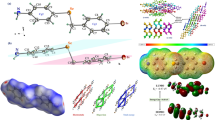
Abstract
The 1:2 ratio salicylic acid/biimidazole co-crystal is studied in this work. I This compound was obtained from a flux with methanol followed by slow evaporation, rendering colorless plates, that crystallize in the monoclinic system, space group P21/n, with unit cell parameters a = 12.0969 (4) Å, b = 5.0714 (1) Å and c = 15.4825 (4) Å, β = 99.007(2)º, and V = 938.11(4) Å3. The experimental characterization is achieved through X-ray diffraction structure analyzes, infrared spectroscopy (FTIR) and thermal analysis (TGA/DSC). This characterization is complemented by theoretical calculations at the DFT/6–31++G(d,p) level, employing the HCTH407 functional. Geometrical optimizations are performed and a detailed Non-Covalent-Index analysis is carried out. It is found that the crystal structure is maintained by conventional and non-conventional hydrogen bonds, of the type N–H···O, O–H···N and C–H···O hydrogen bonds responsible for the formation of planar supramolecular units forming R2 2(9) rings and slip-stacked packing, which, through DFT/HCTH calculations, are shown to stabilize the structure.
Graphical Abstract
The synthesis and crystal structure of the title compound, C10H9N2O3, is reported. The bimidazole and salicylic acid form a co-crystal in a 1:2 ratio. Its crystal structure is maintained by conventional and non-conventional hydrogen bonds. Two conventional hydrogen bonds, of the type N–H···O and O–H···N, form planar supramolecular unit through R2 2(9) rings, the DFT/HCTH calculations shows us that the R2 2(9) rings formation stabilize the supramolecular structure.

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The rational synthesis of co-crystals has captured lately a considerable amount of attention. A fundamental interest in understanding and optimizing the molecular assembly processes is partially responsible for it; however, this is mainly due to the co-crystal potential applications in many areas, particularly in medical and pharmacological ones [1]. Since the co-crystal composition and the three-dimensional arrangement depend on how complementarily molecular moieties interact with each other forming supramolecular synthons, the understanding and characterization of these intermolecular interactions in the synthons is one of the basis in co-crystals designing. Thus, the capability of effectively combining different synthons [2] has proved to be a key tool for a precise construction of supramolecular assemblies, which leads to molecular architectures of considerable complexity.
Among all the types of intermolecular interactions, hydrogen bonds (HBs), thanks to their robust and directional nature, play an important role in designing and synthesizing co-crystals [3]. Therefore, analyzing and understanding these interactions is important in co-crystal characterization. Frequently HBs are described in terms of their geometry; however, in many cases the employed geometrical parameters do not explicitly involve the hydrogen atoms [4]. Moreover, the geometric characterization usually constitutes only a first step in a description of the co-crystals non-covalent interactions, because the dependence of the system geometric properties on the kind or strength of the non-covalent interaction is not free of controversy [5]. Thus, classifying non-covalent interactions as Van der Waals, steric clashes or hydrogen bonds, only based in terms of pairwise distances between atoms, frequently obtained from their Van der Waals radii, could give rise to systematic errors, as generally overestimated HB lengths. Therefore, nowadays more elaborate methods for analyzing non-covalent interactions are employed, with many of these methods based in employing the electronic density properties [6], as is the case of the Atoms in Molecules (AIM) theory [7, 8]. This theory, among other things, considers the fact that upon formation of atomic interactions, the electronic density develops critical points in the interatomic region: bond critical points (BCP). It has been evidenced that for hydrogen bonded complexes, some many electronic density properties evaluated at the BCP, such as the density itself, its Laplacian or the kinetic energy density, correlate with the HB interaction energy [8, 9]. Another alternative approach is based on the analysis of the electron localization function (ELF) [10]. This function depends on the electron and kinetic energy density, and was developed to highlight the regions where electron localization occurs [11]. It has been reported that the ELF bifurcation value also correlates with the HB strength [12]. Moreover, recently a new approach, based on the analysis of the electron density and their reduced gradient, was introduced [13]. This allows simultaneous analysis and visualization of a wide range of non-covalent interactions, leading to a more detailed characterization of these interactions.
