Abstract
The clinical management of anaplastic thyroid carcinoma and follicular thyroid carcinoma is challenging and requires an alternative therapeutic strategy. Although atovaquone is an FDA-approved anti-malarial drug, studies has recently demonstrated its anti-cancer activities. In line with these efforts, our study shows that atovaquone is an attractive candidate for thyroid cancer treatment. We show that atovaquone significantly inhibits growth, migration and survival in a concentration-dependent manner in 8505C and FTC113 cells. Mechanistically, atovaquone inhibits mitochondrial complex III activity, leading to mitochondrial respiration inhibition and reduction of ATP production in thyroid cancer cells. The inhibitory effects of atovaquone is reversed in mitochondrial respiration-deficient 8505C ρ0 cells, confirming mitochondrial respiration as the mechanism of atovaquone’s action in thyroid cancer. In addition, atovaquone suppresses phosphorylation of STAT3 in thyroid cancer wildype but not ρ0 cells, demonstrating that STAT3 phosphorylation inhibition by atovaquone is a consequence of mitochondrial respiration inhibition. Notably, we further demonstrate that atovaquone significantly augments doxorubicin’s inhibitory effects via suppressing mitochondrial respiration and STAT3. Our findings suggest that atovaquone can be repurposed for thyroid cancer treatment. Our work also highlights that targeting mitochondrial respiration may represent potential therapeutic strategy in thyroid cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Thyroid cancer is the most common malignancy of endocrine organs with increasing incidence in the past few decades worldwide (Sipos and Mazzaferri 2010). Thyroid cancers including papillary, follicular, poorly differentiated, and anaplastic thyroid carcinomas show significant differences in genetics, biological and clinical behaviors (Nikiforov and Nikiforova 2011). Although substantial advances have been made on the molecular carcinogenesis and progression of thyroid cancer, specific inhibition of oncogenic pathways is ineffective, in particular in follicular and anaplastic thyroid carcinomas which are undifferentiated, aggressive, and resistant to therapeutic options (Kondo et al. 2006; Cabanillas et al. 2016; Jin et al. 2016). Various recent studies reveal essential roles of mitochondrial metabolism in not only energy production but also macromolecule biosynthesis and activation of signaling pathways, to maintain the growth and survival of tumor and even tumor stem cell (Weinberg and Chandel 2015; Lee et al. 2015). Targeting mitochondria may represent an alternative therapeutic strategy for thyroid cancer.
Atovaquone [trans-2-[4-(4-chlorophenyl) cyclohexyl]-3-hydroxy-1, 4-naphthalene-dione] is a FDA-approved broad spectrum anti-parasitic drug. It kills parasites via inhibiting the cytochrome bc1 complex and mitochondrial respiration, leading to disruption of mitochondrial membrane and blockage of energy supply (Fry and Pudney 1992; Srivastava et al. 1997). Recently, atovaquone has been demonstrated to be an potential anticancer agent targeting both cancer cells and cancer stem cells in preclinical models in vitro and in vivo, and increase radiosensitivity by alleviating tumor hypoxia (Xiang et al. 2016; Fiorillo et al. 2016; Zhou et al. 2009).
In this work, we investigated the effects of atovaquone alone and its combination with doxorubicin in thyroid cancer cell lines, and analysed the mechanism of its action. We show that atovaquone inhibits growth, migration and survival of thyroid cancer cells, and acts synergistically with doxorubicin. We further show that the inhibitory effects of atovaquone are attributed to its inhibition on mitochondrial respiratory complex III and STAT3.
Materials and methods
Cell culture, generation of ρ0 cell line and drugs
Human thyroid cancer cell lines 8505C and FTC133 were obtained from the Institute of Cell Research, Chinese Academy of Sciences, China. The H9c2 cardioblast cell line was obtained from American Type Culture Collection. Cells were cultured as monolayers in a humidified atmosphere (5% CO2, 20% O2, 37 °C) in minimal essential medium (Sigma, US) supplemented with 10% fetal bovine serum (FBS) (Hyclone, UK). To establish ρ0 cell lines lacking of mitochondrial DNA, cells were growing in minimal essential medium containing 2 μg/ml ethidium bromide, 10% FBS, 100 μg/ml sodium pyruvate, 50 μg/ml uridine and antibiotics (Sigma, US) for 40 days and thereafter maintained in above media without ethidium bromide as previously described (Hashiguchi and Zhang-Akiyama 2009). Atovaquone and doxorubicin were purchased from Sigma-Aldrich.
