Abstract
The theory of shapes, as proposed by David Kendall, is concerned with sets of labeled points in the Euclidean space \(\mathbb {R}^d\) that define a shape regardless of translations, rotations and dilatations. We present here a method that extends the theory of shapes, where, in this case, we use the term generalized shape for structures of unlabeled points. By using the distribution of distances between the points in a set we are able to define the existence of generalized shapes and to infer the computation of the correspondences and the orthogonal transformation between two elements of the same generalized shape equivalence class. This study is oriented to solve the registration of large set of landmarks or point sets derived from medical images but may be employed in other fields such as computer vision or biological morphometry.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
A variety of objects can be represented as point sets in \(\mathbb {R}^d\), where \(d\) is usually 2 or 3. One is often presented with the problem of deciding whether two of these point sets, and/or the corresponding underlying objects or manifolds, represent the same geometric structure or not. In the case of correspondence, we are interested in the transformation that relates one form to another. A connected fundamental question is: what conditions must a set of points verify in order to faithfully represent an object? Another question is: what kind of similarity we want to achieve between the objects, i.e. rigid or non rigid? The easiest hypothesis is when the correspondences are known, there is a small amount of noise in the point set representation and the transformation is rigid. The closed form solution was given by Shonemann in [28].
Each set of points that we want to register is called shape and the registration task is to find a way to align two or more shapes. The Procrustes method of shape comparison arose as a way of superimposing point sets with known correspondence. A modern and complete study of the Procrustean metric and shape manifolds was presented by Kendall in [17] and was further extended by the author in [18].
The mathematical aspects of the theory of shapes are most of the times not practical in the implementation of algorithms for object recognition and matching. Point-based methods to register surfaces which brings relatively dense point sets into correspondence have become popular with the introduction of the iterative closest point (ICP) method [4].
This algorithm is designed to work with different representations of surface data: point sets, line segment sets (polylines), implicit surface, parametric curves, triangle sets, implicit surfaces and parametric surfaces. For medical image registration the most relevant representations are likely to be point sets and triangle sets, as algorithms for delineating these features from medical images are widely available.
The algorithm is able to register a data shape \(P\) with \(N_p\) points to a model shape \(X\) with \(N_x\) primitives. For each element of \(P\), the algorithm first identify the closest point on \(X\) shape, then finds the least square rigid-body transformation relating these pairs of point sets. The algorithm then redetermines the closest point set and continues until finds the local minimum match between the two surfaces, as determined by some tolerance threshold.
The optimization can be accelerated by keeping track of the solutions at each iteration. As the algorithm iterates to the local minimum closest to the starting position, it may not find the correct match. The solution proposed in [4] is to start the algorithm multiple times, each with a different estimate of the rotation alignment, and choose the minimum of the minimum obtained.
In the context of the determination of correspondences and transformation in successive steps several authors built extensions or generalizations of this approach. While retaining the basic iterative principle of ICP, Rangarajan et. al in [27] formulated a variant of the Procrustes distance between two discrete sets of points in which the correspondence maps are unknown a priori. Their algorithm alternates between calculating optimal rotations and determining correspondence maps. For every fixed rotation \(\mathbf {R}\), it computes the association matrix \(M\) between the two sets of points \(A =\left\{ A_i\right\} \) and \(B = \left\{ B_j\right\} \), minimizing the average of the square residuals \(\sum _{i,j}M_{ij}\left\| \mathbf {R}A_i - B_j\right\| \), under the soft constraint that \(M\) is indeed a measure of coupling, i.e the values of \(M\) elements are between 0 and 1, each row sums to 1, and the 1 value of an element \(M_{ij}\) means the perfect correspondence between \(A_i\) and \(B_j\). As is the case of the original ICP, this algorithm can also converge to a local rather than a global minimum, and the correspondence maps can still be discontinuous and distorting.
Memoli [23] provides theoretical exposition of a similar functional in the context of Gromov-Hausdorff distances between shapes. Ghosh et. al [14] used a similar framework with a smooth surface deformation mechanism together with closest-point maps to determine both correspondence maps and the transformations in an alternating iterative procedure. Their algorithm requires user initialization which may influence the outcome; the way correspondences are assigned can lead to a deformation mechanism that produces a distorting and/or discontinuous map between the two surfaces.
Shapiro and Brady [30] match feature points on the basis of consistent same-space distances by an eigen-analysis technique, following the inter-image distance-based matching technique of Scott and Longuett-Higgins [29]. The solution presented in [29] has a very elegant implementation founded on a well-conditioned eigen-vector solution which involved no iteration, but does not handle large rotations and may become unstable for some value of the parameters. Conversely, Shapiro et al. in [30] introduce a modal shape description to handle also the rotations and the instability, but their solution lacks of the formal proof and, in our tests, does not always provide the results we would have expected.
Boyer et al. [9] introduced the concept of continuous Procrustes distance and proved that it provides a true metric for two-dimensional surfaces derived from anatomical structures embedded in a three-dimensional space.
Jian and Vemuri in [16] reformulate the task of point set registration as the problem of aligning two Gaussian mixture models (GMM) such that a statistical discrepancy measure between the two corresponding mixtures is minimized. Another probabilistic approach that uses GMM and a closed form solution to establish the correspondences using the expectation maximization algorithm was given by Myronenko and Song in [24].
It is worth to mention the paper of Belongie et al. [3] that introduces the shape context concept used to measure the similarity between shapes in two steps: (1) solving for correspondences, (2) using the correspondences to estimate an aligning transform.
An efficient spectral solution to correspondence problems using pairwise constraints between candidate assignments was presented by [21].
These are only a few of the works dealing with the transformation - correspondence problem and, from the first to the last citation, they are all facing the same ’egg-chicken’ dilemma: find the correspondences to get the transformation or find the transformation to get the correspondences. The best solutions are of course those where the transformation allowed is non-rigid. In this case we may find perfect correspondences using a mathematical transformation, but one question arises: is it not true that if we allow for non-rigid correspondence we may align any two given objects, such as an apple and a pear or a mouse and an elephant? It is important to consider the applications and the input-output of the algorithms dealing with the correspondence-registration combination. The input of such algorithms may represent different pose of the same object or views of an object subject to a set of changes such as deformations. The data describing the input can be images or geometric features such as points or triangle meshes. The correspondence could be global in the case of an image dataset or local in the case of geometric features matching. The output of such algorithms gives the transformation that register the datasets on one hand and the correspondence or the measure of match on the other hand. If the input is given as a set of geometric features to be matched with another set of features, for instance in the point-set registration, once non-rigidity is allowed, there is an infinite number of ways to map one set onto another. The smoothness constraint is necessary because it discourages mapping that are too arbitrary. One of the simplest and most used measures is the space integral of the square of the second order derivatives of the mapping function. This leads to the thin-plate-spline (TPS) function. Introduced by Bookstein in [6] for surface registration in medical imaging and morphometry, and formally described by Wahba in [31] these functions are currently used by most non-rigid registration algorithms of point sets (see also [3, 13, 16, 24]). The TPS function is easy to compute and implement and it has the advantage to decouple the transformation into an affine part and a non-linear deformable part. In situations where there is no shearing and scaling we can constrain the affine transformation to a rigid one. Usually, all the non-rigid algorithms first find a common reference system of the two datasets, then proceed with the deformation of one dataset in order to fit the second dataset. Our main concern is that, once we introduce a non-rigid deformation, even if the initial alignment is not satisfactory, the algorithm will yield a very good alignment because of the freedom of the deformation. By keeping trace of the rigid alignment, as in the TPS case, we may assert the goodness of the alignment and the usefulness of the registration. For instance, Chui and Rangarajan in [13], implemented the registration with a deterministic annealing scheme to optimize the correspondence matrix by updating the transformation parameters. The algorithm is clearly attempting to solve the matching problem using a coarse to fine approach. Global structures such as the center of mass and principal axis are first matched, followed by the non-rigid matching of local structures. This means that the rigid alignment will be given by the alignment of the geometric moments of the two data sets considered. This solution has a number of drawbacks such as the sensibility to outliers, noise, occlusions but also to the deformation which is our main concern.
Lipman and Funkhouser in [22] used a different approach for the computation of correspondences of approximately and/or partially isometric surfaces. They employed the Mobius transformation and random sampling to vote for the best correspondences for each triplets of points extracted from each of the two datasets to be registered. In their approach, the datasets given as 3D meshes, which are genus-0 surfaces, were conformally mapped to a sphere.
All of these approaches work well on synthetic models but our preoccupation is how useful are they in practical applications. For instance, in medical image applications, where there are multiple acquisitions of the same anatomical area, if we use different sensors (e.g. computed tomography CT and ultrasound US) and we want to register some segmented surfaces from these datasets, how can an algorithm distinguish between noise and deformation, or how can a deformable algorithm take into account the outliers? Since the deformation is modeled by a mathematical model, such as TPS, we need to be sure of the correct rigid alignment, which is, in most of the cases, not guaranteed. There is also a concern about the computational cost of these algorithm and the numerous parameters that must be solved/known in advance.