Due to their biological activity and their importance in the crystal engineering field, supramolecular assemblies formed by compounds containing the 2,2′-biimidazole moiety have been widely studied [14]. 2,2-biimidazole possesses five possible forms: di-deprotonated (biim2), mono-deprotonated (Hbiim), neutral (neutral, H2biim), mono-protonated (monocation, H3biim+) and di-protonated (dication, H4biim2+) [15]. Among them, thanks to the fact that neutral H2biim can act as proton donor, as well as proton acceptor, it has become an organic target for construction of new co-crystals. In this manner, the H2biim could be bound to a second component with complementary functional groups, as Carboxylic units, to produce supramolecular systems: a carboxylic acid functional group may act as proton donor–acceptor, binding to H2biim, forming structures, as those reported by. Gao et al. [14, 16], assembled through the formation of O–H···N and N–H···O HB patterns, developing R2 2(9) graph sets building blocks. In this direction, Atencio et al. [17] have shown that the supramolecular R2 2(9) synthon could be a designing tool, leading to modular self-assembly of metal-based architectures with different dimensionality. These authors have reported several biimidazole complexes, where both carboxylate group oxygens may be directly attached to the N–H donors, via a charge assisted R2 2(9) heteromeric synthon. Within this context, in this work we discuss a co-crystal, whose asymmetric units are formed from one salicylic acid molecule and ½ neutral biimidazole molecule, with coplanar imidazole rings. The crystal structure is analyzed in detail and the non-covalent interactions found in this structure are characterized beyond the geometric analysis, employing both, the AIM theory and a new Non-Covalent-Index (NCI).
Experimental
General Procedures and Materials: Co-crystalization of Salicylic Acid and 2,2-Biimidazole
The IR spectra were recorded on a Shimadzu FTIR spectrophotometer, model 8300, as KBr pellets. The multicomponent 2,2-biimidazole:salicylic acid 2:1 co-crystal was prepared by mixing salicylic acid (0.12 mmol, Aldrich, 98%) and 2,2-biimidazole (0.02 mmol). The reagents were separately grounded with an agate pestle and mortar, and dissolved in ethanol (60 mL). The solution was mixed and placed in a reflux system for a period of 15 min at a constant temperature of 338 K. After slow evaporation of the reflux solution, colorless block-shape crystals of approximately 0.9–1.2 mm length were obtained.
Single Crystal X-Ray Data Collection and Structure Determination
Single crystal X-ray diffraction analysis was carried out on a KAPPA APEX II DUO Diffractometer, with graphite monochromator and Mo-Kα radiation (λ = 0.71069 Å) operating at 50 kV and 30 mA. A total of 2181 frames were collected with 0.30° scans in ϕ and ω, with an exposure time of 10 s per frame [18].
The structure was solved and refined using the Bruker SHELXTL Software Package [19]. The non-hydrogen atoms were refined anisotropically, while the hydrogen atoms bound to C atoms were placed geometrically and refined using a riding model, with C–H = 0.93 Å, Uiso(H) = 1.2 Ueq(C) for aryl H; and O–H = 0.82 Å and Uiso(H) = 1.2Ueq(O).
Differential Scanning Calorimeter and Thermogravimetric Analysis
The melting points were measured with a Mettler Toledo TGA/DSC-1 model LF 645 instrument. Accurately weighed samples (~0.015 mmol) were prepared in a covered aluminum crucible, having pierced lids to allow escape of volatiles. The sensors and samples were under nitrogen purge during the experiments, temperature calibration was carried out and a heating rate of 283 K/min was used.
Thermogravimetric analysis also was performed on a Mettler Toledo TGA/DSC-1 model LF 645 instrument. Approximately 3 mg (~0.015 mmol) sample was heated from 278 to 1073 K at 283 K/min under nitrogen purge.
Theoretical Calculations
All geometrical optimizations were performed at the DFT level of calculation, using the HCTH functional [20] and the Gaussian09 program [21] For all the calculations, the basis set 6–31++G(d,p) was employed. All the optimizations were performed using the Berni algorithm; analytical Hessian calculations were carried out along all the optimization process. The thresholds for convergence were 0.000450 (a.u.) and 0.000300 for maximum force and root mean square (rms) force, respectively. Vibrational frequency calculations were performed on the clusters to assure that the calculated structure corresponds to a minimum. Optimization of the biimidazole:salicylic acid complexes, with the 1:1 and 1:2 ratios was performed, with the monomer initial structures built up by using the crystal structure bond distances and angles measured in this work.
The binding energy, was calculated from,
where E AB is the energy of the salicylic acid-biimidazole complex, E A and E B are, respectively, the energies of the optimizated monomers of salicylic acid and biimidazole, and n and m are the number of salicylic acid and biimidazole molecules in the complex. It is important to mention that this binding energy captures all non-covalent interactions, such as HBs or dispersion contributions. Nevertheless, due to the characteristics of the studied systems, it is assumed that the hydrogen bonding is the predominant interaction in the stabilization process. The binding energy per monomer, ΔE/s, can be defined from Eq. (1), where s is the number of molecules in the complex.