Measurement of cell proliferation and apoptosis
5000 cells in 96-well plate were treated with drug alone, or combination for 72 h prior to proliferation analysis using Quick Cell Proliferation Assay Kit II (Abcam, US) according to the manufacturer’s instructions. 20, 000 cells in 24-well plate were treated with drug alone, or comnbination for 24 h prior to apoptosis analysis by measuring cytosolic oligonucleosome-bound DNA using a Cell Death ELISA kit (Roche Diagnostics GmbH, Germany).
Measurement of cell migration
Cell migration was measured by using Boyden Chamber. 10, 000 cells and drug at different concentrations were seeded on gelatin collated cell culture insert with membrane pore at 6.5 mm diameter. After 8 h incubation, cells in the upper chamber move through the pores towards the lower surface of the filter. Immigrated cells on the upper surface of the insert were removed with a cotton swab. Migrated cells were fixed with 4% paraformaldehyde and stained with Giemsa (Sigma, US). The migrated cells were counted and the photos were taken under light microscope.
Western blot analyses
One million cells in 6-well pate were treated with drug alone or combination for 24 h prior to western blot analyses. Cell were trypsinized and suspended in radioimmunoprecipitation assay (RIPA) lysis buffer (Invitrogen, US) containing protease inhibitor cocktails and phosphatase inhibitors (Invitrogen, US). Nuclear and mitochondrial proteins were isolated using the protocols described previously (Wegrzyn et al. 2009). Proteins from extracts of whole cell, nuclear and mitochondria were resolved by SDS-PAGE and blotted with anti-STAT3, p-STAT3(Tyr705) rabbit monoclonal antibody (Cell Signalling, US), Ac-H3, GRIM-19 and β-actin monoclonal antibody (Santa Cruz, US), respectively. Detection was performed by using the enhanced chemical luminescence (Pierce, US) and a LAS-3000 luminescent image analyser.
Measurement of mitochondrial complex III activity
Twenty thousand cells in 24-well plate were treated with drug alone or combination for 24 h. Mitochondria-rich fractions were prepared using a Mitochondrial Isolation Kit (Pierce Biotechnology) prior to measuring mitochondrial complex III activity using Mitochondrial Complex III Activity Assay Kit (BioVision, US). This assay is based on the reduction of cytochrome c through the activity of Complex III. The absorbance of reduced cytochrome c can be measured at 550 nm.
Measurement of mitochondrial respiration
Oxygen consumption rate (OCR), an indicator of mitochondrial respiration, was measured using the Seahorse XF96 analyser (Seahorse Bioscience, US). 10, 000 cells in 96-well plate were treated with drug alone or combination. After 24 h treatment, cells were equilibrated to the un-buffered medium for 30 min at 37 °C in a CO2-free incubator prior to being transferred to the Seahorse X96 analyser. OCR was measured under basal conditions, in the presence of the mitochondrial inhibitors oligomycin (OLI, 0.5 μM) or antimycin A and rotenone (A&R, 0.25 μM), or in the presence of the mitochondrial uncoupler (FCCP, 0.5 μM) to assess maximal mitochondrial respiration. The Seahorse XF-96 software calculated OCR automatically.
Measurement of cellular ATP level
Ten thousand cells in 96-well plate were treated with drug alone or combination. After 24 h treatment, ATP levels were measured by the ATP Bioluminescence Assay Kit HS II (Roche) according to the manufacturer’s instructions.
Statistical analyses
All data are expressed as mean and standard deviation. Statistical analyses were performed by unpaired Student’s t test with p-value <0.05 considered statistically significant.
Results
Atovaquone inhibits growth, survival and migration in thyroid cancer cells.
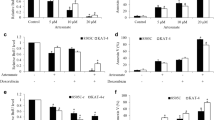
To investigate the possible effects of atovaquone on thyroid cancer, we performed proliferation, apoptosis and migrations assays on thyroid cancer cells exposed to atovaquone at different concentrations. Human thyroid cancer cell lines, 8505C and FTC133, are often used as in vitro cell models representing anaplastic thyroid carcinoma (ATC, the most dedifferentiated, aggressive subtype) and follicular thyroid carcinoma (FTC), respectively (Saiselet et al. 2012). These two cell lines harbor different oncogenic mutations including BRAF, P53 and PTEN (Saiselet et al. 2012). We found that atovaquone at 5, 10 and 20 μM inhibited proliferation and induced apoptosis of 8505C and FTC133 in a dose-dependent manner (Fig. 1a, b). The IC50 of atovaquone in inhibiting proliferation is ~7 μM for FTC133 and ~10 μM for 8505C (Fig. 1a). In addition, atovaquone significantly inhibited thyroid cancer cell migration as assessed by Boyden Chamber assay (Fig. 1c, d). It is noted that FTC133 is more sensitive to atovaquone than 8505C cells.