In the following, we propose a simplification of the hypothesis of the problem in order to have a solution that is completely controllable by the theoretical development. The simplified hypothesis is when we want to register two sets of points represented in different coordinate systems without knowing the homologies; the theoretical part consider the ideal case when no noise is present, while in the practical implementation we study how robust is this assumption in the presence of noise.
2 Overview of Approach
The departing point of our generalized shape method is the the work of Kendall [17] who introduces the theory of shapes. The theory of shapes is concerned about of k labelled points \(\mathbf {x}_\mathbf {1},\ldots ,\mathbf {x}_\mathbf {k}\) or \(k-ad\) in an Euclidean space \(\mathbb {R}^d\), where \(k\ge 2\). Normally, the centroid of the \(k\) points will serve as origin, and the scale will be such that the sum of the squared distance of the points from that origin will be equal to unity. Informally, the shape is ’what is left when the differences which can be attributed to translations, rotations and dilatations have been quotiented out’.
By ignoring the translation, scaling and rotation, the shape space denoted by the symbol \({\varSigma }^k_d\) has the dimension:
This is because the constraints on the total number of degrees of freedom (DOF) are reduced accordingly by the DOF of the translations \(d\), rotations \(\frac{1}{2}d(d-1)\) and scaling. Eq. (1) holds provided that \(k \ge d+1\).
The author introduced a norm and a metric topology on the shape space deriving the shape-manifolds. The distance between two \(k-ad\) on the shape manifold \({\varSigma }^k_d\) is called the procrustes distance.
Starting from the description of the shape by using a set of points, a natural extension of this theory, that we introduce in this paper, is to quotient out also the effect of the labeling of the points. In this work we keep the significance of the scaling. Accordingly, in our approach the definition of the shape or generalized shape becomes: ’a generalized shape is what is left when the differences which can be attributed to translations, rotations and permutations have been quotiented out’.
In this paper we study the implications of such an assumption, we build the theoretical basis and we give some practical results. The invariance of a point set under the action of rotations, translations and permutations will be studied in accordance with the set of distances between each possible pair of points in the set.
The applications we are interested on are in the medical field, especially in the operating room navigation scenario, where closed form solutions of registration algorithms are required to have a clinical validation. Most of the solutions currently employed in this field are based on iterative schemes that alternates correspondence and registration steps or algorithms that use an optimization approach, as shown by Yaniv and Cleary [33]. The solution we propose overcomes the limitations of these methods since it solves the correspondence problem before the registration took place and infers the global registration from the correspondences found.
The idea of using a global descriptor, invariant to the isometric transformation was already introduced in the work of Osada et al. [25], but the authors used the distribution of distances for object classification. In our work, we extend the use of this invariant descriptor under the action of isometries associated with the Euclidean metric and we define a class of equivalent shapes based on this descriptor, in order to recover the correspondences and the registration transformation between two shapes belonging to the same generalized shape class. We therefore study the condition of existence of such an equivalent class and the algorithmic modalities to obtain the isometric transformation and the point to point correspondences between two point sets belonging to the same generalized shape.
The following sections describe our approach. Sects. 3 and 4 presents some known concepts and preliminary results that introduce our method. The core of our approach is given in Sect. 5, where a method to infer the computation of the isometric transformation between two generalized shapes is described, and Sect. 6, where a method to compute the correspondences between two sets of points that represent the same generalized shape is introduced.
Based on this findings, an experimental evaluation for a dataset of pulmonary landmark point derived from 4D CT image data is presented in Sect. 7.
3 Theoretical Foundations
Let us fix a coordinate system in \(\mathbb {R}^d\).
Definition 1
If we denote by \(O(d)\) the group of the \( d \times d\) orthogonal matrices, and by \(S_k\) the group of all permutations of \(\left\{ 1,\ldots ,k\right\} \), a set of \(k\) points \(X = \left\{ \mathbf {x}_\mathbf {1},\ldots ,\mathbf {x}_\mathbf {k}\right\} , \subset \mathbb {R}^d\) is equivalent to \(Y = \left\{ \mathbf {y}_\mathbf {1},\ldots ,\mathbf {y}_\mathbf {k}\right\} \subset \mathbb {R}^d\), in the sense of generalized shape, if and only if there exists \(R \in O(d)\), a vector \(\mathbf {t} \in \mathbb {R}^d\) and a permutation \(\pi \in S_k\) such that:
Notice that the introduced notion is an equivalence relation.
Definition 2
We write \(\left[ X\right] \) the equivalence class of \(X\) and we call it generalized shape.
For any two vectors \(\mathbf {x} = (x^1,\ldots ,x^d)\) and \(\mathbf {y} = (y^1,\ldots ,y^d)\) in \(\mathbb {R}^d\) we denote by \(\left\langle \mathbf {x}, \mathbf {y}\right\rangle = \sum _{i=1}^d x^iy^i\) their scalar product and by \(\left\| \mathbf {x} - \mathbf {y} \right\| = \left\langle \mathbf {x} - \mathbf {y} , \mathbf {x} - \mathbf {y} \right\rangle ^ {1/2}\) the Euclidean distance between them. The Euclidean distance defines a metric space called the Euclidean space.
Definition 3
For any set \(X = \left\{ \mathbf {x}_\mathbf {1},\ldots ,\mathbf {x}_\mathbf {k}\right\} \) of vectors in \(\mathbb {R}^d\), the center of mass is given by the vector \(\bar{\mathbf {x}}= \frac{1}{k}(\mathbf {x}_\mathbf {1} +\ldots +\mathbf {x}_\mathbf {k})\). The set \(\bar{X} = \left\{ \mathbf {x}_\mathbf {1} - \bar{\mathbf {x}},\ldots ,\mathbf {x}_\mathbf {k} - \bar{\mathbf {x}} \right\} \) is called the centered coordinates.
In the following we shall use the notation \(X\) to represent a set of \(k\) points \(\mathbf {x}_\mathbf {1},\ldots ,\mathbf {x}_\mathbf {k} \in \mathbb {R}^d\) describing a generalized shape and, in the same time, a \(d \times k\) matrix with columns \(\mathbf {x}_\mathbf {1},\ldots ,\mathbf {x}_\mathbf {k}\).
If we sum over \(i\) and divide by \(k\) equation (2) we have: \(R \frac{1}{k}\sum _{i=1}^k\mathbf {x}_\mathbf {i} + \mathbf {t} = \frac{1}{k}\sum _{i=1}^k\mathbf {y}_\mathbf {i}\), therefore we can express \(\mathbf {t}\) as \(\bar{\mathbf {y}} - R \bar{\mathbf {x}}\) so we can rewrite equation (2) as \(R \bar{X} = \bar{Y}_{\pi }\), where \(\bar{Y}_{\pi } = \left\{ \mathbf {y}_{\varvec{\pi }(\mathbf{1})} - \bar{\mathbf {y}},\ldots ,\mathbf {y}_{\varvec{\pi }(\mathbf{k})}-\bar{\mathbf {y}}\right\} \).
This formula shows that the only important transformations in the generalized shape definition are the orthogonal transformation, or the rotation, \(R\) and the permutation \(\pi \).
Definition 4
Given \( X =\left\{ \mathbf {x}_\mathbf {1},\ldots ,\mathbf {x}_\mathbf {k}\right\} \subset \mathbb {R}^d\) and \(Y =\left\{ \mathbf {y}_\mathbf {1},\ldots ,\mathbf {y}_\mathbf {k}\right\} \) \(\subset \mathbb {R}^d\), we define the distance between them as:
that is the minimum of the \(L_p\) norm over all the permutations of elements in \(Y\). This distance ranges from \(d_1\) to \(d_\infty \):
Observe that, since \(S_k\) is finite, it makes sense to define (3) as a minimum rather than as an infimum.
We use this definition when the correspondences between the points are not known and the computation of the registration transformation is not required (Sect. 6).
The Hausdorff distance is another measure of distance useful when the correspondences are not known. Unlike the previous distance measure, this distance may be employed only when the registration transformation is known.
Definition 5
The Hausdorff distance is given by:
This distance measures the worst-case deviation between the two point sets, while the \(L_p\) norm measures only average deviation. The Hausdorff distance will be employed in our experimental evaluation (see Sect. 7).
Definition 6
We say that two shapes \(X\) and \(Y\), embedded in the Euclidean space \(\mathbb {R}^d\), are isometric when there exists a bijective mapping \({\varPhi }: X \rightarrow Y\) such that \( d(\mathbf {x}_\mathbf {i}, \mathbf {x}_\mathbf {j}) = d({\varPhi }(\mathbf {x}_\mathbf {i}), {\varPhi }(\mathbf {x}_\mathbf {j})\) for all \(x_i, x_j \in X\). Such \({\varPhi }\) is an isometry between \(X\) and \(Y\).