NCI Analysis
Recently Jhonson et al. [22, 23] proposed a new approach to study and visualize non-covalent interactions, introducing a new NCI, based on the analysis of the electron density and its reduced gradient [13, 24]. This approach allows the description of deviations from homogeneous electron distributions. The key function in the NCI analysis is the reduced gradient density (RGD), which depends on the density ρ and its gradient, as:
This function has the capability to highlight interactions on the low density, low density gradient regimes. In regions where the density decays to zero, i.e. density tails far from the molecules, the RGD will have large positive values; whereas, in regions of uniform density, the RGD will approach to zero. Consequently, the analysis of the RGD in function of the electron density reveals the different kind of density behaviors and chemical interactions existing in the system. For instance, far from the nuclei, the density shows an exponentially decaying tail; thus, in a RGD versus ρ plot, this region appears at low densities with the RGD values tending to diverge. On the other hand, near the nuclei, the density is large and the corresponding RGD values appear at the right edge of the plot. In regions where inter or intramolecular interactions between fragments are present, covalent bonding or non-covalent interactions, the electron density will display critical points, and the RGD will become zero. A covalent bond region has a density critical point; hence, the RGD displays a spike with RGD ≃ 0 at large density values, approximately between 0.2 and 0.4 a.u. In supramolecular complexes, the non-covalent interactions also display critical points; yet, for these cases the interaction occurs at low density regions. Thus, in RGD versus ρ plots the non-covalent interactions appear as spikes or troughs with RGD ≃ 0 in the low density regions. In this manner, the RGD and density values act as indicators of the interaction strength. However, the repulsive interactions also appear in the region of low ρ-low RGD values. It is for this reason that an electron density analysis is necessary to assign the trough nature in the plot (HBs, Van der Walls, etc). The sign of the Laplacian of the electron density, ∇2 ρ, is used to distinguish between attractive and repulsive interactions. It indicates whether the net gradient flux of density is entering (∇2 ρ < 0) or leaving (∇2 ρ > 0) an infinitesimal volume around a reference point, which could be interpreted as a concentration or depletion of electron density in the reference point, relative to the surrounding. To further analyze the density, its Laplacian is decomposed into a sum of contributions along the three principal axes of maximum variation. These contributions correspond to the eigenvalues (λi) of the Hessian matrix: ∇2 ρ = λ1 + λ2 + λ3, (λ1 < λ2 < λ3). These eigenvalues also allow to characterize the electron density critical points, where ∇ρ = 0. Thus, the critical points are classified according to their rank, ω, the number of non-zero curvatures at the critical point, and the algebraic sum of the signs of the Hessian eigenvalues λi, σ: (ω, σ). For the stable critical points, ω = 3. There are four types of stable critical point: (3,−3) nuclear critical point (NCP); (3,−1) bond critical point (BCP); (3,+1) ring critical point, and (3,+3) cage critical point (CCP). At an electron density maximum (nuclei positions) the three eigenvalues are negatives. In the interatomic regions, λ3 varies along the internuclear direction, while λ1 and λ2 change in the plane perpendicular to the λ3. For bonding interactions, characterized by an accumulation of electron density in the internuclear region, λ1 < λ2 < 0 and λ3 > 0. For covalent bonding, the negative contributions dominate and ∇2 ρ < 0, for non- covalent interactions, such as hydrogen bonds or Van der Waals interactions, the positive contribution is dominant and ∇2 ρ > 0. In the case of hydrogen bonds, there is an accumulation of electronic density in the internuclear region, and λ2 < 0, while for Van der Waals interactions, the densities are very low in the internuclear region, and the values of λ2 are negative and very close to 0. On the other hand, for repulsive interactions, such as steric repulsion, there is electron density depletion from the internuclear region, and λ2 > 0. Therefore, analysis of the λ2 sign allows distinguishing between bonding or non-bonding interactions, while the electron density itself enables us to assess the interaction strength.