Atovaquone targets multiple functions of thyroid cancer cells. Atovaquone at 5, 10 and 20 μM significantly inhibits proliferation (a) and inducing apoptosis (b) in 8505C and FTC133 cells. Representative imagers (c) and quantification (d) showing the inhibitory effect of atovaquone on thyroid cancer cell migration. *, p < 0.05, compared to control
Atovaquone inhibits mitochondrial respiration and STAT 3 phosphorylation in thyroid cancer cells
Previous findings suggest that the action of atovaquone on Plasmodium and Pneumocystis is the inhibition of mitochondrial complex III (Birth et al. 2014; Biagini et al. 2008). Given the importance of mitochondrial energy metabolism in thyroid cancer (Lee et al. 2015; Johnson et al. 2015), we investigated whether atovaquone also targets thyroid cancer through inhibiting complex III and thereafter mitochondrial respiration. We measured the oxygen consumption rate (OCR) as an indicator of mitochondrial respiration in thyroid cancer cells after atovaquone treatment. We found that both FTC133 and 8505C cells treated with atovaquone had a significantly reduced baseline OCR and were non-responsive to uncoupling of mitochondrial oxidative phosphorylation via FCCP (Fig. 2a), indicating inhibition of mitochondrial respiration by atovaquone in thyroid cancer cells. Interestingly, we found that atovaquone slightly decreased the rate of glycolysis in 8505C and FTC-133 cells (Supplementary Fig. S1A and B). In addition, the degree of inhibition is less on the rate of glycolysis than mitochondrial respiration, suggesting that ATP reduction may be largely due to the mitochondrial respiration inhibition by atovaquone. Importantly, time course analysis found that glycolytic rate was significantly increased at 2 h and 6 h treatment but then slightly decreased at 12 h and 24 h treatment of 20 μM atovaquone (Supplementary Fig. S1C and D). This suggests that mitochondrial respiration inhibition is the primary effect and glycolysis level change is the secondary effect as a compensatory energy source in atovaquone-treated cells.
Atovaquone inhibits mitochondrial respiration in thyroid cancer cells. Atovaquone inhibits mitochondrial respiration (a), decreases mitochondrial complex III activity (b), and intracellular ATP levels (c) in 8505C and FTC133 cells. OLI (Oligiomycin), FCCP and A&R (Antimycin and rotenone) were injected into the wells at the time indicated by arrows. d Atovaquone dose-dependently and time-dependently inhibits phosphorylation of STAT3 in thyroid cancer cells. e Atovaquone decrease mitochondrial p-STAT3 but not nuclear p-STAT3 level in 8505C cells. 20 μM atovaquone was used for time-course analysis. *p < 0.05, compared to control
Consistent with the previous reports (Birth et al. 2014; Biagini et al. 2008), atovaquone significantly inhibited activity of mitochondrial complex III in thyroid cancer cells (Fig. 2b). Atovaquone also decreased cellular ATP levels in FCT133 and 8505C cells (Fig. 2c). These results demonstrate that atovaquone acts on thyroid cancer via inhibiting mitochondrial respiration.
Interestingly, we observed the time-dependent and concentration-dependent decrease of STAT3 phosphorylation in thyroid cancer cells exposed to atovaquone (Fig. 3d and supplementary Fig. S2A and B). In addition, Atovaquone inhibits mitochondrial but not nuclear p-STAT3 (Fig. 2e and supplementary Fig. S2C). This result is consistent with the Wegrzyn et al.’s work that mitochondrial STAT3 is important for cellular mitochondrial functions (Wegrzyn et al. 2009). JAK, Src and MEK have been shown to regulate STAT-3 phosphorylation in various types of cells (Xiang et al. 2016; Song et al. 2003; Gkouveris et al. 2014). We further analyzed the upstream modulators of STAT-3 phosphorylation by investigating these signaling pathways in 8505C cells after atovaquone treatment. However, we did not observe significant difference on p-JAK2 (Y570), p-Src(Y416) and p-Erk(T202/T204) in control and atovaquone-treated cells (Supplementary Fig. S3), demonstrating that atovaquone inhibits STAT3 activation in thyroid cancer cells in a JAK/Src/MEK independent manner.