In the Euclidean space the isometry group, which are the self-isometries with the function composition operator, is the Euclidean group of rotations, translation and reflections. Since in this work we focus on the extrinsic geometry, we shall use interchangeably the terms isometry and Euclidean transformation. Some of the possible extensions of our work, as suggested in Section 8, could handle also the intrinsic geometry and therefore inelastic deformations.
Furthermore, for practical reasons, we use a natural relaxation of isometry or the almost isometry: a function \({\varPhi }': X \rightarrow Y\) is said an almost isometry if there exists some \(\epsilon \ge 0\) such that \(\left| d(\Phi '(\mathbf {x_i}), \Phi '(\mathbf {x_j}) - d(\mathbf {x_i}, \mathbf {x_j})\right| \le \epsilon \), for all \(x_i, x_j \in X\).
We are interested on whether this isometry exists and how to find it. As the generalized shapes are given as an equivalence class modulo rotations, translation and permutations, the isometry, in case exists, is given by the composition of the three functions. Since it is clear that rotations and translations are always isometries, we are wandering in which case the permutation still lead to isometry. To do this, we shall work directly with the distance distributions between pairwise points in each set.
Definition 7
Given a set of points \(\mathbf {x}_\mathbf {1},\ldots ,\mathbf {x}_\mathbf {k} \in \mathbb {R}^d\), we call distance distribution matrix the \(k \times k\) matrix whose entries are given by the pairwise distances \(D^X_{i,j} = \left\| \mathbf {x}_\mathbf {i} - \mathbf {x}_\mathbf {j}\right\| \)
A reduced form of the distance distribution matrix is the distance distribution vector.
Definition 8
Given a set of points \(\mathbf {x}_\mathbf {1},\ldots ,\mathbf {x}_\mathbf {k} \in \mathbb {R}^d\), we call distance distribution vector the \( \mathbb {R}^{\left( {\begin{array}{c}k\\ 2\end{array}}\right) }\) vector whose entries are given by the pairwise distances:
where \(V^X_{i,j} = \left\| \mathbf {x}_\mathbf {i} - \mathbf {x}_\mathbf {j}\right\| \) with \(1 \le i < j \le k\)
Definition 9
\(\left\{ V_{1,2},\ldots ,V_{1,k},V_{2,1},\ldots ,V_{k-1,k}\right\} \) is called distance distribution set.
Note that the distance distributions are invariant under rigid motions. The permutation of \(k\) indices will yield a permutation of the distance distributions. The next sections will analyze when the distance distributions suffice to characterize the orbit of a generalized shape and how we can recover the rigid transformation and the correspondences between two generalized shapes.
The main question is if the distance distribution matrix characterizes in a unique way the generalized shapes.
In the case of labeled points (i.e. in the sense of Kendall’s shapes) the answer is yes and it is illustrated by the next theorem. A simple proof of this theorem, using the singular value decomposition, can be found in [1].
Theorem 1
If \(X = \left\{ \mathbf {x}_\mathbf {1},\ldots ,\mathbf {x}_\mathbf {k}\right\} \) and \(Y =\left\{ \mathbf {y}_\mathbf {1},\ldots ,\mathbf {y}_\mathbf {k}\right\} \) are sets of points in \(\mathbb {R}^d\) and \(\left\| \mathbf {x}_\mathbf {i} - \mathbf {x}_\mathbf {j} \right\| = \left\| \mathbf {y}_\mathbf {i} - \mathbf {y}_\mathbf {j} \right\| \) , \(\forall 1\le i,j \le k\), then there exist a rigid transformation given by \(R \in O(d)\) and \(\mathbf {t} \in \mathbb {R}^d\) such that \(R\mathbf {x}_\mathbf {i} + \mathbf {t} = \mathbf {y}_\mathbf {i}\) for all \(i\).
So, in the case of labeled points, i.e. when the correspondences are known, the distance distribution matrix is an invariant that completely characterize the equivalence class of a shape. We are interested if the same holds in the case of generalized shapes.
The following examples will show that the answer is no, i.e. even if the distribution of distances is the same, there are configurations that are not related by an isometric transformation. Before giving these examples we note that the distance distribution of a point-set \(X = \left\{ \mathbf {x}_\mathbf {1},\ldots ,\mathbf {x}_\mathbf {k}\right\} \) may be given also as a monotone increasing sequence \(\left( d_1,d_2,\ldots ,d_{\left( {\begin{array}{c}k\\ 2\end{array}}\right) }\right) \), where \(\left\{ d_1,d_2,\ldots ,d_{\left( {\begin{array}{c}k\\ 2\end{array}}\right) } \right\} =\left\{ \left\| \mathbf {x}_\mathbf {i} - \mathbf {x}_\mathbf {j}\right\| \right\} \) with \(1 \le i < j \le k \). This sequence is the same as the distance distribution vector up to a permutation and it will be used when we derive the correspondences of points as we shall later see (sect. 6).
The first example is taken from [5], where homometric sets are defined as finite sets of integers with the same sets of differences: consider two sets of points \(X = \left\{ 0, 1, 4, 10, 12, 17\right\} \) and \(Y = \left\{ 0, 1, 8, 11, 13, 17\right\} \) in \(\mathbb {R}\). Their distance distribution sequence is given by the ordered vector \((1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 16, 17)\), but it is obvious that the point set does not represent the same shape even if we consider the possibility of reflection.
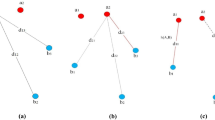
We insert here a more general example in \(\mathbb {R}^d, d \ge 2\) taken from [20] and shown in Fig. 1. Given a triangle \(ABC\), let \(a\) be the midpoint of \(BC\) and \(b\) be the midpoint of \(AC\). Let \(K\) be the line through \(a\) perpendicular to the line \(Aa\). Let \(L\) be the line through \(b\) perpendicular to the line \(Bb\). If \(D\) is the intersection of \(L\) and \(K\), \(E\) is the point of \(L\) such that \(dist(E, b) = dist(D, b)\) , and \(F\) is the point of \(K\) such that \(dist(F, a) = dist(D, a)\) then the shapes \(\left\{ A,B,C,D\right\} \), \(\left\{ A,B,C,E\right\} \), and \(\left\{ A,B,C,F\right\} \) have the same distribution distance matrix up to a permutation but they are not isometric. In fact, it is easy to see from the fact that the triangles \(BDE\) and \(ADF\) are isosceles and \(DAEC\) and \(DBFC\) are parallelograms that \(DA = FA = EC\), \(DB = FC = EB\) and \(DC = FB = EA\), therefore the \(6\) distances that form the distance distribution vector of each of the shapes are identical up to a permutation. The point sets given by \(\left\{ A,B,C,D\right\} \), \(\left\{ A,B,C,E\right\} \), and \(\left\{ A,B,C,F\right\} \) do not represent the same generalized shape and we will formally prove this in the sect. 4.
There are ways to create different shapes with the same distribution of distances, as we have seen in the previous examples, therefore we want to know which distance distributions define in a unique way a shape. In the next section we see that most distribution of distances define uniquely a generalized shape.
4 Distance Distribution and Generalized Shapes: Existence
We introduce in this section the notion of labeling of a point set in the context of the generalized shape theory. Most of the results here may be found in [7] and [8]. The scope of this part is to show which point sets with the same distribution of distances are the same modulo rotations, translations and permutations. More precisely we are interested in the relation between the permutation of the distances as elements of the symmetric group \(S_{\left( {\begin{array}{c}k\\ 2\end{array}}\right) }\) and the permutations between the point sets. Even if the results presented in this section are well known, we have considered appropriate to include these results here, since they are closely connected with the existence of the generalized shape as an equivalence class of shapes.
Denote by \(C\) the set of \(\left( {\begin{array}{c}k\\ 2\end{array}}\right) \) distinct pairs.
\(C = \left\{ (i,j) | i\ne j, i,j = 1,\ldots k\right\} \). This means that \(\forall (i,j)\) and \((i',j') \in C\) distinct, the sets \(\left\{ i,j\right\} \) and \(\left\{ i', j'\right\} \) are also distinct.
Two point sets \(X = \left\{ \mathbf {x_1},...,\mathbf {x_k}\right\} \) and \(Y =\left\{ \mathbf {y_1},...,\mathbf {y_k}\right\} \) have the same distance distribution vector if there exists a permutation \(\theta \in S_{\left( {\begin{array}{c}k\\ 2\end{array}}\right) }\) such that: \(V^X_{(i,j)} = V^Y_{\theta (i,j)}\), \(\forall (i,j) \in C\), where \(V^X_{(i,j)} = \left\| \mathbf {x_i} - \mathbf {x_j}\right\| \) and \(V^Y_{(i',j')} = \left\| \mathbf {y_i} - \mathbf {y_j}\right\| \) are the components of the distance distribution vector and \(\theta (i,j) = (i',j')\).