From the previous discussion, around the critical points, where |∇ρ| ~ 0, the RGD also tends to zero, and its behavior, in particular the topology of the RGD isosurfaces, is dominated by the behavior of the |∇ρ| topology. Bohorquez et al. [13] showed that from the analytical expansion of the electron density around a critical point r 0 = (x 0, y 0 , z 0), the density gradient could be approximated as ∇ρ(r) ≈ (x − x 0) λ1 u 1 + (y − y 0) λ2 u 2 + (z − z 0) λ3 u 3, where {λi} are the eigenvalues of Hessian matrix, {u i} their respective eigenvectors and r = (x, y, z) corresponds to a position vector, satisfying that |r − r 0|→0. After assuming that both r and r 0 are represented in terms of the same basis {u i}, taking the square of the approximate expression for ∇ρ(r) and dividing by |∇ρ|2, the equation of an ellipsoid centered in r 0 and aligned with the local principal directions, {u i}, is obtained,
whose principal axes are given by: a = (|∇ρ|/λ1), b = (|∇ρ|/λ2) y c = (|∇ρ|/λ3). The Eq. 3 shows that the information contained in the Hessian of the system can be translated into 3D RGD graphical representations. Thus, around the nuclei the magnitude of the λi are approximately the same; hence a ≃ b ≃ c and ρ(r) around the nuclei has spherical symmetry, for covalent bonding, λ1, λ2 >> λ3 and consequently, c > a,b and the RGD ellipsoid has cylindrical shape with the axis aligned with the bond path between the bonded nuclei. For non-covalent interactions, as HB, λ1, λ2 << λ3; hence c < a,b, and the RGD isosurfaces have disc shapes, perpendicular to the bond path. For the limiting case of weak interactions, such as Van der Waals interactions, the RGD isosurface has a flat sprawled shape. The information corresponding to the strength of the interaction is provided by coloring the RGD isosurface according their density values.
Results and Discussion
Analysis of FTIR Spectra of Biimidazole:Salicylic Acid 1:2
In the salicylic acid infrared spectrum, an intense signal, due to the C=O vibration, is observed at 1790 cm−1, while a wide and intense band between 2500 and 3000 cm−1, is assigned to the O–H group. For the biimidazole IR spectrum, a band observed in the 3142–2632 cm−1 region, is attributed to the N–H group stretching, whose signal is expected to be between 3200–2500 cm−1, whereas a signal at 1675 cm−1 is assigned to the C=C stretching, and the band centered at 1332 cm−1 is attributed to the C–N vibration. The co-crystal FTIR spectrum shows a broad band between 3000–2400cm−1, which we think corresponds to the overlap between the carboxylic acid O–H stretching signal, and that corresponding to the N–H stretching, which indicates the presence of intermolecular hydrogen bonds. A less intense signal is observed at 1710 cm−1, which we think corresponds to the shifted signal of the C=O stretching of the carboxyl group participating in hydrogen bondings. These results point to the formation of the salicylic acid-biimidazole supramolecular heterosynthon.
Analysis of the Crystal Structure of Biimidazole:Salicylic Acid 1:2
Figure 1 exhibits the co-crystal molecular structure and atomic labeling scheme. The presence of the neutral form of the salicylic acid molecule was confirmed by measuring the C6–O2 [1.255(2) Å] and C6–O1 [1.266(2) Å] bond distances; moreover, it was found that the molecule is almost planar, with maximum deviations from the mean least square molecular plane of 0.011(1) Å on the C6, and −0.006(1) Å on the O3. For this molecule, an intramolecular O3–H3···O1 HB was found, which participates in a S(6) graph set. It is obtained that the biimidazole molecule is also planar, with maximum deviation from the mean least square molecular plane of ±0.003 Å, with its center of mass located at its inversion center. Table 1 displays the detailed crystal data and refinement information, while in Table 2 a representative set of geometrical parameters are presented. Figure 2 shows the HB pattern observed in the co-crystal structure, which described a R2 2(9) graph set, constituted by the HB interactions between the salicylic acid and the biimidazole via two HBs: O2–H2···N2 and N4–H4···O1 [1 − x, −y, 1 − z]. A second salicylic acid molecule, related to the previous one by an inversion center, is also bonded with the central biimidazole through an equivalent HB pattern, establishing a second R2 2(9) ring and generating another planar supramolecular unit. These two units are connected by a non-conventional C5–H5···O3 [½ + x, ½ − y, −½+z] HB, and are stacked in two different directions, with a dihedral angle of 82.28(1)°, forming an infinite supramolecular chain, described by a C2 2(9) graph set (Table 3). Let us emphasize that in this structure, the donor and acceptor hydrogen bonding capacity of the two co-crystal components are fully utilized.
Thermal Analysis
DSC analyses of the co-crystal, obtained after slow evaporation, yields an endothermic peak at 513.54 K. This endothermic peak is different from those corresponding to the melting points of the salicylic acid (441.45 K) or biimidazole (>623.15 K), result that indicates the formation of a co-crystal between biimidazole and salicylic acid. Furthermore, a second peak at 625.4 K is observed, which is attributed to the decomposition of the biimidazole molecule.
In the TGA analysis, thermal stability is observed until 506.15 K, where a decrease in the baseline was observed, with a mass loss of 63.81%. This is associated with the decomposition of the carboxylic acid. A second transition at 625.4 K, with a mass loss of 32.92%, is related with the biimidazole decomposition.