Atovaquone is ineffective in mitochondrial respiration-deficient 8505C ρ0 cells. a mRNA levels of two mitochondrial genome-encoded genes: MT-ND6 and MT-CO2, are 105 lower whereas nuclear genome-encoded gene SDHA is not affected in the ρ0 CML cells. Fold-expression change of the transcript abundance in 8505C ρ0 is first normalized to β-actin expression and then calculated relative to its wildtype counterparts. b 8505C ρ0 cells have minimal basal as well as spare OCR. c 8505C ρ0 cells have significantly slower growth rate than wildtype cells. Equal number of cells was seeded on day 1 and cell density was monitored every day for 6 days. The inhibitory effects of atovaquone on migration (d), apoptosis (e) and STAT3 phosphorylation (f) are reversed in 8505C ρ0 cells. *p < 0.05, compared to wildtype
Atovaquone is ineffective in mitochondrial respiratory chain-deficient 8505C ρ0 cells
To further confirm the inhibition of mitochondrial respiration as the mechanism of the action of atovaquone in thyroid cancer, we generated mitochondrial respiratory chain-deficient ρ0 cells. ρ0 cells are characterized by lacking mitochondrial DNA and thus some critical catalytic subunits of respiratory chain enzymes leading to deficient mitochondrial-respiration (Chandel and Schumacker 1999). We tried to obtain both 8505C and FTC133 ρ0 cells by culturing them in the presence of ethidium bromide as previously reported (Hashiguchi and Zhang-Akiyama 2009). However, 8505C but not FTC133 is able to generate ρ0 cells as shown by the 105 fold lower mitochondrial DNA and minimal mitochondrial respiration (Fig. 3a, b), suggesting that FTC113 is more reliant on mitochondrial respiration compared to 8505C cells. The basal and reserved glycolysis rate is marginally elevated in ρ0 compared to wildtype cells (Supplementary Fig. S4A and B). We further measured ATP level in wildtype and ρ0 cells. Consistent with the previous report (Xiao et al. 2016), the absolute ATP levels are remarkably less in ρ0 compared to wildtype (Supplementary Fig. S4C). In addition, 8505C ρ0 cells also demonstrate a significantly reduced proliferation and migration, indicative of the importance of mitochondrial respiration to thyroid cancer cell growth and migration (Fig. 3c, d). The remarkably less ATP levels in ρ0 cells correlates well with our findings on the minimal growth and migration in ρ0 cells (Fig. 3c, d). Notably, atovaquone became ineffective in inhibiting migration and inducing apoptosis in 8505C ρ0 cells (Fig. 3d, e), confirming that atovaquone acts on thyroid cancer via mitochondrial respiration inhibition. We further found that the inhibitory effect of atovaquone on STAT3 phosphorylation was also abolished in 8505C ρ0 cells (Fig. 3f and supplementary Fig. S5).
Atovaquone augments doxorubicin’s efficacy in thyroid cancer via inhibiting mitochondrial respiration and STAT3
Doxorubicin has been used most often as cytotoxic chemotherapy in anaplastic thyroid cancer. It causes cancer cell death through inducing torsional stress and nucleosome destabilization, leading to inhibition of DNA and RNA synthesis (Yang et al. 2014). We next investigate whether targeting different sites of action by combing doxorubicin and atovaquone can achieve better efficacy than single drug alone in thyroid cancer cells. We found that atovaquone significantly augments anti-proliferation and pro-apoptotic effects of doxorubicin in FTC133 and 8505C cells (Fig. 4a, b). Cardiomyocytes are known to be sensitive to alterations on mitochondrial metabolism (Chistiakov et al. 2017). We therefore investigated the toxicity of atovaquone and its combination with doxorubicin in cardiomyocytes using the same concentrations that were used in thyroid cancer cells. Similarly, atovaquone at 5, 10 and 20 μM induces apoptosis in H9c2 cardiomyoctyes in a dose-dependent manner (Supplementary Fig. S6A). Doxorubicin at 50 nM slightly induces H9c2 apoptosis (Supplementary Fig. S6B). Notably, the combination of doxorubicin and atovaquone does not induce more apoptosis than atovaquone or doxorubicin alone in H9c2 cells (Supplementary Fig. S6B), which is different to thyroid cancer cells. This demonstrates that combination of doxorubicin with atovaquone exhibits preferential toxicity to thyroid cancer cells compared to cardiomyocytes.