Definition 10
\(\theta \) is a labeling of the points if there exists a permutation \(\pi \in S_k\) of the indices such that:
The previous definition links the correspondences between pairs of each sets given by two indices \((i,j) \in C\) for \(X\), \(\theta (i,j) \in C\) for \(Y\) and point correspondences between \(X\) and \(Y\) given by \(\pi (i)\) and \(\pi (j)\), while the next result follows this definition and shows the correspondence with the generalized shapes.
Corollary 1
Given \(X = \left\{ \mathbf {x}_\mathbf {1},\ldots ,\mathbf {x}_\mathbf {k}\right\} \) and \(Y =\left\{ \mathbf {y}_\mathbf {1},\ldots ,\mathbf {y}_\mathbf {k}\right\} \) with the same distribution of distances up to a permutation \(\theta \in S_{\left( {\begin{array}{c}k\\ 2\end{array}}\right) }\) which is a labeling of points, then \(Y \in \left[ X\right] \) in the sense of generalized shape.
This connection between the permutation of \(\left( {\begin{array}{c}k\\ 2\end{array}}\right) \) distances and the permutation of \(k\) points gives the generalized shape equivalence class. We need to know which \(S_{\left( {\begin{array}{c}k\\ 2\end{array}}\right) }\) permutations are good permutations in the sense of Eq. (8).
The next theorem, taken from [8], proves that a permutation over \(C\) is a labeling if it preserves adjacency.
Theorem 2
For \(k \ne 4\), \(\theta \in S_{\left( {\begin{array}{c}k\\ 2\end{array}}\right) }\) is a labeling (i.e. induces equivalent shapes modulo rotations, translations and permutations) if and only if \(\forall i,j,l\) pairwise distinct indices we have
An interesting thing here is that for \(k = 4\), Theorem 2 is not true. In fact, observe that the relation \(\theta (i,j) \cap \theta (i,l) \cap \theta (i,m) = \emptyset \) cannot be contradicted because we cannot choose \(n\) distinct from \(i,j,l,m\). The theorem becomes true in all cases if we impose the additional condition that, for each pairwise distinct \(i,j,l,m \in \left\{ 1,\ldots ,k\right\} \), \(\theta (i,j) \cap \theta (i,l) \cap \theta (i,m) \ne \emptyset \).
A simple counterexample for \(k=4\) case is given by the point sets from the previous Sect. 3, Fig. 1, where the sets of distances \(\left\{ AB, BC, AC, AD, BD, CD\right\} \) and \(\left\{ AB, BC, AC, CE, BE, AE\right\} \) are the same and the condition (ii) from Theorem 2 holds, but the point sets are not isometric.
In case of the example \(X = \left\{ 0, 1, 4, 10, 12, 17\right\} \) and \(Y = \left\{ 0, 1, 8, 11, 13, 17\right\} \) in \(\mathbb {R}\), \(k=6\), the distributions of the ordered vectors of distances of \(X\) and \(Y\) are the same: \((1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 16, 17)\) so we are in the hypothesis of the Theorem 2. If we denote the points of each set with numbers from \(1\) to \(6\), following the natural order, we have: \(V^X_{(2,3)} = V^Y_{\theta (3,4)}\) and \(V^X_{(1,3)} = V^Y_{\theta (5,6)}\) so, from Theorem 2 the two sets do not represent the same generalized shape.
Equipped with this result that allows us to identify good permutations, the next theorem shows that if two point sets are sufficiently close and have the same distribution of distances, then they represent the same generalized shape. This result is weaker than desired, but gives some intuition on which point sets might be determined by their pairwise distance.
Theorem 3
( [7] ) Let \(X = \left\{ \mathbf {x}_\mathbf {1},\ldots ,\mathbf {x}_\mathbf {k}\right\} \subset \mathbb {R}^d\) a set of \(k\) points, then there exists a neighborhood \(V(X) \in (\mathbb {R}^d)^k\) of \(\left( \mathbf {x}_\mathbf {1},\ldots ,\mathbf {x}_\mathbf {k}\right) \) such that \(\left( \mathbf {y}_\mathbf {1},\ldots ,\mathbf {y}_\mathbf {k}\right) \in V(X)\) is a configuration with the same distribution of distances vector of \(X\) if and only if the two point configurations belong to the orbit of the same generalized shape.
Even if necessary for our framework, the results we have seen in this section do not help us to calculate the correspondences or the isometric transformation between two sets of points that represent the same generalized shape. The next section will analyze a method to recover the isometric transformation between elements of the same generalized shape in the ideal case, when the distribution of distances are exactly the same and the existence of the generalized shape is guaranteed.
Finally, Sect. 6 will present a method to compute correspondences between the points of two generalized shapes in the presence of noise.
5 Distance Distribution and Generalized Shapes: Isometric Transformation
In this section we are in the hypothesis of the Theorem 2, i.e. if there is a correspondence \(\theta \) between the distance distributions of two sets, then \(\theta \) is a labeling. We are interested here to compute the rigid part of the transformation without knowing the point to point correspondence.
We begin this section with some known general results from linear algebra that we shall use.
Recall that if \(\pi \in S_n\) is a permutation of \(\left\{ 1,2,\ldots ,n\right\} \), then the \(n \times n\) matrix \(P_{\pi }\):
is the permutation matrix associated to \(\pi \), where \(\mathbf e_j\) denotes a column vector of length \(n\) with 1 in the \(j\)th position and 0 in every other position.
The permutation matrix \(P_{\pi }\) associated to \(\pi \) is orthogonal, its transpose or inverse correspond to \(P_{\pi ^{-1}}\) and \(det P_{\pi } = \pm 1\).
Multiplying a \(d \times n\) matrix \(X\) on the right by \(P_{\pi }\) permutes the columns of \(X\) by \(P_{\pi }\).
Lemma 1
If \(P_{\pi }\) is the permutation matrix associated to \(\pi \in S_n\) and \(X\) and \(Y\) are two \(n \times n\) positive symmetric semidefinite matrices such that \(Y = P_{\pi } X P_{\pi }^{-1}\) then:
-
(i)
\(X\) and \(Y\) have the same set of eigenvalues
-
(ii)
There exist eigenvalue decompositions of \(X \!=\! U_XDU_X^{-1}\) and \(Y= U_YDU_Y^{-1} \), such that \(P_{\pi } = U_YU_X^{-1}\)
The proof of this lemma is is a simple consequence of the definitions.
Observe that not all the eigen-decompositions of \(X\) and \(Y\) lead to \(P_{\pi }\) since the decompositions are not unique. The previous lemma ensure only the existence.
Given a set of points \(X =\left\{ \mathbf {x}_\mathbf {1},\ldots ,\mathbf {x}_\mathbf {k}\right\} \subset \mathbb {R}^d\), we call Gram matrix the \(k \times k\) matrix whose entries are given by the inner products \(G_{ij} = \left\langle \mathbf {x}_\mathbf {i}, \mathbf {x}_\mathbf {j}\right\rangle \). The Gram matrix may also be given as the matrix product \(X^TX\).
A \(n \times n\) matrix \(M\) is called symmetric if \(M_{ij} = M_{ji}\) for all \(i,j = 1,\ldots ,n\). A \(n \times n\) matrix \(M\) is called positive semidefinite if for all \(\mathbf {x} \in \mathbb {R}^n, \mathbf {x}^T M \mathbf {x} \ge 0\).
The Gram matrix is positive semidefinite and symmetric, and every positive semidefinite matrix is the Gram matrix for some set of vectors. Further, in finite-dimensions it determines the vectors up to isomorphism, i.e. any two sets of vectors with the same Gram matrix must be related by a single unitary matrix as stated by the next lemma.
Lemma 2
For any two \(X\) and \(Y\,d \times k\) matrices, if their Gram matrices are equal, i.e. \(X^TX = Y^TY\), then there is a matrix \(A \in O(d)\) such that \(AX = Y\).
Proof
\(X^TX\) is positive semidefinite therefore can be written as \(Q {\varLambda } Q^T\) for \(Q \in O(k)\) and a non-negative diagonal matrix \({\varLambda }\). Using the singular value decomposition of \(X\) we can write \(X = U_X {\varSigma } Q^T\), where \(U_X \in O(d)\) and \( {\varSigma }^T {\varSigma } = {\varLambda }\).
Considering \(X^TX = Y^TY\) we may write also \(Y = U_Y {\varSigma } Q^T\), where \(U_Y \in O(d)\) and \( {\varSigma }^T {\varSigma } = {\varLambda }\).
Then, we can write \(AX = Y\) for orthogonal \(A = U_YU_X^T\). \(\square \)
The next lemma gives useful hints to connect the distance distribution with the Gramian derived from two sets.