Equilibrium Geometry and Binding Energies of the Biimidazole:Salicylic Acid 1:1 and 1:2 Complexes
In order to analyze the non-covalent interactions observed in the co-crystal, we have optimized the molecular structures of the biimidazole, the salicylic acid, and the two biimidazole:salicylic acid complexes, with ratios of 1:1 and 1:2. Table 4 displays the calculated bond distances, corresponding to the monomer optimized geometries, together with the difference between the theoretical and experimental crystallographic results. It was found that the mean difference between the theoretical calculations and the crystallographic results are 0.008 and 0.004 Å for the salicylic acid and the biimidazole, respectively, with no significant differences between the crystallographic and optimized geometries. For the two optimized complexes, Table 5 lists the geometrical parameters characterizing the intermolecular O–H···O, N–H···O and O–H···N interactions. The calculated donor–acceptor distances are larger than those found in the crystal structure. Thus, for the intramolecular O3–H3···O1 HB, the average difference with the crystal structure distances are 0.0127 and 0.0147 Å, for the 1:1 and 1:2 complexes, respectively. For the O2–H2···N2 and N4–H4···O1 intermolecular HBs, these differences are 0.060 and 0.025 Å for the 1:1 complex, and 0.059 and 0.0207 Å, for the 1:2 complex. The binding energies are −15.46 and −30.97 kcal/mol, for the 1:1 and 1:2 complexes, respectively. Since the stabilization of the complexes is mainly due to the O–H···N and N–H···O intermolecular HB formation, these results imply an average stabilization energy per hydrogen bond of approximately −7.7 kcal/mol. Let us point out that due to the complex symmetries, this value is approximately the same for both complexes.
Electron Density Analysis of the Non-covalent Interactions Observed in Biimidazol:Salicylic Acid 1:1 and 1:2 Complexes
The quantum theory of atoms in molecules (QTAIM) [24] is employed to investigate the non-convalent interactions in the biimidazole:salicylic acid supramolecular units. This theory, based in the direct analysis of the electron density (ρ), allows obtaining information on the molecular behavior, and on the atomic and bond properties. As discussed above, the electron density critical points, where ∇ρ = 0, contains information, that permits the system characterization. In particular, the properties of BCPs serve to summarize the nature of the interaction between two atoms. Since the HB definition requires evidence of a bond formation between the interacting fragments, it is clear that the HB existence should be manifested by the presence of a BCP. Along the hydrogen bond path, ρ decreases from the nuclei towards the BCP, where it has a local minimum, positive λ3. Also at the BCP, the electron distribution shows a maximum in the plane perpendicular to the bond path, such that their negative curvatures λ1 and λ2 increase with the value of ρ. This growth in the magnitude of λ1 + λ2 is accompanied by an increase in the positive amplitude, λ3, satisfying λ3 > |λ1 + λ2| and; therefore, ∇2 ρ BCP,HB > 0. Thus for HBs, a sharp electron concentration in the plane where ρ is maximum, leading to an increase in ρ BCP,HB , is accompanied by a larger ρ depletion along the direction of the bond path.
For the biimidazole:salicylic acid complex, Fig. 3 shows the AIM molecular graph, exhibiting the critical points present in the system, NCP (3, −3), RCP (3, +1) and BCP (3, −1), and the bond path between the NCP pairs. For the two complexes studied here, Table 6 displays the BCP properties of the intramolecular O3–H3···O1 HB and the intermolecular O–H···N and N–H···O HBs, together with their bond order. From there, one can observe that, as should be expected, all the considered HB interactions are of the close shell type, ∇2 ρ BCP,HB > 0. Also, comparison of the BCP topological properties leads to conclude that there are no meaningful changes between the topological properties of the HBs involved in the 1:1 and 1:2 complexes. Only a slight increase in the ρ BCP,HB and bond order values is observed for the H4···O1 HB in the 1:2 complex respect to those in the 1:1 one. This result indicates that there is no significant difference between the cooperative effects present in both complexes [9, 25]. Moreover, since it has been reported that the hydrogen bond strength correlates with accumulation of electron density between the hydrogen and the acceptor atoms, measured by the ρ BCP,HB value [25, 26], the strength of the considered HBs follows the order, O2–H2···N2 > O3–H3···O1 > N4–H4···O1. Notice that the two weakest HBs are those where the O1 participates as a bifurcated acceptor, with the weakest interaction corresponding to that where N4 participates as an H-donor. We also performed a bond order calculation within the fuzzy atom formalism introduced by Hirshfeld [27, 28]. In contrast to QTAIM, where molecular space is divided by sharp boundaries, the fuzzy atom formalism takes into account the chemically intuitive idea of shared electronic density, belonging simultaneously to a pair of bonded atoms, i.e. it takes explicitly into account the electronic density polarization. Thus, within this formalism, the molecular space is divided in atomic regions with no sharp boundaries that instead vary continuously from one region to another. For the considered HBs, the results in Table 6 show that the bond orders follow the trend found for ρ BCP . For the strongest O2–H2···N2 HB, the bond order is approximately the same for both complexes; however, the results show that the intramolecular O3–H3···O1 HB bond order slightly decreases from the 1:1 to the 1:2 complex, whereas for the intermolecular N4–H4···O1 HB, the opposite behavior is observed, with the bond order increasing by approximately 10%. Thus, these results indicate that for the H4···O1 HB the electronic delocalization increases for the 1:2 complex, which has been shown to correlate with the HB stability, and with an increase in the system cooperativity [9, 25].