Synergistic inhibition by atovaquone and doxorubicin in thyroid cancer cells. Atovaquone at 5 μM significantly enhances anti-proliferative (a) and pro-apoptotic (b) effects of doxorubicin (50 nM). The effects atovaquone (5 μM) and doxorubicin (50 nM) alone, and combination of atovaquone and doxorubicin in basal OCR (c), maximal OCR (d), cellular ATP production (e) and STAT3 phosphorylation (f) in 8505C and FTC133 cells. Atovaquone but not doxorubicin inhibits all of them. Basal OCR is calculated as the first three measurement. Maximal OCR is calculated as the three measurement after FCCP injection. *p < 0.05, compared to control
Unlike atovaquone, doxorubicin did not affect mitochondrial respiration, ATP production and STAT3 phosphorylation in FTC133 and 8505C cells (Fig. 4c, d, e and f). We did not observe further reduction on mitochondrial respiration, ATP levels and STAT3 phosphorylation in thyroid cancer cells neither (Fig. 4c, d, e and f and supplementary Fig. S7), suggesting that the synergy between doxorubicin and atovaquone are due to concurrently inhibiting DNA/RNA synthesis, mitochondrial respiration and STAT3 phosphorylation.
Discussion
Thyroid carcinogenesis and progression involves multiple and complicated genetic and epigenetic alterations, including somatic mutations, alteration/dysregulation in gene expression patterns and methylation, leading to the activation of various oncogenic pathways (Nikiforov and Nikiforova 2011). Molecular therapeutic strategy by specific inhibition of oncogenic pathways did not provide a significant survival benefit in advanced progressive thyroid cancer (Jin et al. 2016). An alternative approach would be to focus on targeting what is common rather than on what is divergent between different molecular subclasses of thyroid cancer. In this work, we demonstrate that atovaquone effectively targets thyroid cancer cells and acts cooperatively with doxorubicin, via suppressing mitochondrial complex III and subsequent mitochondrial energy crisis and STAT3 inhibition.
We firstly demonstrated that atovaquone as a single drug alone effectively inhibited proliferation and migration, and induced apoptosis, with IC50 at 7~10 μM (Fig. 1). The anti-proliferative and pro-apoptotic effects of atovaquone at micromolar concentration has been reported in breast and colon cancers, and leukemia (Xiang et al. 2016; Fiorillo et al. 2016; Ashton et al. 2016). Our results are consistent with previous studies and support the anti-cancer properties of atovaquone. In addition, we are the first to demonstrate that atovaquone inhibits thyroid cancer cell migration (Fig. 1c, d), suggesting the possible negative role of atovaquone in cancer metastasis. When patients with immunodeficiency virus infection received atovaquone suspension with food at the standard regimen of 750 mg twice daily, the average steady-state plasma concentration and minimum plasma concentration was ~ 21.0 μg/mL and ~16 μg/mL, respectively (Falloon et al. 1999). This indicates that the IC50 of atovaquone are clinically achievable in thyroid cancer patients.
Although doxorubicin remain a mainstay therapy for various cancers, it causes cardiotoxicity (Qureshi et al. 2015). Our findings demonstrate that combination of atovaquone and doxorubicin at the same concentrations that acts synergistically in thyroid cancer cells does not further decrease viability of cardiomyocytes compared to single drug alone (Fig. 4 and supplementary Fig. S6), suggesting that combination therapy achieves greater efficacy in targeting cancer cells while minimizes toxicity of normal cells. Importantly, atovaquone at similar concentrations is not toxic to fibroblast cells (Fiorillo et al. 2016). Besides diarrhea, nausea, rashes and fatigue, atovaquone suspension is also well tolerated (Falloon et al. 1999). This is supported by the previous work that the administration of oral atovaquone to mice inhibited tumor growth and prolonged survival in a murine model of multiple myeloma without significant toxicity to mice (Xiang et al. 2016). It is noted that atovaquone administration is associated with improved outcomes in patient with leukemia (Xiang et al. 2016). Our in vitro preclinical findings together with published studies demonstrate that atovaquone is well tolerated and has anti-cancer activities not only in pre-clinical models but also in patients. Future studies in patients derived xenograft models and human clinical trials are warranted to demonstrate the translational potential of atovaquone in thyroid cancer treatment.