Lemma 3
Given the sets \(X = \left\{ \mathbf {x}_\mathbf {1},\ldots ,\mathbf {x}_\mathbf {k}\right\} \) and \(Y =\left\{ \mathbf {y}_\mathbf {1},\ldots ,\mathbf {y}_\mathbf {k}\right\} \), the following statements are equivalent:
-
(i)
\(\left\| \mathbf {x}_\mathbf {i} - \mathbf {x}_\mathbf {j} \right\| = \left\| \mathbf {y}_\mathbf {i} - \mathbf {y}_\mathbf {j} \right\| \), \(\forall 1\le i,j \le k\).
-
(ii)
\(\forall n = 1,\ldots ,k\) fixed \(\left\langle \mathbf {x}_\mathbf {i} - \mathbf {x}_\mathbf {n}, \mathbf {x}_\mathbf {j} - \mathbf {x}_\mathbf {n} \right\rangle = \big \langle \mathbf {y}_\mathbf {i} - \mathbf {y}_\mathbf {n}, \mathbf {y}_\mathbf {j} - \mathbf {y}_\mathbf {n} \big \rangle \), \(\forall 1\le i,j \le k\).
-
(iii)
\(\left\langle \mathbf {x}_\mathbf {i} - \bar{\mathbf {x}}, \mathbf {x}_\mathbf {j} - \bar{\mathbf {x}} \right\rangle = \left\langle \mathbf {y}_\mathbf {i} - \bar{\mathbf {y}}, \mathbf {y}_\mathbf {j} - \bar{\mathbf {y}} \right\rangle \), \(\forall 1\le i,j \le k\).
This lemma may be easily demonstrated by applying multiple times the well known result:
for all \(\mathbf {p}, \mathbf {q} \in \mathbb {R}^d\).
We are ready to give a stronger result about the connection between the permutations of the distance distribution matrix and the generalized shapes.
Lemma 4
Given \(X = \left\{ \mathbf {x}_\mathbf {1},\ldots ,\mathbf {x}_\mathbf {k}\right\} \) and \(Y =\left\{ \mathbf {y}_\mathbf {1},\ldots ,\mathbf {y}_\mathbf {k}\right\} \) two point sets in \(\mathbb {R}^d\), the following statements are equivalent:
-
(i)
\(Y \in \left[ X\right] \)
-
(ii)
\(\exists P_{\pi }\) a permutation matrix, such that \(D^X = P_{\pi }^TD^YP_{\pi }\). Moreover, \(P_{\pi } = U_{Y}U_X^T\), where \(U_Y\) and \(U_X\) are orthogonal matrices from an eigenvalue decomposition of \(\bar{Y}^T\bar{Y}\) and \(\bar{X}^T\bar{X}\).
-
(iii)
\(\exists P_{\pi }\) a permutation matrix and \(R \in O(d)\), such that \(R \bar{X} = \bar{Y} P_{\pi }\). Moreover, \(P_{\pi } = U_{Y}U_X^T\), where \(U_Y\) and \(U_X\) are orthogonal matrices from an eigenvalue decomposition of \(\bar{Y}^T\bar{Y}\) and \(\bar{X}^T\bar{X}\).
Proof
Lemma 3 says that the correspondences between the elements of the distance distribution matrix are the same as the correspondences between the Gram matrix of the centered coordinates ((i) \(\Leftrightarrow \) (iii)), therefore if we show that there exists \(P_{\pi } \in S_k\) such that \(\bar{X}^T\bar{X} =P_{\pi }^T \bar{X}^T\bar{X}P_{\pi }\), the same relationship will hold between \(D^X\) and \(D^Y\).
\(Y \in \left[ X\right] \Leftrightarrow \) exists \(R \in O(d)\), a vector \(\mathbf {t} \in \mathbb {R}^d\) and a permutation \(\pi \in S_k\) such that:
\( R\mathbf {x}_\mathbf {i} + \mathbf {t} = \mathbf {y}_{\varvec{\pi }(\mathbf{i})}\) for all \(i = 1,\ldots ,k\).
Let \(P_{\pi }\) be the permutation matrix associated with \(\pi \).
If we denote \(Y_{\pi } = \left\{ \mathbf {y}_{\varvec{\pi }(\mathbf{1})},\ldots ,\mathbf {y}_{\varvec{\pi }(\mathbf{k})}\right\} \), we have \(Y_{\pi } = YP_{\pi }\) and
From hypothesis \(\left\| \mathbf {x}_\mathbf {i} - \mathbf {x}_\mathbf {j}\right\| = \left\| \mathbf {y}_{\varvec{\pi }(\mathbf{i})} - \mathbf {y}_{\varvec{\pi }(\mathbf{j})}\right\| \forall i,j = 1,\ldots ,k\), if and only if, by Lemma 3, \(\left\langle \mathbf {x}_\mathbf {i} - \bar{\mathbf {x}}, \mathbf {x}_\mathbf {j} - \bar{\mathbf {x}} \right\rangle = \left\langle \mathbf {y}_{\varvec{\pi }(\mathbf{i})} - \bar{\mathbf {y}}, \mathbf {y}_{\varvec{\pi }(\mathbf{j})} - \bar{\mathbf {y}} \right\rangle \), \(\forall 1\le i,j \le k\).
This is equivalent to \(\bar{X}^T\bar{X} = \bar{Y}_{\pi }^T\bar{Y}_{\pi }\) or, using (12), \(\bar{X}^T\bar{X} = P_{\pi }^T\bar{Y}^T\bar{Y}P_{\pi }\).
The formula of \(P_{\pi }\) derives from Lemma 1.
We can rewrite (ii) as \(\bar{X}^T\bar{X} = (\bar{Y}P_{\pi })^T\bar{Y}P_{\pi }\), therefore by Lemma 2 this is true if and only if there exists an orthogonal matrix \(R\) such that \(R\bar{X} = \bar{Y}P_{\pi }\).
We have so far a relation between centered coordinates \(\bar{X}\) and \(\bar{Y}\) together with the matrix \(R \in O(d)\) and \(\pi \in S_k\).
The equivalence as generalized shapes between \(X\) and \(Y\) follows the same reasoning as in Theorem 1, putting \(\mathbf {t} = \bar{\mathbf {y}} - R\bar{\mathbf {x}}\). \(\square \)
Notice again that the eigenvalue decomposition is not unique, therefore the previous result ensures only the existence of the matrix \(P_{\pi }\) and does not help in finding the permutation or the orthogonal matrix that relates the two point sets.
In our assumption, the permutation that realizes the correspondence between the point sets is not known, so we want to find the orthogonal matrix that relates the two point sets without knowing the correspondences. The next theorem will introduce a way to find this matrix. The complete solution of this problem will be given as an algorithmic method.
The \(d \times d\) matrix \(\bar{Y}\bar{Y}^T\) does not depend on the permutation of the elements of \(Y\), since for all \(\pi \in S_k\), \(\bar{Y}_{\pi }\bar{Y}_{\pi }^T = \bar{Y}P_{\pi }P_{\pi }^T\bar{Y}^T = \bar{Y}\bar{Y}^T\). This allows us to establish the next result that will be the base of the algorithm that finds the rigid transformation between two sets representing the same generalized shape without knowing the correspondences between the point sets.
Theorem 4
Given \(X = \left\{ \mathbf {x}_\mathbf {1},\ldots ,\mathbf {x}_\mathbf {k}\right\} \) and \(Y =\left\{ \mathbf {y}_\mathbf {1},\ldots ,\mathbf {y}_\mathbf {k}\right\} \) two point sets in \(\mathbb {R}^d\) with \(Y \in \left[ X\right] \), then the matrices \(\bar{X}\bar{X}^T\) and \(\bar{Y}\bar{Y}^T\) have the same eigenvalues, including their algebraic multiplicities. If the eigenvalues are all distinct we denote by \(S\) the \(d \times d\) matrix of the \(\mathbf {s}_i\) eigenvectors of \(\bar{X}\bar{X}^T\) written as columns, \(T\) the \(d \times d\) matrix of the \(\mathbf {t}_i\) eigenvectors of \(\bar{Y}\bar{Y}^T\) written as columns and \(\pi \in S_k\) the permutation determined from \(\bar{Y}_{\pi } = R\bar{X}\). Then we have:
for all \(j=1,\ldots ,k, i=1,\ldots ,d\) and \(\delta _i = \pm 1\).
Moreover, if \(\left\langle \mathbf {x}_\mathbf {j} -\bar{\mathbf {x}}, \mathbf {s}_\mathbf {i}\right\rangle \ne 0 \) and we denote
\({\varDelta } = Diag \left( \delta _1,\ldots ,\delta _d\right) \), where
than \(R = T{\varDelta } S^T\).
Proof
From Lemma 4 it follows that if \(Y \in \left[ X\right] \) then \(\exists R \in O(d)\) such that \(\bar{Y}_{\pi } = R\bar{X}\).