Besides the three previously discussed conventional hydrogen bonds, a possible non-conventional HB, of the type C–H···O was observed. To gain insight in the characteristics of this interaction, we have performed several optimizations, starting from systems whose geometries correspond, or are close, to that observed in the crystal. However, for all these cases, none of the optimized geometries showed any stable C–H···O interaction. As an alternative approach, we calculated the electron density keeping fixed the crystallographically obtained geometry. The QTAIM analysis performed from this density has revealed the existence of a BCP between H5 and O3 (Fig. 5a). This result may be evidence for the existence of a C5–H5···O3 interaction. Again, its topological properties and the bond order are listed in Table 6, showing that ρ BCP,HB is an order of magnitude lower than the conventional HBs considered, and the bond order is the lowest for all the HBs found.
NCI Analysis of the Biimidazol:Salicylic Acid Non-covalent Interactions
For the biimidazole and salicylic acid molecules, and for the 1:2 biimidazole:salicylic acid complex, we have studied their non-covalent interactions through a RGD function analysis. For the biimidazole molecule, Fig. 4a shows the RGD results as a function of Sig(λ2)ρ. Three spikes are clearly observed. The one at the largest density, at approximately ρ ≃ 0.06 and with RGD = 0, is associated with a molecular (3, +1) RCP. For the remaining spikes, RGD ≠ 0; hence, they are not related with any critical point. Nevertheless, these RGD sharp variations should be consequences of changes in the intramolecular interactions between molecular fragments. Thus, we have studied the RGD isosurfaces, which provide a visualization of the regions in real space where non-covalent interactions take place. The obtained isosurfaces are colored according to their interaction kind and strength, ranging from large negative values of Sig(λ2)ρ, colored in red, indicative of attractive interactions, to large positive values of Sig(λ2)ρ, colored in blue, associated with non-bonding or repulsive interactions. For the biimidazole molecule, the isosurface results, corresponding to RGD = 0.45 a.u., are displayed at the bottom of Fig. 4a. Two blue isosurfaces are observed inside the rings, one is associated with the RCP, discussed earlier, whereas the other one, near the ring C–C bond, is related to a weak repulsion between the nitrogen atoms electron lone pairs. A third kind of isosurface, colored in gray, corresponds to a weak attractive N···H interaction. For the salicylic acid molecule, the NCI interaction results are displayed in Fig. 4b. The RGD vs Sig(λ2)ρ plot shows a more complex pattern than that found in the previous case. Two attractive intramolecular interactions are observed. One of them is associated with a (3, −1) BCP, corresponding to the O3–H3···O1 hydrogen bond. The second spike, at smaller density and RGD ≠ 0, is associated with a weak attractive interaction between the oxygen of the hydroxyl group and the hydrogen in the ring orto position. The RGD results for positive Sig(λ2)ρ values, show two close spikes, which are associated with two RCPs, one corresponding to the benzene ring, and the other one to the hydrogen bond ring including O3–H3···O1 HB. A third spike, with RGD ≠ 0, observed at larger Sig(λ2)ρ value, may be related with a a weak interaction C12–H12···O2. All these interactions may be visualized in the 3D RGD isosurface plot, shown at the bottom of Fig. 4b. A red disk is observed between the H3 and O1 atoms, arising from the O3–H3···O1 HB, and another pink disk is observed, related to the C12–H12···O2 weak interaction. This figure clearly shows two blue rings, corresponding to RCPs, one at the benzene center and the other one enclosed by the intramolecular hydrogen bond O3–H3···O1. A fifth light blue isosurface, quite close to the pink disk, may be associated with the electronic redistribution arising from the formation of the weak interaction C12–H12···O2. Figure 4c displays the RGD results for the biimidazole:salicylic acid 1:2 complex. A comparison of the complex NCI data plot with those of the monomers