We next demonstrate that the mechanism of the action of atovaquone in thyroid cancer is the inhibition of mitochondrial complex III, leading to blockage of mitochondrial respiration and ATP production (Fig. 2a, b and c). This is further demonstrated by the rescue of atovaquone’s effects in mitochondrial respiration deficient-8505C ρ0 cells (Fig. 3a, b, c, d and e). Our findings and other work (Srivastava et al. 1997; Fiorillo et al. 2016; Birth et al. 2014) suggest that the common mechanisms of atovaquone’s action on both human cancer cells and parasites are inhibition of mitochondrial respiration, disruption of mitochondrial membrane potential and blockage of mitochondrial functions. However, our work cannot exclude the possibility that atovaquone might also inhibit thyroid cancer growth through an effect on TCA cycle substrate level phosphorylation. Interestingly, other anti-helminthic drugs killing parasites by targeting mitochondrial electron transport chain, such as pyrvinium, have also been shown to have potent anti-cancer activities (Xiao et al. 2016; Momtazi-Borojeni et al. 2017; Xiang et al. 2015). Targeting mitochondrial metabolism has been recently revealed to be useful in the management of various cancers, including thyroid cancer (Lee et al. 2015; Johnson et al. 2015). Our findings on the remarkably compromised growth and migration in mitochondrial respiration deficient ρ0 cells by specifically depleting mitochondrial DNA (Fig. 3a, b, c and d) provides the fundamental evidences to show that mitochondrial metabolism is essential for aggressive thyroid cancer.
The ability of atovaquone on STAT3 inhibition has been recently reported in leukemia cells (Xiang et al. 2016). We demonstrate that atovaquone inhibits STAT3 activation in a JAK/Src/Erk-independent manner (Fig. 2e). In contrast, our study is consistent with and further extend the previous work that STAT3 phosphorylation is inhibited by atovaquone in thyroid cancer cells as a consequence of mitochondrial dysfunction (Figs. 2d and 3f). Wegrzyn et al.’s work demonstrates that STAT3 −/− cells have significant deficits of Complex I and II activities, suggesting that STAT3 is the upstream modulator of proper mitochondrial functions. Our findings are consistent with Wegrzyn et al.’s work on the point that STAT3 and mitochondrial functions are closely associated. Our finding further extends Wegrzyn et al.’s work that STAT3 deactivation can also be the consequence of mitochondrial dysfunction, which is supported by Yasuo et al.’s work that mitochondrial dysfunction leads to inhibition of STAT3 phosphorylation (Harada et al. 2012). In addition, Qureshi et al.;‘s finding that STAT3 activation protects cardiomyocytes from apoptosis induced by doxorubicin (Qureshi et al. 2015) is similar to our finding, demonstrating that STAT3 activation may be a survival mechanism in response to various stress conditions in cells.
In conclusion, our work demonstrate that an antheliminthic atovaquone inhibits mitochondrial III and subsequent mitochondrial respiration and STAT3 Tyr705 activation, leading to potent inhibition of growth, survival and migration in thyroid cancer cells. Our work also highlights targeting mitochondrial metabolism as a useful sensitizing strategy for thyroid cancer response to chemotherapy.
References
Ashton TM, Fokas E, Kunz-Schughart LA, Folkes LK, Anbalagan S, Huether M, Kelly CJ, Pirovano G, Buffa FM, Hammond EM, Stratford M, Muschel RJ, Higgins GS, McKenna WG (2016) The anti-malarial atovaquone increases radiosensitivity by alleviating tumour hypoxia. Nat Commun 7:12308. https://doi.org/10.1038/ncomms12308
Biagini GA, Fisher N, Berry N, Stocks PA, Meunier B, Williams DP, Bonar-Law R, Bray PG, Owen A, O'Neill PM, Ward SA (2008) Acridinediones: selective and potent inhibitors of the malaria parasite mitochondrial bc1 complex. Mol Pharmacol 73(5):1347–1355. https://doi.org/10.1124/mol.108.045120
Birth D, Kao WC, Hunte C (2014) Structural analysis of atovaquone-inhibited cytochrome bc1 complex reveals the molecular basis of antimalarial drug action. Nat Commun 5:4029. https://doi.org/10.1038/ncomms5029
Cabanillas ME, Zafereo M, Gunn GB, Ferrarotto R (2016) Anaplastic thyroid carcinoma: treatment in the age of molecular targeted therapy. Journal of oncology practice / American Society of Clinical Oncology 12(6):511–518. https://doi.org/10.1200/JOP.2016.012013.