We have \(\bar{Y}\bar{Y}^T = R\bar{X}P_{\pi }^TP_{\pi }\bar{X}^TR^T = R(\bar{X}\bar{X}^T)R^T\).
If we decompose the real, symmetric matrix \(\bar{X}\bar{X}^T\) using the eigen-decomposition \(\bar{X}\bar{X}^T = S {\varLambda } S^T\) then:
\(RS\) is orthogonal as product of orthogonal matrices so (15) shows that \(\bar{X}\bar{X}^T\) and \(\bar{Y}\bar{Y}^T\) have the same eigenvalues, including their algebraic multiplicities and \(RS\) is a matrix of eigenvectors for \(\bar{Y}\bar{Y}^T\).
Since the eigenvalues are distinct and knowing that the geometric multiplicity of an eigenvalue is less than the algebraic multiplicity, the dimension of \(Null\left( \bar{X}\bar{X}^T-\lambda _i I_d\right) \) is one therefore, for all \(i=1,\ldots ,d\) if \(\mathbf {t}_i\) is an eigenvector of \(\bar{Y}\bar{Y}^T\) then \(\mathbf {t}_i = \pm R \mathbf {s}_i\).
In the case \(\left\langle \mathbf {x}_\mathbf {j} -\bar{\mathbf {x}}, \mathbf {s}_\mathbf {i}\right\rangle \ne 0 \) denoting \(\delta _i = \frac{\left\langle \mathbf {x}_\mathbf {i} -\bar{\mathbf {x}}, \mathbf {s}_\mathbf {i}\right\rangle }{\left\langle \mathbf {y}_{\varvec{\pi }(\mathbf{j})} -\bar{\mathbf {y}}, \mathbf {t_i}\right\rangle }\) and \({\varDelta } = Diag \left( \delta _1,\ldots ,\delta _d\right) \), we can write \(R = T{\varDelta } S^T\).
Since the inner product is invariant to isometries and \(\bar{Y}_{\pi } = R\bar{X}\), with \(R\) orthogonal we can write:
\(\left\langle \mathbf {x}_\mathbf {j} -\bar{\mathbf {x}}, \mathbf {s}_\mathbf {i}\right\rangle = \left\langle R(\mathbf {x}_\mathbf {j} -\bar{\mathbf {x}}),R\mathbf {s}_\mathbf {i}\right\rangle =\pm \left\langle \mathbf {y}_{\varvec{\pi }(\mathbf{j})} -\bar{\mathbf {y}}, \mathbf {t_i}\right\rangle ,\)
and the proof is completed. \(\square \)
Remark that if \(\left\langle \mathbf {x}_\mathbf {j} -\bar{\mathbf {x}}, \mathbf {s}_\mathbf {i}\right\rangle = 0\) for all \(j=1,\ldots ,k\) , \(\delta _i\) cannot be determined from (14). In this case \(\bar{X}^T\mathbf {s}_i = 0\), therefore \(\bar{X}\bar{X}^T\mathbf {s}_i = 0\). This means \(\mathbf {s}_i\) is the eigenvector corresponding to the eigenvalue \(0\). This remark allows us to give the following corollary.
Corollary 2
If the eigenvalues are all distinct and non-zero, then \(\exists j=1,\ldots ,k\) such that \(\left\langle \mathbf {x}_\mathbf {j} -\bar{\mathbf {x}}, \mathbf {s}_\mathbf {i}\right\rangle \ne 0\), and therefore:
Following this corrolary, the matrix \({\varDelta } \!=\! Diag \left( \delta _1,\ldots ,\delta _d\right) \) is completely determined only when \(\pi \) is known, while we set out to achieve the computation of \(R\) without knowing \(\pi \). The next corollary will solve this problem by giving a way to compute \({\varDelta }\).
Let’s first denote, for each \(i =1,\ldots ,d\), \(A_i^{-}\) the sets:
In a similar way we define \(A_{i_-}^{Y}\) and \(A_{i_+}^{Y}\).
Corollary 3
If \(Y \in \left[ X\right] \) and none of the eigenvalues of the matrix \(\bar{X}\bar{X}^T\) is zero, then, for each \(i=1,\ldots ,d\) one of the two cases happens:
-
(i)
\(A_{i_-}^{X} = A_{i_-}^{Y}\) and \(A_{i_+}^{X} = A_{i_+}^{Y}\) and thus \(\delta _i=1\)
-
(ii)
\(A_{i_-}^{X} = A_{i_+}^{Y}\) and \(A_{i_+}^{X} = A_{i_-}^{Y}\) and thus \(\delta _i=-1\)
In this way the matrix \({\varDelta } = Diag \left( \delta _1,\ldots ,\delta _d\right) \) is determined without knowing the permutation \(\pi \).
We conclude this section with the following algorithm that, in most cases, solves the rigid transformation between two elements of the same generalized shape, in the case where the point to point correspondences are not known.

In general, the eigenvalues of a matrix cannot be computed exactly, as they are roots of a polynomial, this making our algorithm impractical. The next section will introduce a method to find point to point correspondences between two point sets representing the same shape. However, if the distribution of distances of two point sets is the same up to a threshold, we can compare the eigenvalues of the Gramian matrices of the centered coordinates by using a very small threshold \(\epsilon \). We can do the same to compare the sets \(A_{i_-}^{X}, A_{i_+}^{X}, A_{i_-}^{Y},A_{i_+}^{Y}\).
Complexity
The Algorithm 1 requires the computation of \(\bar{X}\bar{X}^T\) (complexity at most \(O(d^2k)\), its eigenvalue decomposition (complexity \(O(d^3)\), the computation of \(\delta _i\) (complexity \(O(dk)\)), the computation of \(R\) (complexity at most \(O(d^3)\)). Since \(d << k\) the resulting complexity is at most \(O(dk^2)\).
6 Distance Distribution and Generalized Shapes: Correspondences
6.1 Distance Distribution Permutation
So far, we have seen how to verify the existence of the generalized shape when we know the distance distribution matrix or the distance distribution vector and how to find the rigid transformation between two elements of the same generalized shape, without knowing the correspondences.
Algorithm 1 shows how to find the registration matrix when we know the point sets and the fact that they belong to the same generalized shape. This result is not guaranteed in the presence of a high level of noise making it not usable for most practical applications.
In this section we give a method to compute the correspondences between two point sets belonging to the same generalized shape in the presence of noise and a method to derive the rigid transformation that relates the two point sets using the correspondences.
The problem to solve is, given \(X = \left\{ \mathbf {x}_\mathbf {1},\ldots ,\mathbf {x}_\mathbf {k}\right\} \) and \(Y =\left\{ \mathbf {y}_\mathbf {1},\ldots ,\mathbf {y}_\mathbf {k}\right\} \) two point sets in \(\mathbb {R}^d\) with \(Y \in \left[ X\right] \), to find the permutation \(\pi \in S_k\) such that:
where \(R \in O(d)\), \(\mathbf {t} \in \mathbb {R}^d\) and \(P_{\pi }\) is the permutation matrix associated to \(\pi \).
Since \(O(k!)\) different correspondence permutations are possible amongst the two point sets, brute force search is intractable.
As before, the invariant we chose, to find the right correspondences, is given by the distance distribution vector as defined in section 3.
Theorem 2 proved in what cases the distance distribution vector defines a generalized shape in a unique way.
We shall derive the permutation \(\pi \) from the permutation \(\gamma \in S_{\left( {\begin{array}{c}k\\ 2\end{array}}\right) }\) that ’align’ the two distribution distance vectors \(V^X = (V_{1,2}^X,\ldots ,V_{k-1,k}^X)\) and \(V^Y = (V_{1,2}^Y,\ldots ,V_{k-1,k}^Y)\), such that:
Equation (17) holds in the ideal case, when no noise is present. In the practical application we may relax the exact correspondence and search for a permutation matrix \(P_{\gamma }\) of dimension \(k(k-1)/2\) such that:
The use of the Manhattan distance or \(L_1\) norm in (18) is coherent with the definition of the distance between generalized shapes given in the sect. 3 by the Eq. (3). Through the rest of the paper, when there are no ambiguities, we shall use the notation \(\left\| \cdot \right\| _1 =\left| \cdot \right| \).
The \(L_1\) norm works better in the presence of noise and is equivalent with \(L_2\) norm, in the sense of equivalence between norms. Considering \(\left\| \cdot \right\| _2 \le \left\| \cdot \right\| _1\), the Eq. (18) gives an upper bound also for the \(L_2\) norm.
Since there are \((k(k-1)/2)!\) ways to arrange a shape vector, we need an efficient mode to solve (18). We shall further see that this process takes \(O(NlogN)\), where \(N = k(k-1)/2\), by ordering each of the shape vectors.