reveals an even more complicated pattern than those observed for each of the monomers. First, the negative side of Sig(λ2)ρ (attractive interactions) shows four spikes. Three of them are associated with conventional hydrogen bonds, whose strengths decrease as O2–H2···N2 > O3–H3···O1 > N4–H4···O1. These interactions may be visualized in 3D RGD isosurface plots, displayed at the bottom of Fig. 4c and in Fig. 5b. In these Figures, the previous complex attractive interactions correspond to red discs. Also, the C12–H12···O2 and C5–H5···O3 weak interactions are observed as light pink disks; moreover, from Fig. 4c is possible to assign a (3,−1) BCP to the interaction C5–H5···O3 (Fig. 5a), with ρBCP ≈ 0.0083 a.u. It is interesting to compare this value with that found for the intermolecular N4–H4···O1 hydrogen bond, where ρBCP ≈ 0.0318 a.u., which quantitatively evidences that even the weakest conventional hydrogen bond is significantly stronger than the non-conventional C5–H5···O3 hydrogen bond. Finally, Fig. 6 presents the RGD isosurfaces for the interaction between two 1:2 biimidazole:salicylic entities, as observed in the crystal. This Figure clearly shows zones of attractive interactions intercalated with others of repulsive interactions. It can be easily visualized from all the RGD isosurface plots (Figs. 4, 5) that the hydrogen bond interactions are local, whereas π–π interactions (Fig. 6) are delocalized, this would give rise to cooperativity effects on the stabilization of the crystal [29].
Plots of RGD (ρ) versus sing (λ2)ρ and RGD isosurfaces for a bimidazole, b salicylic acid and c 1:2 biimidazole:salicylic acid complex. The isosurfaces correspond to RGD = 0.45 a.u., over the range of −0.04 a.u < sing(λ2) ρ < 0.04 a.u., colored as explained in the text (from red indicating attractive and blue repulsive interactions). (Color figure online)
a QTAIM molecular graph for the biimidazole:salicylic acid complex. The critical points are colored according to their type (3, −3) = purple, (3, +1) = yellow and (3, −1) = orange. The bond paths are the gold lines between two (3, −3) critical points. b RGD isosurfaces corresponding to RGD = 0.40 a.u., over the range of −0.04 a.u < sing (λ2) ρ < 0.04 a.u, colored as explained in the text (Red indicate attractive and blue repulsive interaction). (Color figure online)
Results for the RGD isosurfaces, with RGD = 0.50 a.u., in a system constituted by two layers of the planar biimidazole:salicylic hydrogen bonded entities as observed in the crystal. The RGD values are in the −0.03 a.u < sing(λ2) ρ < 0.03 a.u. range, and are colored as explained in the text (from red indicating attractive to blue repulsive interactions). (Color figure online)
Summary
The crystallographic results show that bimidazole and salicylic acid form a co-crystal in a 1:2 ratio. The crystal structure features conventional and non-conventional hydrogen bonds, including hydrogen bonds of the type N–H···O and O–H···N, forming planar R2 2(9) ring units that are further linked via π stacking, and by a non-conventional hydrogen bonding of the type C–H···O. The DFT/HCTH calculations reproduced the main features of the crystal structure, including the hydrogen bond patterns. Through these calculations was possible to obtain the stabilization energy per molecule. It is noteworthy that in spite of hydrogen bond network evidenced throughout the structure, the theoretical calculations have not revealed the existence of significant cooperative enhancement in the 1:2 co-crystal formation. Finally, the analysis of the topological properties of the electron density, together with the formulation of a NCI, as the reduced gradient density function employed here, have proven to be a powerful tool to study the nature and characteristics of non-convalent interactions responsible for the formation of many crystallographic structures. In particular, the NCI allows quite effectively visualizing and classifying non-covalent interactions existing in supramolecular systems.
Supporting Information Available
X-ray crystallographic data for this structure has been deposited at the Cambridge Crystallographic Data Center under code CCDC 1012902.