Chandel NS, Schumacker PT (1999) Cells depleted of mitochondrial DNA (rho0) yield insight into physiological mechanisms. FEBS Lett 454(3):173–176. https://doi.org/10.1016/S0014-5793(99)00783-8 [pii]
Chistiakov DA, Shkurat TP, Melnichenko AA, Grechko AV, Orekhov AN (2017) The role of mitochondrial dysfunction in cardiovascular disease: a brief review. Ann Med 50:1–7. https://doi.org/10.1080/07853890.2017.1417631
Falloon J, Sargent S, Piscitelli SC, Bechtel C, LaFon SW, Sadler B et al (1999) Atovaquone suspension in HIV-infected volunteers: pharmacokinetics, pharmacodynamics, and TMP-SMX interaction study. Pharmacotherapy 19(9):1050–1056
Fiorillo M, Lamb R, Tanowitz HB, Mutti L, Krstic-Demonacos M, Cappello AR, Martinez-Outschoorn UE, Sotgia F, Lisanti MP (2016) Repurposing atovaquone: targeting mitochondrial complex III and OXPHOS to eradicate cancer stem cells. Oncotarget 7(23):34084–34099. https://doi.org/10.18632/oncotarget.9122.
Fry M, Pudney M (1992) Site of action of the antimalarial hydroxynaphthoquinone, 2-[trans-4-(4′-chlorophenyl) cyclohexyl]-3-hydroxy-1,4-naphthoquinone (566C80). Biochem Pharmacol 43(7):1545–1553
Gkouveris I, Nikitakis N, Karanikou M, Rassidakis G, Sklavounou A (2014) Erk1/2 activation and modulation of STAT3 signaling in oral cancer. Oncol Rep 32(5):2175–2182. https://doi.org/10.3892/or.2014.3440
Harada Y, Ishii I, Hatake K, Kasahara T (2012) Pyrvinium pamoate inhibits proliferation of myeloma/erythroleukemia cells by suppressing mitochondrial respiratory complex I and STAT3. Cancer Lett 319(1):83–88. https://doi.org/10.1016/j.canlet.2011.12.034 S0304-3835(11)00793-2 [pii]
Hashiguchi K, Zhang-Akiyama QM (2009) Establishment of human cell lines lacking mitochondrial DNA. Methods Mol Biol 554:383–391. https://doi.org/10.1007/978-1-59745-521-3_23
Jin S, Borkhuu O, Bao W, Yang YT (2016) Signaling pathways in thyroid Cancer and their therapeutic implications. Journal of clinical medicine research 8(4):284–296. https://doi.org/10.14740/jocmr2480w
Johnson JM, Lai SY, Cotzia P, Cognetti D, Luginbuhl A, Pribitkin EA, Zhan T, Mollaee M, Domingo-Vidal M, Chen Y, Campling B, Bar-Ad V, Birbe R, Tuluc M, Martinez Outschoorn U, Curry J (2015) Mitochondrial metabolism as a treatment target in anaplastic thyroid Cancer. Semin Oncol 42(6):915–922. https://doi.org/10.1053/j.seminoncol.2015.09.025
Kondo T, Ezzat S, Asa SL (2006) Pathogenetic mechanisms in thyroid follicular-cell neoplasia. Nat Rev Cancer 6(4):292–306. https://doi.org/10.1038/nrc1836
Lee J, Chang JY, Kang YE, Yi S, Lee MH, Joung KH, Kim KS, Shong M (2015) Mitochondrial energy metabolism and thyroid cancers. Endocrinol Metab 30(2):117–123. https://doi.org/10.3803/EnM.2015.30.2.117
Momtazi-Borojeni AA, Abdollahi E, Ghasemi F, Caraglia M, Sahebkar A (2017) The novel role of pyrvinium in cancer therapy. J Cell Physiol 233:2871–2881. https://doi.org/10.1002/jcp.26006
Nikiforov YE, Nikiforova MN (2011) Molecular genetics and diagnosis of thyroid cancer. Nat Rev Endocrinol 7(10):569–580. https://doi.org/10.1038/nrendo.2011.142
Qureshi R, Yildirim O, Gasser A, Basmadjian C, Zhao Q, Wilmet JP, Désaubry L, Nebigil CG (2015) FL3, a synthetic Flavagline and ligand of Prohibitins, protects cardiomyocytes via STAT3 from doxorubicin toxicity. PLoS One 10(11):e0141826. https://doi.org/10.1371/journal.pone.0141826