Lemma 5
If \(a_1 \le a_2 \le \ldots \le a_N\), \(b_1 \le b_2 \le \ldots \le b_N\) and \(\pi :\left\{ 1, \ldots , N\right\} \rightarrow \left\{ 1, \ldots , N\right\} \) is a permutation of the indices, then we have:
This lemma may be easily demonstrated by induction on \(N\).
Corollary 4
For any \(\pi _A\) and \(\pi _B\) permutation of \(N\) indices and \(a_1 \le a_2 \le \ldots \le a_N\), \(b_1 \le b_2 \le \ldots \le b_N\) it holds:
Theorem 5
The solution of (18) is given by:
where \(P_{\pi _Y}, P_{\pi _X}\) are the permutations matrices that order the vectors \(V^Y\) and \(V^X\) respectively.
Proof
From the hypothesis that vectors \(V^X P_{\pi _X}\) and \(V^Y P_{\pi _Y}\) are ordered, it follows the Corollary 4 holds:
for every and all permutations \(P_{\pi _X}^{'}, P_{\pi _Y}^{'}\).
Multiplying on the right with \(P_{\pi _Y}^T\) the expression \( V^X P_{\pi _X} - V^Y P_{\pi _Y}\) we have:
from which the conclusion follows. \(\square \)
We are now able to find the minimum distance between two shape vectors that represent in an unique mode two point sets given in two different coordinate systems regardless of the point ordering.
Once we have the correspondence between the shape vectors the next step is to find the correspondences between the points. This will be described in an algorithmic form in the next section.
6.2 Determining Correspondences
6.2.1 Point-Set Correspondences in a Closed form Solution
The correspondences between the indices of the point sets \(X\) and \(Y\), when \(Y \in \left[ X\right] \) will be given again as a permutation matrix. To do this we have to associate the two indices \(i,j\) that give the distance \(D_{i,j}^X = \left\| \mathbf {x}_i - \mathbf {x}_j \right\| \) in the distance distribution matrix (or alternatively \(D_{i,j}^Y\)) to a unique index \(l\) in the distance distribution vector and vice versa, to each of the indexes of the distance distribution vector two indexes in the point-set. In this second case we have two solutions considering the order of the two vectors that compose the distance.
From Eq. (7), considering that \(i\) represents the index of a row and \(j\) represents the offset inside the \(i\)-th row in the distance distribution matrix, the mapping \((i,j) \longmapsto l\) is given by:
Conversely, if we denote the distance distribution vector index \(l\), the problem is to find the correspondent \((i,j)\) couple of indexes, where again \(i\) represents the row and \(j\) represents the off-set in the distance distribution matrix.
In this case, \(i\) is given by the smallest positive number that satisfies:
Transforming (23) in a second order inequality:
we observe that the sum and the product of the roots are positive, therefore the solution for the \(i-th\) index is given by the smallest integer bigger than the smallest root of the associated second order equation:
From (25) and (22) it follows:
We are now able to write the algorithm that finds the correspondences of two point sets having the same cardinality, regardless of the coordinate system where each of the sets are represented and of their ordering.

Observe that the line 9 of Algorithm 2 is guaranteed by the Theorem 2.
6.2.2 Point-Set Correspondences with Noise
The Algorithm 2 gives the closed form solution of the correspondence of the two point sets \(X\) and \(Y\) representing the same generalized shape.
This solution holds in the ideal case, when no noise is present. In the case of noisy data there is no guarantee to find a common index \(i_c\) in the row 9-th of the Algorithm 2. To solve this we use a technique of voting, associating to each couple \((\mathbf {x}_i, \mathbf {y}_j)\) an increasing vote for each possible correspondence, therefore we build an association matrix. To find the correspondence matrix \(M\) we simply extract the maximum value in each row of the association matrix.
The detailed algorithm is as following:

6.3 Correspondence Test and Results
In this section we present the results of the correspondence algorithm. We run tests with different levels of noise and we compute, for each of the tests, the percentage of good correspondences.
The point sets used for testing are depicted in the Figs. 2 and 3.
Testing 2D data for empirical robustness evaluation. On each of the images the points marked with \(blue\) \(asterisk\) represent the model (joined by a continuous line obtained through interpolation) meanwhile the points marked with \(red\) \(circle\) represent the model transformed by a random permutation and different levels of noise. The fish model on the left column follows [16, 24]. (Color figure online)
Figure 2 depicts the 3D model dataset denoted by the ’*’ blue points. The dataset was permuted and random noise was added (red circles) before the correspondence algorithm was launched. Figure 3 shows the original point-set aligned along a continuous line obtained by interpolation, while the ’\(\circ \)’ points are obtained by adding random noise to a permutation of the original dataset.
In both 2D and 3D tests, since we compute the correspondences relying only on the shape vector, which is invariant to isometries, the permutations of the noisy dataset suffice to test the robustness of our algorithm.
Even if the tests done are very simple and synthetic, we reiterate here that we are addressing applications where there is noise and small deformations without a clear distinction between the two (considered that data usually represent the surface or the border of an object in the real world and is acquired as unstructured set of points). Our main interest in recovering correspondences is to get the best rigid alignment between the data sets.
The noise we added for the purposes of this work is composed by pseudorandom values drawn from the standard normal distribution with mean 1 and different standard deviations (see Table 1). The noise level is given by the standard deviation \(\sigma _i, i \in \left\{ 1, ..,5\right\} \) and represents a percentage fraction of the maximum extension of the point set \(X\), that is \(\sigma _i = i\max \limits _{\mathbf {x}_i, \mathbf {x}_j \in X}\left\| \mathbf {x}_i - \mathbf {x}_j\right\| /100\).
The computation of the rigid transformation in the case of noisy data will be addressed in the next section. In this section we compute the correspondences between two point sets \(X\) and \(Y = XP_{\pi } + N(\mu , \sigma ^2)\), where \(P_{\pi }\) is a random permutation of \(\left| X\right| \) elements, \(N\) is the normal distribution of mean \(\mu = 0\) and variance \(\sigma ^2\) (see Figures 3 and 2).
As we can see from the results (Table 1), the number of wrong correspondences increases with the amount of noise. This is an obvious observation but we are more concerned weather we are able to recover the right registration transformation from the correspondences we found. The answer is yes as we see in the next section and, in this case, we can update the correspondences after the registration using the nearest neighbor technique. Another observation is that good correspondences are strongly related to the highest value in the correspondences matrix \(M\) from the Algorithm 3. Keeping in mind that we only need 3 correspondences to find the parameters for the rigid alignment we may keep only a part of the good correspondences to find the best rigid alignment. The selection order of this correspondence will be given by the highest value in the correspondences matrix.
7 Correspondences and Registration Evaluation Using Pulmonary Landmark Points Derived from 4D CT Image Data
Thoracic 4D computed tomography (CT) image data abound of high-contrast, anatomical landmarks such as vessel and bronchial bifurcations. The fourth dimension is given by the movement during the respiratory cycle.
Castillo et al. in [11] extracted a large number of landmark point pairs for the evaluation of image registration spatial accuracy (Figs. 4 and 5). We use this dataset to test the performance of our correspondence algorithm, then we register the landmarks using the rigid and non-rigid transformations.
The dataset from [11] includes 4D CT images from five patients free of pulmonary disease who were treated for esophageal cancer. Each patient uderwent treatment planning in which 4D CT images of the entire thorax and upper abdomen were acquired at 2.5 mm spacing. In this study a high number of landmarks were manually extracted by an expert in thoracic imaging in 5 different breathing phases for each of the patient. The localization error of these landmarks is around 1 mm.
For the purpose of our study we compared for each patient the initial breathing phase against the other 4 breathing phases by registering the landmarks without prior knowledge of the correspondences. After the computation of the landmarks correspondences, we registered the landmarks and we computed the Hausdorff distance, as in the previous section.
Table 2 summarizes the results of the correspondences expressed as a percentages.
Estimates of the Hausdorff distance between the registered set of landmarks are summarized in Table 3. The rigid registration is based on the correspondences found before. Since the number of good correspondences is greater than 50 % of the total number of points, we have used an extraction of \(p = 0.3\) of the best correspondences. For the non-rigid registration, we used the TPS interpolation. The input for the TPS interpolation is given by the same percentage of correspondences.
8 Conclusions and Future Work
The solution of the absolute orientation (Procrustes problem) is given by efficient methods in closed form, but to solve the registration of arbitrary point sets that represent the same shape acquired with different sensors and/or in different moments, there is no closed form solution. As summarized in the introduction, the solutions known so far range from iterative methods, where the optimization take place on each step toward a local minimum (as the ICP algorithm), capable to handle a certain amount of noise, to very complex methods that can handle also outliers and deformations, where the solution is given after a complicate process of optimization and using a large set of parameters.