References
Cromer D, Ryan R, Storm C (1987) Structure of 2,2′-biimidazole. Acta Cryst 43:1435–1437
Etter M (1990) Encoding and decoding hydrogen-bond patterns of organic-compounds. Acc Chem Res 23:120–126
Desiraju G (1989) Crystal engineering: the design of organic solids. Elservier, Amsterdam
Desiraju G, Steiner T (1999) The weak hydrogen bond. Oxford University Press, Oxford
Lakshmi, B., Samuelson, A. G., Jovan Jose, K. V., Gadre, S. R., Arunan E. (2005) Is there a hydrogen bond radius? Evidence from microwave spectroscopy, neutron scattering and X-ray diffraction results. New J Chem 29:371–377
Scoles G (ed) (1992) Atomic and molecular beam methods. Oxford University Press, New York
Bader RFW (1990) Atoms in molecules: a quantum theory. Oxford University Press, Oxford
Grabowski SJ (2007) Relationships between QTAIM and the decomposition of the interaction energy—comparison of different kinds of hydrogen bond. In: Matta CF, Boyd RJ (eds) The quantum theory of atoms in molecules: from solid state to DNA and drug design, Wiley, Darmstadt
Rincon L, Almeida R, García-Aldea D, Diez y Riega H (2001) Hydrogen bond cooperativity and electron delocalization in hydrogen fluoride clusters. J Chem Phys 114:5552–5562
Becke AD, Edgecombe KE (1990) A simple measure of electron localization in atomic and molecular systems. J Chem Phys 92:5397–5403
Alikhani ME, Fuster F, Silvi B (2005) What can tell the topological analysis of ELF on hydrogen bonding? Struct Chem 16:203–210
Seijas LE, Lunar A, Rincón L, Almeida R (2017) On the electron density localization in HF cyclic clusters. J Comput Methods Sci Eng 17:5–18
Bohórquez HJ, Boyd RJ, Matta CF (2011) Molecular model with quantum mechanical bonding information. J Chem Phys A 115:12991–12997
Gao, X., Zhu, M. (2010) Benzoic acid-2,2′-biimidazole (2/1). Acta Cryst E66:o3124
Tadokoro M, Nakasuji K (2000) Hydrogen bonded 2,2′-biimidazolate transition metal complexes as a tool of crystal engineering. Coord Chem Rev 198:205–218
Gao X, Bian L, Wei Guo S (2014) Crystal structure of 1 H,10 H-[2,20-biimidazol]-3-ium hydrogen tartrate hemihydrates. Acta Cryst E70:o1221–o1222
Reinaldo Atencio R, Chacón M, González T, Briceño A, Agrifoglio G, Sierraalta A (2004) Robust hydrogen-bonded self-assemblies from biimidazole complexes. Synthesis and structural characterization of [M(biimidazole)2(OH2)2] 2+ (M = Co2+, Ni2+) complexes and carboxylate modules. Dalton Trans 4:505–513
Bruker (1998) SAINT: SAX area-detector integration, Version 5.01, Bruker AXS, Madison
Sheldrick G (2008) A short history of SHELX. Acta Cryst A 64:112–122
Hamprecht FA, Cohen A, Tozer DJ, Handy NC (1998) Development and assessment of new exchange-correlation functional. J Chem Phys 109:6264–6271
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Rob MA, Cheeseman JR, Montgomery JA Jr, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, P. M. W. Gill, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA (2003) Gaussian 03. Gaussian, Wallingford
Jhonson ER, Keinan S, Mori-Sánchez P, Contreras-García J, Cohen A, Yang W (2010) Revealing non-covalent interactions. J Am Chem Soc 132:6498–6506
Contreras-García J, Jhonson ER, Keinan S, Chaudret R, Piquemal JP, Beratan DN, Yang W (2011) NCIPLOT: a program for plotting noncovalent interaction regions. J Chem Theory Comput 7:625–632
Hohenberg P, Kohn W (1964) Inhomogeneous electron gas. Phys Rev B 136:864–871
Rincon L, Almeida R, García-Aldea D (2005) A many-body energy decomposition analysis of cooperativity in hydrogen fluoride. Int J Quant Chem 102:443
Espinosa E, Alkorta I, Elguero J, Molins E (2002) From weak to strong interactions: a comprehensive analysis of the topological and energetic properties of the electron density distribution involving X–H···F–Y systems. J Chem Phys 117:5529–5532
Hirshfeld FL (1977) Bonded-atom fragments for describing molecular charge densities. Theory Chim Acta 44:129
Mayer I, Salvador P (2004) Overlap populations, bond orders and valences for ‘fuzzy’ atoms. Chem Phys Lett 383:368–375
Alonso M, Woller T, Martín-Martínez FJ, Contreras-García J, Geerlings P, De Proft F (2014) Understanding the fundamental role of π/π, σ/σ, and σ/π dispersion interactions in shaping carbon-based materials. Chem Eur J 20:4931–4941
Acknowledgements
The authors thank the financial support to CDCHTA-ULA (Grant No. C-1921-15-08-AA).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bruno-Colmenarez, J., Atencio, R., Quintero, M. et al. Crystal Structure Analysis and Topological Study of Non-covalent Interactions in 2,2-Biimidazole:Salicylic Acid 2:1 Co-crystal. J Chem Crystallogr 47, 47–58 (2017). https://doi.org/10.1007/s10870-017-0679-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10870-017-0679-x