Saiselet M, Floor S, Tarabichi M, Dom G, Hebrant A, van Staveren WC et al (2012) Thyroid cancer cell lines: an overview. Front Endocrinol 3:133. https://doi.org/10.3389/fendo.2012.00133
Sipos JA, Mazzaferri EL (2010) Thyroid cancer epidemiology and prognostic variables. Clin Oncol (R Coll Radiol) 22(6):395–404. https://doi.org/10.1016/j.clon.2010.05.004
Song L, Turkson J, Karras JG, Jove R, Haura EB (2003) Activation of Stat3 by receptor tyrosine kinases and cytokines regulates survival in human non-small cell carcinoma cells. Oncogene 22(27):4150–4165. https://doi.org/10.1038/sj.onc.1206479
Srivastava IK, Rottenberg H, Vaidya AB (1997) Atovaquone, a broad spectrum antiparasitic drug, collapses mitochondrial membrane potential in a malarial parasite. J Biol Chem 272(7):3961–3966
Wegrzyn J, Potla R, Chwae YJ, Sepuri NB, Zhang Q, Koeck T et al (2009) Function of mitochondrial Stat3 in cellular respiration. Science 323(5915):793–797. https://doi.org/10.1126/science.1164551
Weinberg SE, Chandel NS (2015) Targeting mitochondria metabolism for cancer therapy. Nat Chem Biol 11(1):9–15. https://doi.org/10.1038/nchembio.1712
Xiang W, Cheong JK, Ang SH, Teo B, Xu P, Asari K et al (2015) Pyrvinium selectively targets blast phase-chronic myeloid leukemia through inhibition of mitochondrial respiration. Oncotarget 6(32):33769–33780. https://doi.org/10.18632/oncotarget.5615.
Xiang M, Kim H, Ho VT, Walker SR, Bar-Natan M, Anahtar M, Liu S, Toniolo PA, Kroll Y, Jones N, Giaccone ZT, Heppler LN, Ye DQ, Marineau JJ, Shaw D, Bradner JE, Blonquist T, Neuberg D, Hetz C, Stone RM, Soiffer RJ, Frank DA (2016) Gene expression-based discovery of atovaquone as a STAT3 inhibitor and anticancer agent. Blood 128(14):1845–1853. https://doi.org/10.1182/blood-2015-07-660506
Xiao M, Zhang L, Zhou Y, Rajoria P, Wang C (2016) Pyrvinium selectively induces apoptosis of lymphoma cells through impairing mitochondrial functions and JAK2/STAT5. Biochem Biophys Res Commun 469(3):716–722. https://doi.org/10.1016/j.bbrc.2015.12.059
Yang F, Teves SS, Kemp CJ, Henikoff S (2014) Doxorubicin, DNA torsion, and chromatin dynamics. Biochim Biophys Acta 1845(1):84–89. https://doi.org/10.1016/j.bbcan.2013.12.002
Zhou J, Duan L, Chen H, Ren X, Zhang Z, Zhou F, Liu J, Pei D, Ding K (2009) Atovaquone derivatives as potent cytotoxic and apoptosis inducing agents. Bioorg Med Chem Lett 19(17):5091–5094. https://doi.org/10.1016/j.bmcl.2009.07.044
Acknowledgements
This work was supported by the Research Grant from the Health Department of Guangxi Zhuang Autonomous Region (Grant No. 216).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare no conflicts of interests.
Electronic supplementary material
ESM 1
(DOC 369 kb)
Rights and permissions
About this article
Cite this article
Lv, Z., Yan, X., Lu, L. et al. Atovaquone enhances doxorubicin’s efficacy via inhibiting mitochondrial respiration and STAT3 in aggressive thyroid cancer. J Bioenerg Biomembr 50, 263–270 (2018). https://doi.org/10.1007/s10863-018-9755-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10863-018-9755-y