In this paper, we built the basis for a closed form solution, robust to a small amount of noise, of the Procrustes problem in the case when no matching correspondences are given a priori. The solution we have proposed makes use of the distribution of the distances. We have first analyzed, from a theoretical point of view, when the distribution of distances completely characterizes the shape of a point set and how to recover the isometric transformation between two sets of points, when the distribution of distances is given but the correspondences are not. Beside this theoretical contribution, we have developed algorithms to find the correct alignment of shapes given as unstructured point sets. The registration took place after the correspondences were found and the iteration is used only to refine the distance between the aligned shapes. We have seen in the introduction that there are no efficient methods that can handle the variability of the data (i.e. noise, outliers, deformation) that can guarantee the goodness of the result and, in the same time, the possibility of the validation.
Some of the research areas, where the registration requires a completely validated method, range from the medical applications (registration in the OR of surfaces extracted from the 3D reconstruction of organs) to robotic applications, where the accuracy and the real-time response are fundamental.
In the case of the medical applications the registration rely on datasets representing most of the time deformed surfaces but in order to map the instruments in the imaging space (e.g. biopsy needle, endoscopic camera, laparoscope etc.) we still need to isolate the rigid component of the registration.
With the solution we propose in this paper, we are able to register noisy data represented by sets of points with the same cardinality.
The experimental test we have performed employed a thoracic dataset from which a set of landmarks was extracted on different breathing phases. The algorithm we have introduced was able to recover a high number of correspondences and to extract the rigid transformation between the point sets without having the homologies between the landmarks before. A further refinement was obtained by applying a non-rigid transformation to the result. The next step will be to extend the method to handle incomplete data and outliers.
To replace the ICP algorithm with a closed form variant, in the case of dense point sets we may subsample to provide the candidates for potential correspondences. The main question is to subsample the points in a way that such that subsample points are still matchable. The joint clustering-matching algorithm presented by Chui in [12] can be a valid alternative for this extension.
If we replace the Euclidean distances in the shape matrix with geodesics we may handle also a larger class of isometries that include also the so called inelastic deformations, i.e. deformations that do not stretch or tear the object (see also [10]).
The results we have presented may be used also to extend the theory of shapes as defined by [18]. The equivalence class of k-ad points in \(\mathbb {R}^d\) will handle in this case also the permutations. It will be interesting to study how the topological properties of the new formed equivalence classes will change.
Another application of our method for the computation of correspondences could be in the case of matching a collection of shapes. In this case, once we compute the correspondences between each couple of shapes by using the generalized shapes method, we may invoke a cycle-consistency criterion - the fact that composition of maps along a cycle of shapes should approximate the identity map. The solutions of this problem, at the state of the art, range from fuzzy correspondences [19], semidefinite programing [15] to permutation synchronization, which finds all the matchings jointly, in one shot, via a relaxation to eigenvector decomposition [26]. The same reasoning can be applied to the synchronization of the rotation matrices in the registration of a shape represented in different coordinate systems, once we have an initial guess to all couples of coordinate systems using the generalized shapes context. Some examples of the solution to the synchronization problem over the special orthogonal group \(SO(d)\) is by using a penalty function involving the sum of unsquared deviations toward a convex optimization in [32], or using the semidefinite programing based relaxation which approximates the maximum likelihood estimator in [2].
References
Arun, K.S., Huang, T.S., Blostein, S.D.: Least-squares fitting of two 3-d point sets. IEEE Trans. Pattern Anal. Mach. Intell. 9(5), 698–700 (1987)
Bandeira, A.S., Charikar, M., Singer, A., Zhu, A.: Multireference alignment using semidefinite programming. In: ITCS, pp. 459–470 (2014).
Belongie, S., Malik, J., Puzicha, J.: Shape context: A new descriptor for shape matching and object recognition. In. In NIPS, vol. 54, pp. 831–837. Citeseer, Citeseer (2000).
Besl, P.J., McKay, N.D.: A method for registration of 3-D shapes. IEEE Trans Pattern Anal Mach Intell 14(2), 239–256 (1992)
Bloom, G.S.: A counterexample to a theorem of s. piccard. J. Comb. Theory, Ser. A 22(3), 378–379 (1977)
Bookstein, F.L.: Principal warps: thin-plate splines and the decomposition of deformations. IEEE Trans Pattern Anal Mach Intell 11(6), 567–585 (1989)
Boutin, M., Kemper, G.: On reconstructing n-point configurations from the distribution of distances or areas. arxiv:0304192 (2003).
Boutin, M., Kemper, G.: Which point configurations are determined by the distribution of their pairwise distances? Int. J. Comput. Geometry Appl. 17(1), 31–44 (2007)
Boyer, D.M., Lipman, Y., St Clair, E., Puente, J., Patel, B.A., Funkhouser, T., Jernvall, J., Daubechies, I.: Algorithms to automatically quantify the geometric similarity of anatomical surfaces. Proceedings of the National Academy of Sciences of the United States of America 108(45), 18,221–18,226 (2011).
Bronstein, A., Bronstein, M., Kimmel, R.: Numer Geom Non-Rigid Shapes, 1st edn. Springer Publishing Company, Incorporated (2008)
Castillo, R., Castillo, E., Guerra, R., Johnson, V.E., McPhail, T., Garg, A.K., Guerrero, T.: A framework for evaluation of deformable image registration spatial accuracy using large landmark point sets. Phys Med Biol 54(7), 1849 (2009)
Chui, H.: Non-rigid point matching: Algorithms, extensions and applications. Ph.D. thesis, Yale University (2001).
Chui, H., Rangarajan, A.: A new point matching Algorithm for non-rigid registration. Comput Vis Image Underst 89(2–3), 114–141 (2003)
Ghosh, D., Sharf, A., Amenta, N.: Feature-driven deformation for dense correspondence. Medical Imaging: Visualization, Image-Guided Procedures, and Modeling (Proc. SPIE) 7261 (2009).
Huang, Q.X., Guibas, L.J.: Consistent shape maps via semidefinite programming. Comput. Graph. Forum 32(5), 177–186 (2013)
Jian, B., Vemuri, B.C.: Robust Point Set Registration Using Gaussian Mixture Models (2011).
Kendall, D.G.: Shape manifolds, procrustean metrics, and complex projective spaces. Bull Lond Math Soc 16(2), 81–121 (1984)
Kendall, D.G., Barden, D., Carne, T.K., Le, H.: Shape and Shape Theory. Wiley, New York (1999)
Kim, V.G., Li, W., Mitra, N.J., DiVerdi, S., Funkhouser, T.: Exploring collections of 3D models using fuzzy correspondences . ACM Trans. Graph. 31(4), 1–11 (2012).
Lemke, P., Skiena, S., Smith, W.: Reconstructing sets from interpoint distances . Discrete Comput. Geom. 25, 597–631 (2003).
Leordeanu, M., Hebert, M.: A spectral technique for correspondence problems using pairwise constraints. In: ICCV, pp. 1482–1489 (2005).
Lipman, Y., Funkhouser, T.: Mobius voting for surface correspondence. ACM Transactions on Graphics (Proc. SIGGRAPH) 28(3) (2009).
Mémoli, F., Sapiro, G.: A theoretical and computational framework for isometry invariant recognition of point cloud data. Found Comput Math 5(3), 313–347 (2005)
Myronenko, A., Song, X.S.X.: Point Set Registration: Coherent Point Drift. IEEE Trans Pattern Anal Mach Intell 32(12), 2262–2275 (2010)
Osada, R., Funkhouser, T.A., Chazelle, B., Dobkin, D.P.: Shape distributions. ACM Trans. Graph. 21(4), 807–832 (2002)
Pachauri, D., Kondor, R., Singh, V.: Solving the multi-way matching problem by permutation synchronization. In: NIPS, pp. 1860–1868 (2013).
Rangarajan, A., Chui, H., Bookstein, F.L.: The Softassign Procrustes Matching Algorithm. Inf Process Med Imaging 1230, 29–42 (1997)
Schönemann, P.H.: A generalized solution of the orthogonal procrustes problem. Psychometrika 31(1), 1–10 (1966)
Scott, G.L., Longuet-Higgins, H.C.: An Algorithm for associating the features of two images. Proc Royal Soc B 244(1309), 21–26 (1991)
Shapiro, L.S., Brady, J.M.: Feature-based correspondence: an eigenvector approach. Image Vis Comput 10(5), 283–288 (1992)
Wahba, G.: Spline Models for Observational Data, CBMS-NSF Regional Conference Series in Applied Mathematics, vol. 59. SIAM (1990).
Wang, L., Singer, A.: Exact and stable recovery of rotations for robust synchronization. CoRR abs/1211.2441 (2012).
Yaniv, Z., Cleary, K.: Image-Guided Procedures: A Review (2006).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Maris, B.M., Fiorini, P. Generalized Shapes and Point Sets Correspondence and Registration. J Math Imaging Vis 52, 218–233 (2015). https://doi.org/10.1007/s10851-014-0538-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10851-014-0538-8