackai E, Sullivan K. Secondary immunologic consequences in chromosome 22q11.2 deletion syndrome (DiGeorge Syndrome/Velocardiofacial Syndrome). Clin Immunol Orlando Fla. 2010;136:409–18.
Giardino G, Radwan N, Koletsi P, Morrogh DM, Adams S, Ip W, et al. Clinical and immunological features in a cohort of patients with partial DiGeorge syndrome followed at a single center. Blood. 2019;133:2586–96.
Cuss AK, Avery DT, Cannons JL, Yu LJ, Nichols KE, Shaw PJ, et al. Expansion of functionally immature transitional B cells is associated with human-immunodeficient states characterized by impaired humoral immunity. J Immunol. 2006;176:1506–16.
Chen M, Yang Y-S, Shih J-C, Lin W-H, Lee D-J, Lin Y-S, et al. Microdeletions/duplications involving TBX1 gene in fetuses with conotruncal heart defects which are negative for 22q11.2 deletion on fluorescence in-situ hybridization: TBX1 microdeletions/duplications in fetal CTD. Ultrasound Obstet Gynecol. 2014;43:396–403.
Gibney ER, Nolan CM. Epigenetics and gene expression. Heredity. 2010;105:4–13.
Zhang Z, Shi L, Song L, Maurer K, Zhao X, Zackai EH, et al. Chromatin modifications in 22q11.2 deletion syndrome. J Clin Immunol. 2021;41:1853–64.
Bartik LE, Hughes SS, Tracy M, Feldt MM, Zhang L, Arganbright J, et al. 22q11.2 duplications: expanding the clinical presentation. Am J Med Genet A. 2021;188A:779–87.
Acknowledgements
The authors gratefully acknowledge the study staff and the patients. We also acknowledge Dr. Aaqib Zaffar Banday for his support in statistical analysis and figure graphics.
Funding
DMM and KES were funded by Children’s Hospital of Philadelphia. DB was funded by an educational grant from ESID. Wallace Chair of Pediatrics (KES), ESID Mid-term fellowship to DB, MCHRI Uytengsu-Hamilton 22q11 Neuropsychiatry Research Award to KES, and NIH GM125757 to BE.
Author information
Authors and Affiliations
Contributions
DB: acquisition of data, data analysis, data interpretation, drafting manuscript, editing and critical revision. DMM, TBC, BSE and EHZ: oversight of general phenotyping, sample acquisition and duplication sizing. DEM, TBC, VG and KG: data acquisition, abstraction and preliminary analysis. DS, JL, JH and SJ: oversight of patient enrollment and patient data, data acquisition and analysis. KES: concept and design of the study, acquisition of data, data interpretation, editing, critical revision and approval of final version.
Corresponding author
Ethics declarations
Ethics Approval
The Institutional Review Board of Children’s Hospital of Philadelphia approved this study, and patients provided consent. All samplings and procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Consent for Publication
Informed consent for possible publication of clinical data is included in the IRB protocol and consent.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary materials 1:
Supplemental Table S1 Basic laboratory parameters. Supplemental Table S2. Lymphocyte subset analyses by age group. Supplemental Table S3. Clinical details of the most immunodeficient patients. Supplemental Table S4. Antibody titers in different age groups. Supplemental Figure 1. Scatter plots with linear regression line for observed percentages of HLADR+ T & B cells, CD132+ T cells, CD5+ B cells, CD8+CD28- anergic T cells, and gamma-delta T cells in different age groups. (PDF 615 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bhattarai, D., McGinn, D.E., Crowley, T.B. et al. Immunologic, Molecular, and Clinical Profile of Patients with Chromosome 22q11.2 Duplications. J Clin Immunol 43, 794–807 (2023). https://doi.org/10.1007/s10875-023-01443-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10875-023-01443-5